Abstract
Jumonji domain-containing protein 3 (JMJD3/KDM6B) demethylates lysine 27 on histone H3 (H3K27me3), a repressive epigenetic mark controlling chromatin organization and cellular senescence. To better understand the functional consequences of JMJD3 its expression was investigated in brain tumor cells. Querying patient expression profile databases confirmed JMJD3 over-expression in high-grade glioma. Immunochemical staining of two glioma cell lines, U251 and U87, indicated intrinsic differences in JMJD3 expression levels that were reflected in changes in cell phenotype and variations associated with cellular senescence, including senescence-associated β-galactosidase (SA-β-gal) activity and the senescence associated secretory phenotype (SASP). Over-expressing wild type JMJD3 (JMJD3wt) activated SASP-associated genes, enhanced SA-βgal activity, and induced nuclear blebbing. Conversely, over-expression of a catalytically inactive dominant negative mutant JMJD3 (JMJD3mut) increased proliferation. In addition, a large number of transcripts were identified by RNA-seq as altered in JMJD3 over-expressing cells, including cancer- and inflammation-related transcripts as defined by IPA analysis. These results suggest that expression of the SASP in the context of cancer undermines normal tissue homeostasis and contributes to tumorigenesis and tumor progression. These studies are therapeutically relevant because inflammatory cytokines have been linked to homing of neural stem cells and other stem cells to tumor loci.
Keywords: JMJD3, KDM6B, H3K27 demethylase, histone code, epigenetic regulation, senescence-associated secretory phenotype, SASP, inflammation, tumor dissemination, glioblastoma, cancer, neural stem cell, tumor tropism
Introduction
The histone H3K27 demethylase jumonji domain-containing protein 3 (JMJD3 or KDM6B) is a molecular switch controlling cellular programs of immune activation, injury repair responses including regeneration, and cellular senescence (1–5). JMJD3 exerts these actions through H3K27 demethylase activity and control of polycomb repressive complexes by removal of tri- and di-methyl groups, as well as by direct effects on chromatin organization (4).
Cellular senescence is a program of irreversible growth arrest initiated by stress and dysfunction [a related phenomenon being oncogene-induced senescence (OIS)]. An important component of the cellular senescence program is the senescence-associated secretory phenotype (SASP), a group of secreted proteins that includes growth factors, inflammatory cytokines, and proteases (the senescence messaging secretome) (6,7). Recent findings place JMJD3 central to emergence of the SASP, the consequences of which on surrounding tissues depend on context (8). These may be positive (suppression of tumorigenesis, promotion of tissue repair and regeneration) or deleterious [induction of pro-inflammatory cells, stimulation of tumor progression, undermining of normal tissue homeostasis and repair (9)]. Thus, while the biology of cancers (including glioma) involves aberrant genetic changes, there is accumulating evidence that inflammatory processes associated with aging (10,11), along with epigenetic modifications and chromatin remodeling, play key roles in the neoplastic process (12,13).
In the present study we examined JMJD3 expression in glioma cells and its relation to emergence of senescence-associated phenotypes. We observed that higher levels of JMJD3 expression are associated with characteristic markers including lysosomal senescence-associated β-galactosidase (SA-β-gal) activity, changes in cell and nuclear morphology, and activation of the SASP. Our findings thus indicate that JMJD3, through stimulating secretion of the inflammatory cytokines and proteases of the SASP (1,14,15) and other mechanisms, may contribute to tumorigenesis and tumor progression (16,17).
We specifically relate these studies to treatment of glioma, the most common type of primary malignant brain tumor in adults (18). Because of the invasive nature of glioma, traditional treatment options such as surgery, chemotherapy and radiation, are not curative, and survival averages only 2–3 years following initial diagnosis (19). For well over the past decade, cell-based therapies to target otherwise inaccessible neoplastic and chronic diseases associated with inflammation have increasingly been pursued. Prominent among them are neural stem cells (NSCs) and other stem cells that intrinsically home to sites of tumors and other inflammatory areas (20). Many of the inflammatory cytokines and proteases of the SASP have also been linked to the homing of therapeutically-useful NSCs to tumor foci. Cell-based therapies targeting pathologies associated with inflammation such as cancer and ischemia are currently in clinical trials. Our study is thus particularly relevant because it suggests that the intrinsic biology underlying secretion of these factors is an important component of therapeutic disease targeting.
Materials and Methods
Microarray analysis of JMJD3 expression in patient cohorts
Tumor versus normal gene expression was calculated using 429 primary patient samples and 31 normal samples taken from combined Sun et al. (21) and Gravendeel et al. (22) data sets, and evaluated by 2-way ANOVA (to correct for differences in expression between cohorts) was used to compare data sets using Partek® Genomics Suite™ (Partek, Inc., St. Louis, MO). Fold-change values were calculated based upon the least-squares mean using Partek® Genomics Suite™. Prior to statistical analysis, data were normalized using robust multichip average (RMA) normalization (23). The correlation between JMJD3 and IL6 comes from pre-processed expression data from Freije et al. (24). Pearson correlation coefficient and p-value was calculated using the cor.test function in R (25).
Cell culture
U251 and U87 human glioma cell lines (purchased from ATCC and a gift from Dr. Michael C. Jensen, City of Hope Beckman Research Institute) and normal human astrocytes (NHA; T0250, purchased from Applied Biological Materials, Richmond, BC V6V 2G2, Canada) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and penicillin (100 units/ml)/streptomycin (100 μg/ml). Human fetal mesenchymal stem cells (MSCs) immortalized by transduction with human telomerase reverse transcriptase (26) were cultured in alpha-MEM (Invitrogen, 12571), 100 μM ascorbic acid (Sigma, A8960), 2 mM L-glutamine, 15% FBS, 1% Pen/Strep. All cell cultures were incubated in a humidified chamber containing 6% CO2 at 37 °C.
RNA isolation, cDNA synthesis, and RT-qPCR
RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. A total of 2 μg of RNA was used for cDNA synthesis using the BioRad cDNA synthesis kit. Reaction conditions for cDNA synthesis were as follows: 5 min at 25 °C, 30 min at 42 °C, 5 min at 85 °C, hold at 4 °C. Real-time quantitative PCR (RT-qPCR) was performed on a BioRad CFX96 cycler using BioRad SYBR Green Mix in accordance with the manufacturer’s protocol. Reaction conditions for RT-qPCR were as follows: 3 min at 95°C, followed by 39 cycles of 10 s at 95 °C, 10 s at 55 °C, 30 s at 72 °C. Delta Ct values were calculated using GAPDH or HPRT as a reference. The following oligonucleotides were used:
JMJD3: 5′-TGCTCCGTCAACATCAACATTGGC-3′ and 5′-TGCACGAAGCGGTACACAGGAATA-3′;
IL-6 5′-TTCTGCCAGTGCCTCTTTGCTG-3′ and 5′-AGACAGCCACTCACCTCTTCAG-3′,
IL-8: 5′-CACAACCCTCTGCACCCAGTTT-3′ and 5′-GAGAGTGATTGAGAGTGGACCAC-3′,
MCP-1: 5′-AGAATCACCAGCAGCAAGTGTCC-3′ and 5′-TCCTGAACCCACTTCTGCTTGG-3′,
Gro-β: 5′-GGCAGAAAGCTTGTCTCAACCC-3′and 5′-CTCCTTCAGGAACAGCCACCAA-3′,
Gro-γ: 5′-TTCACCTCAAGAACATCCAAAGTG-3′ and 5′-TTCTTCCCATTCTTGAGTGTGGC-3′,
uPA: 5′-TTGCTCACCACAACGACATT-3′ and 5′-GGCAGGCAGATGGTCTGTAT-3′,
HPRT: 5′-CTTGAGCACACAGAGGGCTACA-3′ and 5′-CATTATGCTGAGGATTTGGAAAGG-3′,
GAPDH: 5′-ACCACCCTGTTGCTGTAGCCAA-3v and 5′-GTCTCCTCTGACTTCAACAGCG-3′
DNA constructs and generation of stable cell lines
All plasmid DNA was prepared using a Qiagen Maxi kit. pMSCV-JMJD3mut and pMSCV-JMJD3wt constructs were purchased from Addgene (Cambridge, MA). A vector control plasmid was generated by excising the JMJD3 sequence from pMSCV-JMJD3mut construct using the XhoI restriction enzyme (New England Biolabs, Inc., Ipswich, MA), followed by T-4 mediated (New England Biolabs) ligation of the vector backbone with itself.
U251 glioma cells were transfected with 4 μg of DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in accordance with the manufacturer’s protocol. Seventy-two hours post-transfection, puromycin (1 μg/ml) was added to culture media to select for transfected cells. A monoclonal stably-transfected U251 cell line for each condition (vector, JMJD3mut, and JMJD3wt) was generated by limiting dilution. Puromycin (1 μg/ml) was added to the culture media during the maintenance of the stable cell lines. U87 glioma cells were transfected with DNA following identical procedures, except that Lipofectamine LTX with plus reagent (Invitrogen, Carlsbad, CA) was used in accordance with the manufacturer’s protocol, and these cells were maintained as a batch culture polyclonal population.
Immunofluorescence and microscopy
Cells were grown on Lab-Tek II glass chamber slides (154526, Nunc, Rochester, NY), rinsed in PBS, fixed with 4% paraformaldehyde (PFA) for 10 min, rinsed in PBS, and permeabilized by incubation in PBS containing Triton X-100 (0.25% v/v; 10 min), and blocked using 5% bovine serum albumin. Samples were then incubated with primary and then secondary antibodies. Cell nuclei were stained using DAPI (0.1 μg ml−1) for 2 min before mounting (DAKO). All images were taken using a Nikon TE2000-U fluorescence microscope, Zeiss LSM 510 confocal microscope, or Zeiss AxioObserver fluorescence microscope, as indicated in the text. Quantitative determinations of immunofluorescence intensity for individual cells were made using CellProfiler (27) (www.cellprofiler.org) with DAPI fluorescence marking cells for subsequent measurements.
Immunoblotting
Protein lysates were made using RIPA buffer (Thermo Scientific) containing protease (cOmplete ULTRA Tablets Mini EASY pack; Roche Applied Bioscience, Indianapolis, IN) and phosphatase inhibitors (PhosSTOP tablets; Roche Applied Bioscience) per manufacturer’s guidelines (1 tablet of each / 7 ml RIPA buffer). Protein concentrations were determined by bicinchoninic acid protein assay (Thermo Scientific) and then adjusted to equal concentrations using lysis buffer, followed by addition of lithium dodecylsulfate loading buffer and 2-mercaptoethanol. Lysates (20 μg) were electrophoresed in Novex 4–12% Tris-glycine pre-cast gels (Invitrogen) in MOPS running buffer. Gel contents were transferred to a PVDF membrane using a BioRad semi-dry transfer system. Membranes were blocked in 5% non-fat milk, followed by addition of primary and secondary antibodies. Membranes were then incubated in Pierce ECL Western blotting substrate reagent and were then imaged using X-ray film.
Antibodies and reagents
The following commercially available antibodies were used at the indicated concentrations for Western blot (WB) immunofluorescence (IF), and chromatin immunoprecipitation (ChIP) analyses: JMJD3 (GTX124222, GeneTex, 1:250 IF, 1:500 WB); H3K27me3 (GTX121184, GeneTex, 1:1000 WB, 1–2 μg/ChIP); histone H3 (AB1791, Abcam, 1–2 μg/ChIP); β-actin (A2228, Sigma, 1:2000 WB); lamin A/C (GTX101126, GeneTex, 1:400 IF); γH2AX (GTX61796, GeneTex, 1:400 IF); and Alexa Fluor 488 Phalloidin (A12379, Molecular Probes, 1:40).
The following commercially available antibodies were used at the indicated dilutions for flow cytometry (FC) analyses: VEGFR-PE (FAB357P, R&D Systems, 1:50); IL-6Rα-PE (352803, BioLegend, 1:50); CXCR4-PE (FAB170P, R&D Systems, 1:50); uPAR-FITC (3936CJ, American Diagnostics, 1:50); CXCR1-Alexa647 (335201, BioLegend, 1:50); CXCR2-PE (32075, BioLegend, 1:50); CCR1-PE (335201, BioLegend, 1:50); CCR3-PE (310705, BioLegend, 1:50); CCR5-FITC (313705, BioLegend, 1:50); GM-CSFa-FITC (306906, BioLegend, 1:50); IgG-PE (IC002P, R&D Systems, 1:50); and IgG-fluorescein (IC002F, R&D Systems, 1:50).
Flow cytometry
Flow cytometry was performed using a Guava EasyCyte flow cytometer (Guava Biotechnologies, Hayward, CA) equipped with software functions for analyzing cell viability and cell size as previously described (28). To measure cell viability, cells were trypsinized from a T175 flask and incubated with Guava ViaCount reagent. Cell viability was determined using the Guava ViaCount assay function. Data were analyzed and scatter plots prepared using Flowjo software (TreeStar; Ashland, OR).
Detection of SA-β-gal activity
X-gal staining was performed using the Senescence β-Galactosidase Staining Kit (9860; Cell Signaling, Danvers, MA) in accordance with the manufacturer’s protocol. Briefly, cells were grown in a T25 flask, rinsed one time with PBS and fixed with 2% formaldehyde, 0.2% glutaraldehyde before incubating with β-Galactosidase Staining Solution (40 mM citric acid/Na phosphate buffer, 5 mM K4[Fe(CN)6] 3H2O, 5 mM KCl], 150 mM NaCl, 2 mM MgCl2 and 1 mg/ml X-gal in distilled water) overnight at 37°C. Wash steps were performed with PBS. Cells were manually counted (n > 200) and averages are presented as mean ± s.d.
Cell proliferation assay
U251 cell lines were cultured in 6-well dishes, and viable cells were counted daily using a Guava EasyCyte flow cytometer (Guava Biotechnologies) over seven days.
Electron microscopy
Cells grown in T175 flasks were detached by 0.25% trypsin/EDTA treatment, pelleted by centrifugation at 1,200 rpm for 5 min, resuspended in fixative buffer (2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2), and then stored at 4 °C until processed using a standard electron microscopy sample preparation protocol (29). Electron microscopy was performed on an FEI Tecnai 12 transmission electron microscope equipped with a Gatan Ultrascan 2K CCD camera.
Enzyme-linked immunosorbent assay (ELISA) and cytokine/MMP antibody arrays
ELISA kits (ELC-IL6-001, ELC-IL8-001) and human cytokine array C5 and human MMP array C1 (AAH-CYT-5 and AAH-MMP-1) were purchased from RayBiotech, Inc. (Norcross, GA). All assays were performed according to the manufacturer’s protocols. 24-h serum-free conditioned media were collected from 1 × 106 cells plated in T-75 flasks. Cytokine and MMP arrays were analyzed by densitometry using BioRad Quantity One software (Hercules, CA). Expression levels were then converted to log2-fold values and displayed as a heat map using Microsoft Excel software.
JMJD3 inhibition
JMJD3 inhibitor GSK-J4 (30) was purchased from Tocris (4594; Bristol, UK). U251.JMJD3wt cells were treated with 20 μM GSK-J4 for 3 days, added every 24 hours along with fresh media.
Chromatin immunoprecipitation assay (ChIP)
ChIP was performed using ChIP-IT Express Kit (102026, Active Motif, Carlsbad, CA) according to the manufacturer’s protocol. Cells were grown in 10 cm dishes to 80% confluence and fixed by adding 260 μl of 37% formaldehyde directly to 10 ml of culture media covering the cells. Fixation was at room temperature for 2 min for histone ChIP assay or 10 min for H3K27me3 ChIP assay. Chromatin samples were prepared by sonication using a Misonex Sonicator 3000 for a total of 4 cycles; 40% power output for 10 s. Sheared chromatin was incubated with rabbit polyclonal antibodies and then precipitated using magnetic Protein G-coupled beads. DNA was purified using the Chromatin IP DNA Purification Kit (103500, Active Motif). Primer sequences used were: Upstream of the IL-6 transcriptional start site, 5′-TGCTTGCTTTCACCAGAGGA-3′ and 5′GCCAGGAGGCTGACTCTTTT-3′; and at the IL-8 transcriptional start site 5′-GGGCCATCAGTTGCAAATC-3′ and 5′-TCCTTCCGGTGGTTTCTTC-3′.
Additional primer sequences to find the optimal site for the demethylation of H3K27me3 up to 90 kb upstream of the IL-6 transcriptional start site were:
5′- AACTATAGTCACCATGCTGT -3′ and 5′- AACTGAGAGTAAAACGGTG -3′;
5′- GTCCTTTTATTGCAGCTTGA -3′ and 5′ - AAGACTTTCCCAGATAAACA -3′;
5′- AAGTGAAATGTAACAGGTTC -3′ and 5′- GGATTTAACTGCACTTTATC -3′;
5′- CTAAGGGCTGAATGTTAAAA -3′ and 5′- TCAACACATCAGAATTGGTC -3′;
5′- AGTAGGCATGCTAGACGAGA -3′ and 5′- GGTATTCCTTGTCACTTTGA -3′;
5′- TGCTTGCTTTCACCAGAGGA -3′ and 5′- GCCAGGAGGCTGACTCTTTT-3′;
5′- AAACATCAGCAAACGTCTGC -3′ and 5′- AGTGCTTCACAGGCTGTAGG -3′;
5′- GTGCTTATGAATAATCATGAAGGTG -3′ and 5′- AATACCCCTGCTAACCAGAA -3′;
5′- TATTCATAGGTTTCTTGAGC -3′ and 5′- TTAGAACAGAGGATGTCAAA -3′.
RNA-seq data analysis
Reads were aligned to the human genome (build hg19) using TopHat (31). Reads Per Kilobase per Million (RPKM) mapped reads (32) were calculated for RefSeq (33) transcripts using an expectation-maximization algorithm in Partek® Genomics Suite™ (Version 6.6, Partek, Inc., St. Louis, MO) based upon the method of Xing et al. (34). RPKM values were then further normalized via log2 transformation following addition of a scaling factor of 0.1 (35). Fold-change values were calculated on a linear scale based upon the least-squares mean, p-values were calculated 1-way ANOVA using the normalized RPKM values, and false discovery rate (FDR) values were calculated using the method of Benjamini and Hochberg (36). Transcripts were defined as differentially expressed if they showed a |fold-change| value > 1.5 and FDR < 0.25. Potential functional significance of JMJD3 over-expression versus vector control was assessed by Ingenuity Pathway Analysis (IPA; Ingenuity® Systems; www.ingenuity.com). IPA analysis was based upon gene-level expression statistics.
Cell invasion assay
In vitro cell invasion assays were performed using BD BioCoat™ Matrigel™ Invasion Chambers (354480; BD Biosciences, Bedford, MA). The upper surface of each transwell chamber is coated with Matrigel matrix to block non-invasive cells from migrating through 8 μm membrane pores. 2.5 × 104 cells/0.5 ml DMEM medium (0.25% FBS) were placed in the upper chamber and incubated at 37 °C, 6% CO2 for 24 h. The bottom wells were filled with 10% FBS which served as a chemoattractant. Invasive cells on the bottom surface of the insert were detached using 0.25% trypsin-EDTA (25200-056; Gibco) and counted by flow cytometry. Assays were performed in triplicate.
Boyden chamber cell migration assay
The Boyden chamber cell migration assay was performed as previously described (37). Briefly, HB1.F3.CD NSCs or MSCs were resuspended in 5% bovine serum albumin (BSA) and added to the top chambers of 8 μm-pore Millicell cell culture inserts (Millipore, P18P01250). As a chemoattractant, serum-free conditioned media was harvested from 106 glioma cells grown in T-75 flasks and placed in the bottom of each transwell. As a negative control, 5% BSA was added to the bottom of the transwell. After 4 hours of incubation, cells that had migrated were removed from the bottom using Accutase (eBioscience, Inc., 00-4555-56) before centrifuging cells in a 96 v-well plate for 5 min at 1200 rpm. The supernatant was aspirated and cell pellets were resuspended in a solution containing Guava ViaCount reagent and PBS 1X (1:1). Total viable cells were counted using a Guava EasyCyte flow cytometer.
NF-κB inhibition
5 × 105 U87 or U251 JMJD3wt cells were plated in T-25 flasks in DMEM media and allowed to adhere for 4 hours. DMEM-C was aspirated and washed twice with PBS. To inhibit NF-κB, U251 JMJD3.wt or U87 cells were exposed to 2.5, 5, or 10 μM Bay 11-7082 (Calbiochem) in DMEM for 60 min. Media was then aspirated and washed twice with PBS before adding serum-free DMEM media. As a vehicle control, we added 10 μM of DMSO (Sigma-Aldrich) to serum-free media. After 24 h, conditioned media was collected and centrifuged for 4 min at 4,000 rpm. Conditioned media was stored at −80 °C until later use in migration assays.
Statistical analysis
Student’s t-test was performed using Microsoft Excel. Two-tailed p values are reported. Statistical significance was set at: *p <0.05; ** p <0.01; *** p <0.001.
Results
JMJD3 expression in patient samples and glioma cell lines
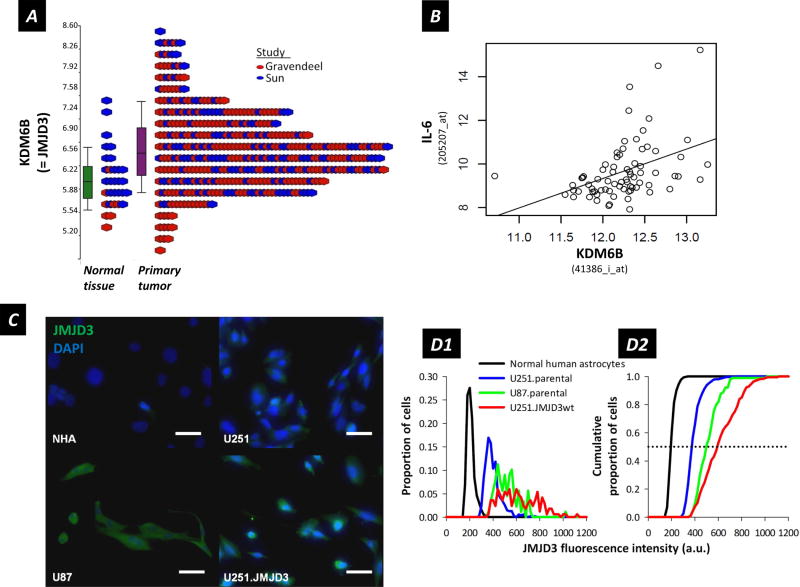
To examine whether JMJD3 is over-expressed at significant levels in patient glioma samples, we first performed analysis for JMJD3 expression using published microarray data available online (22,38). In patient samples we observed 1.37-fold up-regulation (Fisher’s Exact test p-value = 1.5 × 10−5; FDR = 5.8 × 10−5) for JMJD3 expression in primary tumor versus normal tissue (Figure 1A). Further, BRAVO (Biomarkers Recognition and Validation Online) (39) analysis of glioma patient samples (Figure 1B) showed a significant correlation of JMJD3 with expression of interleukin-6 (IL-6), a chemokine linked to inflammation and neural stem cell (NSC) migration to tumor sites (24,37) (Fisher’s Exact test p-value = 2.6 × 10−4; r = 0.41)
Figure 1.
(A) Analysis of KDM6B (=JMJD3) expression in patient tumor samples versus normal tissue (probe 41386_i_at). Fold change = 1.37; by 2-way ANOVA, P-value = 1.5 × 10−5; FDR = 5.8 × 10−5. (B) Correlation between JMJD3 and IL-6 expression in patient glioma samples; n = 74; P-value = 0.000256; r = 0.41. (C) Immunofluorescence detection of JMJD3 in normal human astrocytes (NHA), U251 (parental), U251.JMJD3wt and U87 glioma cells. JMJD3 = green, DAPI = blue. Scale bar = 50 μm. (D1) Histogram of proportions of cells exhibiting JMJD3 staining intensity as indicated (arbitrary units along a 12-bit intensity scale of 0–4095) for the full field images from which the panels in (C) are extracted. (D2) Cumulative proportions of cells plotted against fluorescence intensity to determine the median (0.5, dotted line) JMJD3 staining intensity for the different cell populations. Median staining intensities: 193.5 (n = 392) for normal human astrocytes, 369.6 (n = 551) for U251.parental, 590.7 (n = 168) for U251.JMJD3wt, and 496.5 (n = 89) for U87.parental cells. Cell populations were grown, immunostained and evaluated in parallel under identical conditions.
We then examined endogenous JMJD3 expression in normal human astrocytes and two human glioma cell lines (U251 and U87) by immunofluorescence. Images are presented in Figure 1C, and quantitative summaries of cell JMJD3 immunoreactivity in Figure 1D1 and 1D2. JMJD3 was not detected in normal human astrocytes, expressed at low levels in parental U251 glioma cells, and at relatively higher levels in parental U87 glioma cells and in U251 cells over expressing JMJD3 (U251.JMJD3wt).
Taken together, these observations suggest association of JMJD3 expression with brain tumor cells, as indicated by glioma patient samples and glioma cell lines.
Association of JMJD3 with markers of cellular senescence
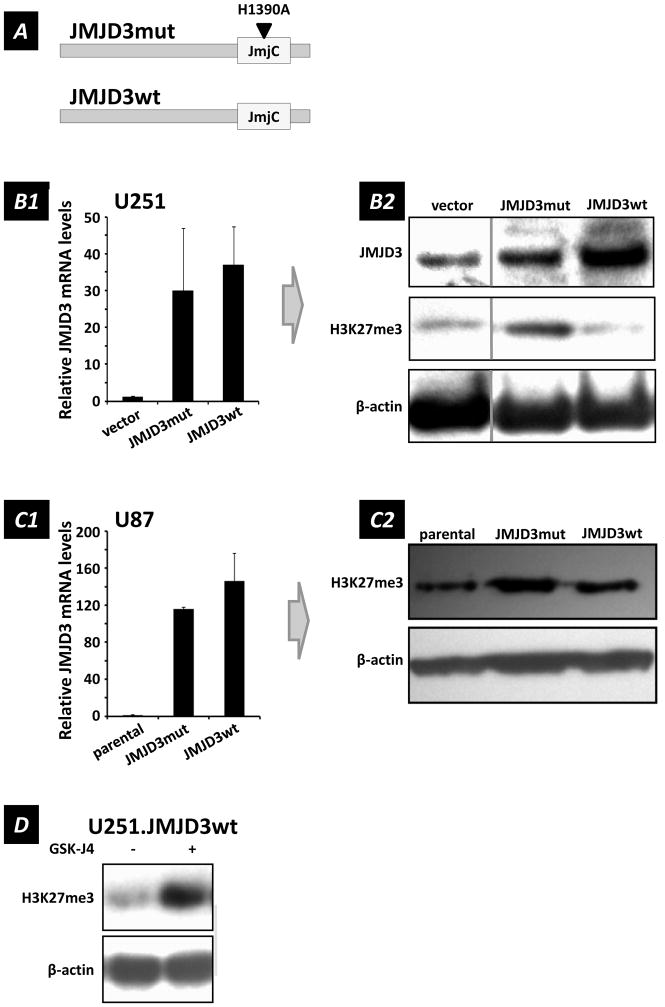
We developed genetically-modified U251 and U87 lines as tools to examine the relationships between JMJD3 expression and display of a series of biomarkers characteristic of senescent cells (40): cell morphology, senescence-associated-β-galactosidase (SA β-gal) activity, proliferation, chromatin remodeling, and the senescence-associated secretory phenotype (SASP). To create these molecular tools to manipulate JMJD3 activity levels, we used functional wild-type and single point mutated (H1390A) catalytically-inactive JMJD3 constructs: JMJD3wt and JMJD3mut (Figure 2A).
Figure 2.
Analyses of U251 human glioma cells transfected with JMJD3mut and JMJD3wt constructs. (A) Schematic of JMJD3mut and JMJD3wt constructs. JMJD3wt contains the full JMJD3 wild type sequence, and has functional H3K27 demethylase activity. JMJD3mut contains a H1390A point mutation located within its JmjC catalytic domain converting histidine to alanine, which renders it enzymatically inactive. (B1) JMJD3 transcript levels detected by RT-qPCR in U251 cells transfected with the JMJD3 constructs indicated (mean ± s.e.m., n = 6). (B2) Western blot showing variation of H3K27me3 levels in U251 cells with expression of JMJD3 constructs (relative to β-actin loading controls). Note that U251 glioma cells transfected to over-express JMJD3wt showed global reduction in H3K27me3 levels, thus confirming that the histidine at position 1390 in the JmjC domain of JMJD3 was essential for H3K27 demethylase activity. (C1) JMJD3 mRNA levels detected by RT-qPCR in U87 cells transfected with the indicated constructs (mean ± s.e.m., n = 3). (C2) Western blot for H3K27me3 in U87 cells transfected with JMJD3mut or JMJD3wt constructs. β-actin served as loading control (parental = untransfected U87 cells). (D) JMJD3 inhibitor GSK-J4 (20 μM; 3 days) antagonizes H3K27me3 methylation in JMJD3 over-expressing U251.JMJD3 cells.
Synthesis of these transcripts in U251 and U87 glioma cells is demonstrated in Figure 2B1 and 2C1. As illustrated in Figure 2B2, over-expression of wild-type JMJD3 demethylase in U251 cells (U251.JMJD3wt) reduced methylation of H3K27 (H3K27me3) below the basal levels shown by the empty vector (U251.vector) control, and methylation levels were increased above baseline by dominant negative JMJD3 (U251.JMJD3mut). Similarly, in U87 cells expressing higher basal levels of JMJD3, H3K27 methylation was increased by expression of inactive JMJD3mut but not appreciably affected by JMJD3wt expression. Use of the JMJD3mut dominant-negative construct allowed us to attribute changes in gene expression and cell morphology to JMJD3 demethylase activity rather than chromatin remodeling effects (41). Additionally, as a control, the empty vector backbone of the JMJD3mut construct (without the JMJD3mut sequence portion) permitted detection of effects potentially related only to vector over-expression.
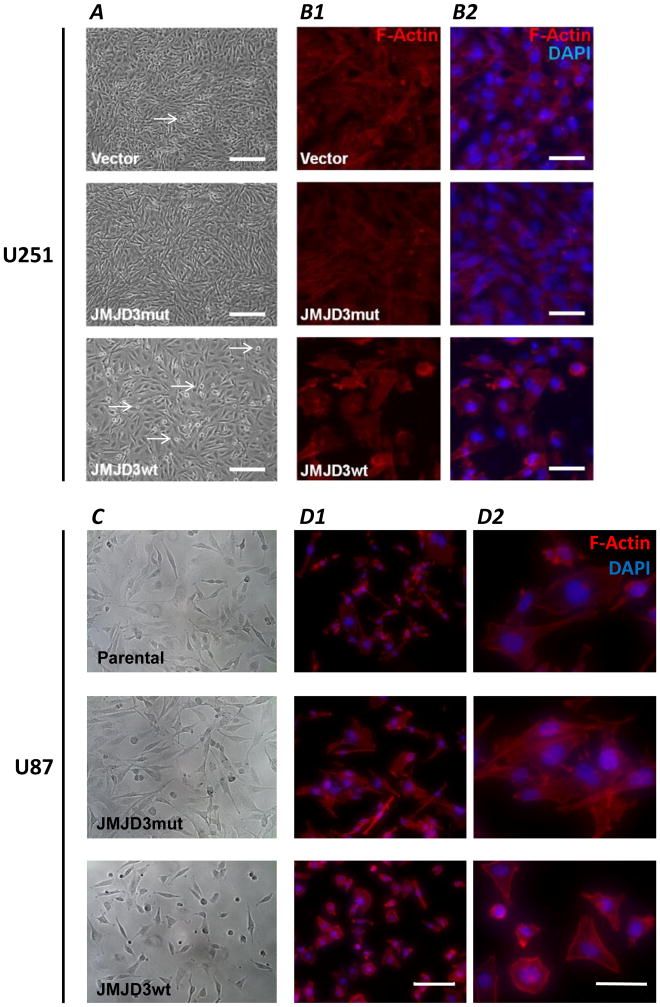
i. Cell morphology
U251.JMJD3wt cells grew as adherent monolayers and displayed contact inhibition, extruding cells from the monolayer when grown beyond confluence (Figure 3A). These extruded or detached cells remained viable and grew as adherent monolayers when transferred at non-confluent density into fresh culture medium (not shown). In contrast, U251.vector and U251.JMJD3mut cells exhibited multilayer growth indicating loss of contact inhibition, and only a few cells displayed anchorage independence (Figure 3A).
Figure 3.
Effects of JMJD3 expression on cell morphology, cytoskeleton, and growth characteristics. (A) Phase-contrast images of U251.vector, U251.JMJD3mut, and U251.JMJD3wt cells. Note the differences in cell morphology, growth density and numbers of cells with anchorage independence growing above the monolayer (arrows). Percentages of cells in each field showing cell-cell contacts and presence of cortical F-actin: U251.vector, 78.3% contacts, 12.3 cortical actin; U251.JMJD3mut, 98.8% contacts, 2.6% cortical actin; U251.JMJD3mut, 23.7% contacts, 38.3% cortical actin. Scale bar = 200 μm. (B) Alexa488-conjugated phalloidin visualization of F-actin (red) in U251 cells transfected with the indicated constructs. In B2, nuclei are counterstained with DAPI (blue). Note the decreased cell contact and presence of cortical (plasma membrane-associated) F-actin in the U251.JMJD3wt cells. Scale bar = 50 μm. (C) Phase-contrast microscopy of parental (untransfected) U87 cells, and U87.JMJD3mut and U87.JMJD3wt cells. (D) Alexa488-conjugated phalloidin visualization of F-actin (red) in U87 cells transfected as indicated. Scale bar = 200 μm. D2 shows enlarged areas of D1 (scale bar = 50 μm).
Tumor cells were stained with Alexa488-conjugated phalloidin to visualize cell morphology and F-actin filaments. In JMJD3-overexpressing U251 (U251.JMJD3wt; Figure 3B1, B2) and U87 (U87.JMJD3wt; Figure 3C, D1, D2) glioma cells, cortical F-actin filaments were prominently associated with the plasma membrane, and these cells displayed fewer cell-cell contacts. In contrast, cultures of U251.vector, U251.JMJD3mut, U87.parental, and U87.JMJD3mut cells with lower levels of JMJD3 expression had networks of cell-cell contacts, and phalloidin staining was more evenly distributed throughout their cytoplasm.
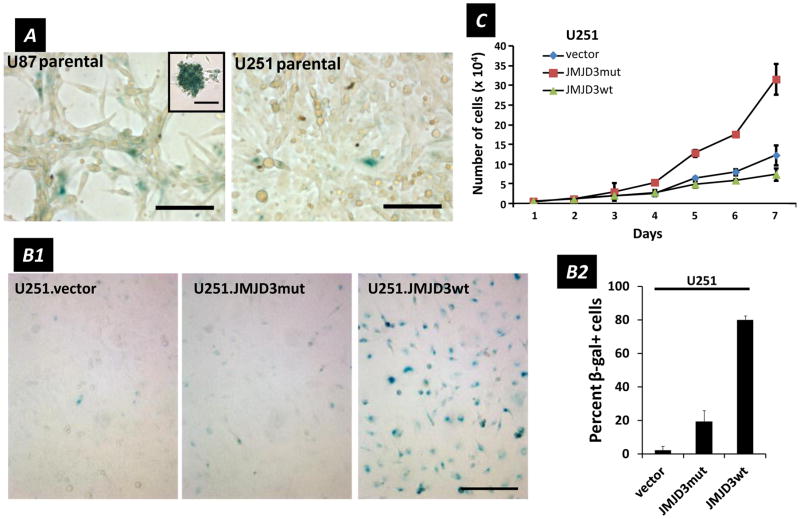
ii. Senescence-associated β-galactosidase (SA-β-gal) activity
In parental glioma cell lines we observed higher SA-β-gal activity (as indicated by blue X-gal staining) in U87 than in U251 cells (Figure 4A), a pattern consistent with their endogenous levels of JMJD3 expression (Figure 1C). Further, over-expression of JMJD3 in U251 cells (U251.JMJD3wt) induced SA-β-gal activity not seen in inactive JMJD3 (U251.JMJD3mut) or U251.vector cells (Figure 4B1). Overall, percentages of SA-β-gal positive cells varied from 0–6% in U251.vector control cells, 15–32% in U251.JMJD3mut cells (the difference with vector control perhaps reflecting incomplete dominant negative suppression), and reached 77–83% in U251.JMJD3wt cells (Figure 4B2).
Figure 4.
SA-β-gal activity and proliferation in glioma cells. (A) Parental U87 and U251 cells stained with X-gal (blue) indicating SA- β-gal activity. Scale bars = 100 μm. (Inset) U87.parental cell sphere stained for SA-β-gal (scale bar = 50 μm). (B1) SA-β-gal activity (blue) of U251 cell lines, vector, JMJD3mut and JMJD3wt, illustrating increase with JMJD3 over-expression. (B2) Percent of SA-β-gal active cells counted in culture. Data are mean ± s.d., n = 3. (C) Increase in cell number over 7 days for U251 lines: vector, JMJD3mut, JMJD3wt. Data are mean ± s.d., n = 3.
iii. Proliferation
The tumor suppressor p16, encoded by the INK4a locus, is regulated by the methylation status of H3K27 under specific control by polycomb group proteins (42–44). In fibroblasts, recruitment of JMJD3 to the INK4a locus leads to expression of p16, which in turn results in the growth arrest observed during normal cellular senescence (1,15). U251.JMJD3wt cells displayed changes in cell growth characteristics that were consistent with, but not as complete as, the G1 arrest seen with cellular senescence in non-tumor cells. Rather, while SA-β-gal+ U251.JMJD3wt cells were mitotically active and could be cultured through multiple passages, we observed differences in cell proliferation rates -- increase for U251.JMJD3mut and decrease for U251.JMJD3wt cells -- when compared to control U251.vector cells (Figure 4C). These data suggest that JMJD3 is modulating proliferation of U251 cells, with over-expression (U251.JMJD3wt) slowing, and the inactive form (U251.JMJ3mut) being permissive, for cell division.
iv. Chromatin remodeling and loss of nuclear envelope integrity
We next looked at whether JMJD3 expression in U251 cells was related to nuclear morphology [a phenotype that may not be linked to demethylase activity (41)], because chromatin organization can influence the size and shape of nuclear structures (45,46). We thus examined nuclear architecture and morphology of JMJD3 over-expressing U251.JMJD3wt cells, and compared these to nuclei in U251.vector and U251.JMJD3mut cells. By DAPI staining of DNA and immunofluorescence imaging of nuclear envelope filament lamin A/C, nuclei of U251.JMJD3wt cells were blebbed and lobular as compared to rounder U251.vector cells and smooth/ovoid U251.JMJD3mut cells (Figures S1A1, S1A2). More detailed examination using transmission electron microscopy (Figure S1B) and confocal microscopy Z-stack reconstruction (Figure S1C) showed that the nuclear envelopes of U251.JMJD3wt cells had ruffled architectures suggesting compromise of membrane integrity (arrows in Figure S1A, S1B, S1C) (47) while nuclear envelopes of U251.vector and U251.JMJD3mut cells were smoother.
v. Senescence-associated secretory phenotype (SASP)
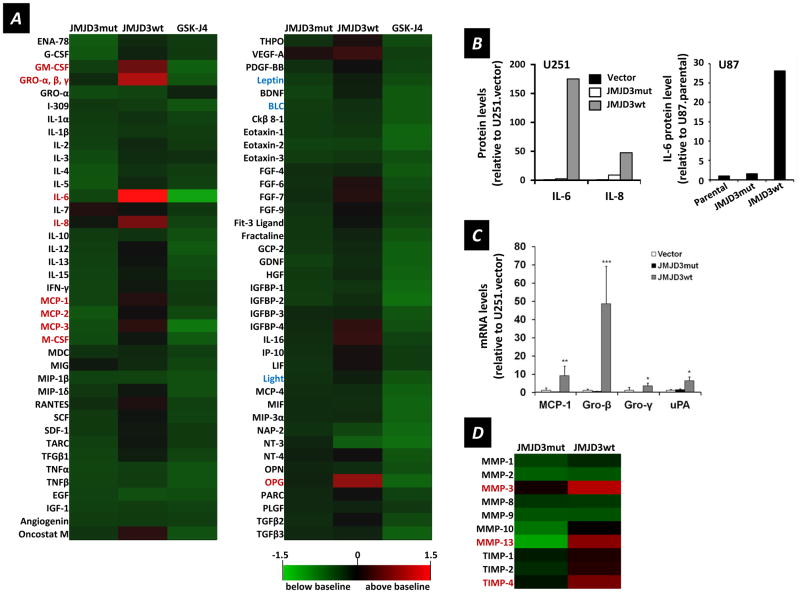
The SASP defines a set of genes encoding soluble factors associated with inflammation and also with malignancy (11,48–50). We used a cytokine antibody array to determine differential secretion profiles relative to control U251.vector cells for U251.JMJD3wt and U251.JMJD3mut cells, as well as for U251.JMJD3wt cells exposed to the JMJD3 inhibitor GSK-J4 (30) (which blocks JMJD3-induced H3K27me3 demethylation activity; Figure 2D). Of the 78 secreted factors analyzed (Figure 5A), U251.JMJD3wt cells secreted higher levels of 22 cytokines (red); notably, GM-CSF, GRO (-α, -β, -γ), IL-6, IL-8, MCP-1 and osteoprotegerin (OPG). This cytokine secretory profile is similar to that reported for senescent fibroblasts and other cells (11,51). Importantly, factors not considered part of the SASP remained near baseline levels (noted in blue): leptin, Light, and BLC. ELISA was used to confirm enhanced secretion of IL-6 and IL-8 in U251.JMJD3wt cells as compared to U251.vector and U251.JMJD3mut cells (Figure 5B), and a similar augmentation of IL-6 production by U87.JMJD3wt cells (Figure 5B).
Figure 5.
Secretory profiles of U251 cells transfected with JMJD3 constructs as indicated and relationship to the SASP. (A) Levels of 78 secreted factors, relative to those of U251.vector cells, for U251.JMJD3mut, U251.JMJD3wt and U251.JMJD3wt plus JMJD3 inhibitor GSK-J4 cells. The heat map key (lower right) indicates log2-fold changes from U251.vector cell baseline levels or U251.JMJD3wt cells treated with vehicle (DMSO) (higher = red, lower = green). (B) Left, IL-6 and IL-8 levels determined by ELISA, relative to those of U251.vector controls, for conditioned media from U251.JMJD3mut and U251.JMJD3wt cells (mean, n = 2). Right, similar data for U87 cells for IL-8 levels relative to those of U87.parental cells for U87 cells transfected with the constructs indicated. (C) RT-qPCR analysis of SASP cytokine mRNA levels relative to U251.vector controls for U251 cells transfected with the constructs indicated (data are mean ± s.d., n = 6). (D) Secreted MMPs and TIMPs determined by antibody array for U251.JMJD3mut and U251.JMJD3wt cells relative to U251.vector control cells. Same heat map scale as in A.
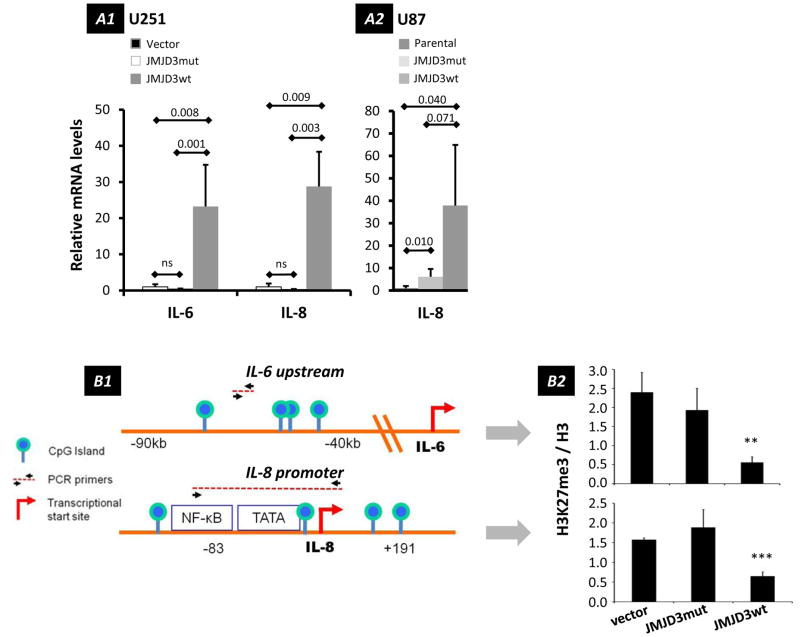
In a complementary approach, RT-qPCR analysis indicated significantly higher IL-6, IL-8, MCP-1, Gro-β, Gro-γ, and uPA mRNA levels in JMJD3 over-expressing U251.JMJD3wt cells compared to U251.vector and U251.JMJD3mut cells (Figures 5C and 6A1), as well as IL-8 in U87 cells (Figure 6A2), confirming transcriptional control of these secreted factors.
Figure 6.
Influence of JMJD3 on expression of IL-6 and IL-8. (A) RT-qPCR analyses of changes in IL-6 (U251 cells) and IL-8 (U251 and U87 cells) mRNA levels after over-expression of JMJD3wt and dominant negative JMJD3mut constructs. (A1) IL-6 and IL-8 mRNA levels in U251 cells relative to vector-only controls (mean ± s.d., n = 6). (A2) IL-8 mRNA levels in U87 cells relative to parental controls (mean ± s.d., n = 3). (B) Methylation status of H3K27 at IL-6 and IL-8 genes (B1) Schematic depicting genomic landmarks and primer annealing sites near IL-6 and IL-8 genes used in ChIP assays. (B2) Results of quantitative ChIP assays showing changes of H3K27me3 relative to histone H3 in U251 cells transfected with the constructs indicated (mean ± s.d., n = 3). JMJD3 demethylates H3K27me3 near IL-6 (p = 0.0013) and IL-8 (p = 0.00019) genes. IL-6 location: 7p21; IL-8 location: 4q13-q21.
Another aspect of the SASP involves the secretion of metalloproteinases (MMPs) and tissue inhibitor of metalloproteinases (TIMPs) (7,52) functioning in extracellular matrix remodeling. Using an antibody array specific for MMPs and TIMPs, we found that conditioned media from U251.JMJD3wt cells contained elevated MMP-3, MMP-10, MMP-13, TIMP-1, TIMP-2, and TIMP-4 relative to U251.JMJD3mut and U251.vector (control) cells, a profile typical of the SASP (Figure 5D).
To better understand JMJD3 targeting of IL-6 and IL-8 genes, we performed a chromatin immunoprecipitation (ChIP) assay to determine whether JMJD3 over-expression reduces H3K27me3 levels specifically at these gene loci. We analyzed CpG islands and transcription factor-binding regions that have been previously reported to have regulatory roles in IL-6 and IL-8 expression (53,54) (Figure 6B1). In U251.JMJD3wt cells, we detected significantly lower levels of H3K27me3 at these sites, compared to U251.vector and U251.JMJD3mut cells, suggesting that these regions are specific targets for JMJD3 (Figure 6B2). These findings suggest that the methylation status of H3K27 determines the expression of IL-6 and IL-8.
iv. Independence from DNA damage
Further, our data suggest that emergence of the SASP in U251.JMJD3wt cells was not a consequence of persistent DNA damage. Because DNA damage triggers SASP independently of p16 and p53 pathways (51,55), phosphorylated histone 2AX (γH2AX) would indicate that the SASP in U251.JMJDwt cells was triggered by persistent DNA damage. However, we found that SASP-expressing U251.JMJD3wt cells lacked γH2AX foci (Figure S2), and only formed γH2AX foci in response to a 10 Gy radiation dose.
Pathway analysis
We studied the potential of JMJD3 as a global regulator of gene expression by using RNA-seq to examine the expression profile of JMJD3 over-expressing cells (U251.JMJD3wt) relative to vector-only controls. Over-expression of JMJD3 mRNA in U251.JMJD3 cells was confirmed by RT-qPCR (Figure 2B1).
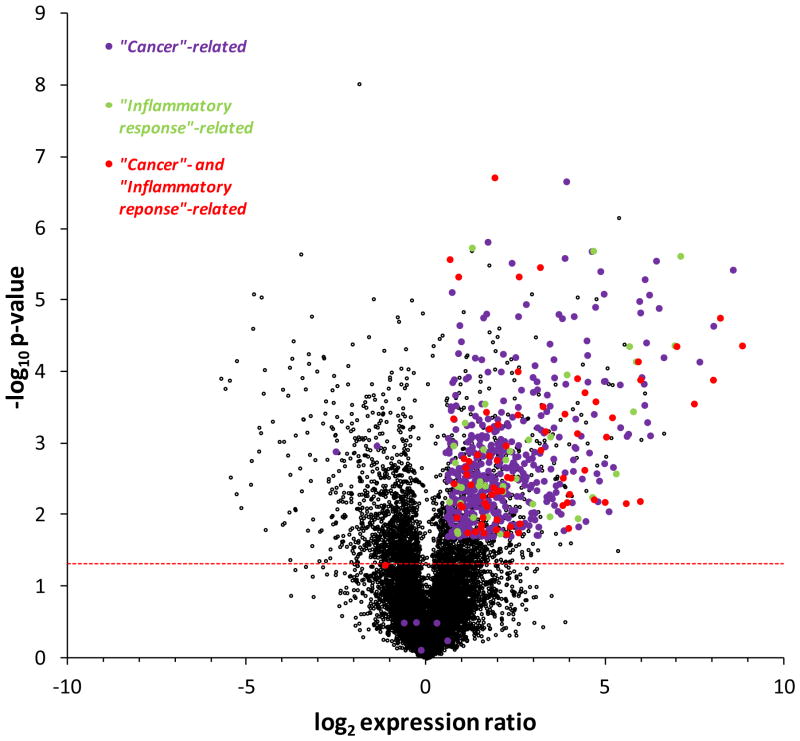
Expression ratio values for the 20,661 genes affected by JMJD3 over-expression in U251 cells are given in Table S1 and plotted in Figure 7. Of these, 4027 with p < 0.05 are above the horizontal line provided for reference in Figure 7. IPA (Ingenuity Pathway Analysis) was then used to assess the potential functional impact of the genes affected by JMJD3 over-expression, and associated these changes with two diseases and disorders relevant to this study: 516 genes to “Cancer” (511 with p < 0.05) and 110 genes to “Inflammatory Response” (109 with p < 0.05). The 73 genes common to “Cancer” and “Inflammatory Response” by IPA are also indicated in Figure 7 (72 with p < 0.05) (Table S2). Overall, 547 of 4027 genes with p < 0.05 (13.6%) were “Cancer”- and/or “Inflammatory Response”-related, and, with three exceptions, all showed enhanced expression in U251.JMJD3 glioma cells.
Figure 7.
RNA-seq of U251.JMJD3wt cells (in comparison to vector control) yielded a total of 37,814 transcripts, 20,661 genes after eliminating duplicate transcripts, and 4027 genes with p < 0.05 (Table S1). Shown are the ratio (log2) in expression of these 20,661 genes plotted against p-value (-log10). The horizontal line for p = 0.05 is provided for reference. By IPA analysis 516 of these genes were associated with “Cancer” (purple), 511 with p < 0.05 (p = 1.90 × 10−7 to 0.02), and 110 genes with “Inflammatory response” (green), 109 with p < 0.05 (p = 1.90 × 10−7 to 0.02). Seventy three genes were common to “Cancer” and “Inflammatory response” (red), 72 with p < 0.05. Of 4027 genes with p < 0.05, 547 (13.6%) were “Cancer”- and/or “Inflammatory response”-related, and with three exceptions showed enhanced expression in U251.JMJD3 glioma cells.
Functional assays
i. Expression by HB1.F3.NSCs of receptors for cytokines present in the SASP phenotype
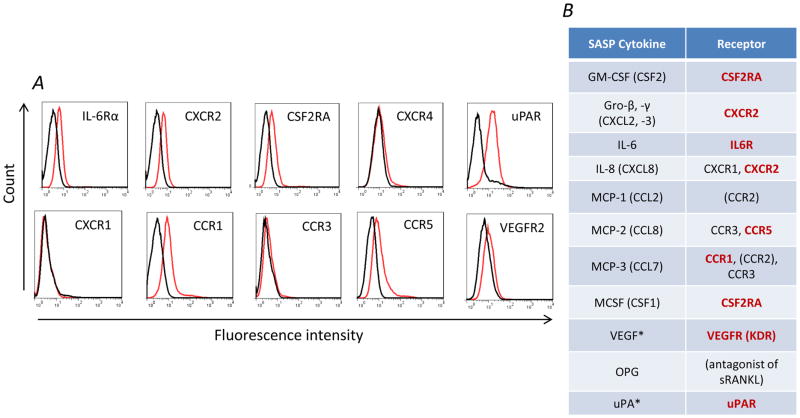
The SASP cytokine expression signature includes inflammatory cytokines that have been shown in other studies to promote HB1.F3.CD NSC migration (37,56). We therefore next evaluated HB1.F3.CD cells for expression of cytokine and chemokine receptors potentially responding to secreted factors of the SASP phenotype. By flow cytometry, we detected VEGFR, IL-6Rα, CXCR2, CCR5, and CSF2RA, as well as uPAR (Figure 8A). Figure 8B shows a summary of receptors expressed by HB1.F3.CD NSCs and their cognate ligands.
Figure 8.
(A) Flow cytometry measurements for surface expression of cytokine receptors by HB1.F3.CD cells (black: isotype control; red: receptor-specific antibody). (B) SASP cytokines assayed by the cytokine array presented in Figure 5A and their corresponding receptors. Receptors marked in bold red are cognate to these SASP factors and expressed by HB1.F3.CD NSCs (CCR2 expression was not examined). *uPA and VEGF are SASP cytokines not assayed by the human cytokine array but whose receptors uPAR and VEGFR are expressed by HB1.F3.CD NSCs.
ii. U251 glioma cell invasion
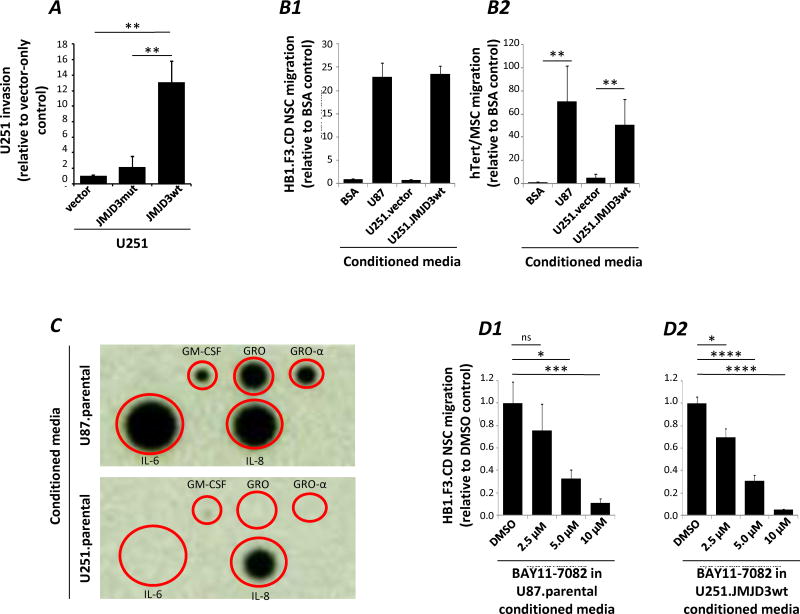
Because JMJD3 expression has been associated with enhanced migration (57), we tested the invasiveness of U251.JMJD3wt, compared to U251.vector and U251.JMJD3mut cells using a Matrigel Boyden chamber invasion assay. U251.JMJD3wt cells were significantly more invasive than U251.vector and U251.JMJD3mut cells (Figure 9A), suggesting enhanced responsiveness to chemoattractants.
Figure 9.
In vitro functional assays. (A) Boyden chamber invasion assay for U251.vector, U251.JMJD3mut, and U251.JMJD3wt cells through Matrigel over a 24 hour period relative to U251.vector control. Data are mean ± s.d., n = 3. (B) Boyden-chamber migration assay for HB1.F3.CD NSC (B1) or hTert/MSC (B2) migration towards conditioned media collected from U87 parental, U251.vector, and U251.JMJD3wt cells, relative to 5% BSA. (C) Cytokine array comparing the secretion of SASP factors from parental U251 and U87 cells. (D) Boyden-chamber migration assay using conditioned media collected from DMSO or NF-κB inhibitor BAY 11-7082-treated parental U87 (D1) or U251.JMJD3wt cells (D2).
iii. Migration of NSCs to media conditioned by glioma cells varying in JMJD3 expression
We then used media conditioned by glioma cell lines varying in JMJD3 expression to test whether soluble factors linked to JMJD3 promoted HB1.F3.CD migration in an in vitro Boyden-chamber cell migration assay. As illustrated in Figure 9B1, HB1.F3.CD migration was greater towards media conditioned by U251.JMJD3wt cells as compared to BSA and U251.vector controls. HB1.F3.CD cell migration towards U251.JMJD3wt conditioned media was also comparable to that seen with media conditioned by U87.parental cells, consistent with their higher basal levels of JMJD3 expression. Enhanced migration associated with JMJD3 over-expression was not limited to NSCs; a similar pattern was observed for mesenchymal stem cells (hTert/MSC) (Figure 9B2).
iv. Comparison of media conditioned by parental U251 and U87 tumor cells
Prompted by the association of JMJD3 with IL-6 expression in patient samples (Figure 1B) and its association with JMJD3wt over-expression in cytokine arrays (Figure 5) we compared levels of cytokines associated with JMJD3 expression in media conditioned by U251.parental and U87.parental cells. U87.parental conditioned media contained higher levels of IL-6 and IL-8 compared to media conditioned by U251.parental cells (Figure 9C), a pattern consistent with higher levels of JMJD3 expression observed in U87.parental cells and a possible mechanism underlying the enhanced migration of HB1.F3.CD NSCs (and hTert/MSCs) to U87 conditioned media.
v. JMJD3 signaling through NF-κB to regulate HB1.F3 NSC migration
NF-κB signaling is thought to drive emergence of the SASP (5). BAY11-7082 is a small molecule inhibitor of NF-κB (58). We observed that HB1.F3.CD NSC migration towards media conditioned U87.parental or U251.JMJD3wt cells showed dose-dependent inhibition by prior BAY11-7082 treatment of the tumor cells (Figure 9D1, D2).
Discussion
Classical characteristics of cellular senescence include β-galactosidase activity (SA-β-gal), proliferation arrest at G1, and the emergence of the senescence-associated secretory phenotype (SASP) (8,40). How processes underlying senescence intersect the intrinsic biology of tumors is not well understood. Here we show that in human patient samples of high grade glioma, JMJD3 is over-expressed and correlated with IL-6 expression. We also took advantage of native differences in JMJD3 expression in two glioma cell lines, low in U251 and high in U87 lines, to manipulate JMJD3 activity by over-expression of wild-type (JMJD3wt) or mutated dominant-negative (JMJD3mut) constructs. In these studies we observed reciprocal effects on H3K27me3 methylation and senescence-associated cell phenotypes: SA-β-gal activity, secretion of SASP cytokines including IL-6 and IL-8 as well as MMPs and TIMPs, growth kinetics, distributions of the actin cytoskeleton, and chromatin organization and nuclear integrity. Further, following a similar pattern, of 4027 genes showing significant change (p < 0.05) with JMJD3 over-expression in U251 cells, expression of 544 (of 547 total) “Cancer”- and/or “Inflammatory Response”-related genes was enhanced. Taken together, these observations place JMJD3 in a central position regulating senescence- and cancer/inflammation-associated phenotypes.
From another perspective, these results provide further evidence implicating senescence-related mechanisms in tumor progression. SASP factors, for example, are increasingly being recognized as regulators of cancer promotion and outcome (59). In this context, the SASP will contribute to tumor progression by providing a microenvironment conducive to angiogenesis, growth, and extracellular matrix remodeling (7). Here, we observed functional roles of JMJD3 in regulating expression of SASP factors including IL-6 and IL-8, and that over-expression of JMJD3 enhanced U251 glioma cell invasiveness associated with increased expression of proteases typical of the SASP. In a possibly related observation, the relationship between JMJD3 expression and cytoskeletal structures, with over expression of JMJD3 in U251 and U87 cells enhancing the presence of cortical actin closely associated with the plasma membrane, may affect the distribution and dynamics of cytokine receptors and responses to cytokines.
In addition, JMJD3 over-expression (U251.JMJD3wt) was accompanied by disorganization in the nuclear lamina within the nuclear envelope and by nuclear blebbing. Lamin influences the size and shape of the nucleus, and serves as an anchoring point for heterochromatin (60). In Hutchinson-Gilford Progeria Syndrome (HGPS), a disease that manifests as premature aging, laminopathy attributed to mutations in the LMNA gene (61) results in disorganization of the nuclear envelope. HGPS cells also have elevated levels of JMJD3, decreased levels of H3K27me3 and loss of heterochromatin integrity (62,63), similar to what we observed in U251.JMJD3wt cells. Thus, JMJD3 function in the nucleus may not be strictly limited to chromatin, but may extend to organization of nuclear filaments such as lamin. Disruption in the nuclear lamina constitutes a state of “chromosomal chaos” in which cancer cells develop aneuploidy (64,65).
Curiously, while previous reports involving normal fibroblasts show that JMJD3 inhibits cell proliferation, our findings demonstrate continued proliferation, albeit at reduced rates, accompanying JMJD3 expression in glioma cells. As a high percentage of gliomas, including U251 and U87 glioma cells, have mutations and deletions in INK4a (44), glioma cells may be responding to senescence stimuli, but incompletely due to loss of effectors responsible for growth arrest.
Our data indicate that over-expression of JMJD3 in U251 glioma cells triggers expression of the SASP in the absence of γH2AX foci marking DNA damage, suggesting that JMJD3-induced chromatin remodeling may be sufficient to trigger emergence of the SASP. JMJD3 over-expression resulted in changes in nuclear shape, and profound effects on chromatin organization. Our data are consistent with previous reports suggesting that chromatin remodeling plays an integral part in triggering the SASP (66). At the same time, because it is not well understood how chromatin changes are instituted and maintained in cellular senescence, and because SAHF formation is cell-type and context dependent (67), absence of γH2AX foci may not necessarily be indicative of an improper senescence program.
We suggest that JMJD3 activity in cancer may be related to the activation of senescence-like programs overlapping with wound repair and with regeneration. Previous work in zebra fish and mice has shown that JMJD3 is necessary for tissue regeneration (68,69). Epigenetic control over gene expression programs is fundamental to wound healing, and tumors have been described as “wounds that do not heal” since many of the characteristics of cancer overlap with wound healing and chronic inflammation (70). For example, both wound healing and tumorigenesis induce cell proliferation, angiogenesis and extracellular remodeling, and up-regulate the expression and secretion of select soluble factors directing host cell migration and recruiting tissue specific stem cells for repair (71).
In concordance with the results from this study, JMJD3 has been shown to regulate expression of IL-6 in mice (3). Linkage of JMJD3 to inflammation is provided by association with stimulation of NF-κB pathway expression in macrophages, as M2 macrophage genes are epigenetically regulated by the methylation status of H3K27 (72). Upon LPS treatment, JMJD3 has been shown to directly promote inflammatory cytokine production by demethylation of H3K27 at the promoter regions of inflammatory cytokine genes (3,14) (see Figure 9). A similar aberrant stimulation of canonical pathways might also be controlled by JMJD3 in cancer. Epigenetic stimulation of the NF-κB pathway is a mechanism of cancer-related chronic inflammation (73), leading to the recruitment of host inflammatory and stem cells typical of tumor pathology (74,75). Thus JMJD3 likely triggers migration of HB1.F3.CD neural stem cells by activating target genes in the NF-κB signaling pathways that overlap with the SASP.
Taken together, our data suggests that epigenetic changes mediated by JMJD3 cause a wide range of effects on gene expression and behavior of glioma cells, including activation of the SASP and concomitant stimulation of nearby cells in the tumor microenvironment. JMJD3 may thus serve as a novel target for inhibiting inflammatory cytokine production in glioma.
In another cancer-related context, NSC tumor-tropism, HB1.F3.CD cell migration to tumor areas is influenced by soluble factors secreted by tumor cells, including IL-6, IL-8, HGF, VEGF, and EGF (28,37,56,76,77), many of which are part of the SASP phenotype for which NSCs express cognate receptors (Figure 8). Because tumor tropism of NSCs is partially dependent upon release of soluble factors by cancer cells, the development of effective NSC-mediated cancer therapies may be further advanced by understanding the molecular mechanisms underlying production of these factors by tumors.
Supplementary Material
Implications.
This glioma study brings together actions of a normal epigenetic mechanism (JMJD3 activity) with dysfunctional activation of senescence-related processes, including secretion of SASP pro-inflammatory cytokines and stem cell tropism towards tumors.
Acknowledgments
Grant support: This work was supported by funding from the California Institute of Regenerative Medicine (DR1-01421), the National Cancer Institute (P30 CA033572), The Rosalinde and Arthur Gilbert Foundation, and STOP Cancer.
The authors acknowledge the technical support of the City of Hope Electron Microscopy Core (Drs. Ricardo Zerda Noriega, Zhuo Li and Marcia Miller), Analytical Cytometry Core (Lucy Brown, Alexander Spalla, Claudio Spalla), Light Microscopy and Digital Imaging Core (Dr. Brian Armstrong), Dr. Purnima Singh for assistance with ChIP assays, Dr. Richard Frank and Fawn Sammy (GeneTex) for technical support on antibodies, Dr. Alexander Annala for comments on the manuscript, Dr. Keely L. Walker for editorial assistance, and Dr. Christine Brown for comments on the manuscript.
Footnotes
Authors’ contributions
Conception and design: J. Najbauer, P.M. Perrigue.
Acquisition of data: N.L. Feng, C.A. Glackin, L. Joseph, D. Mota, P.M. Perrigue, M.A. Reid, M.E. Silva, Y. Tian.
Analysis and interpretation of data: M.E. Barish, J. Najbauer, P.M. Perrigue, M.E Silva, C. Warden.
Writing, review, and/or revision of the manuscript: M.E. Barish, M. Gutova, J. Najbauer, P.M. Perrigue, M.E. Silva.
Study supervision: K.S. Aboody, M.E. Barish, J. Najbauer.
Disclosure of potential conflicts of interest:
KSA is co-founder of TheraBiologics, Inc., a company that supports the development of neural stem cell-mediated treatments for cancer. The authors declare no other conflicts of interest.
References
- 1.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes & development. 2009;23(10):1171–6. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 3.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. The EMBO journal. 2009;28(21):3341–52. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salminen A, Kaarniranta K, Hiltunen M, Kauppinen A. Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. Journal of molecular medicine. 2014;92(10):1035–43. doi: 10.1007/s00109-014-1182-x. [DOI] [PubMed] [Google Scholar]
- 5.Salminen A, Kauppinen A, Kaarniranta K. Emerging role of NF-kappaB signaling in the induction of senescence-associated secretory phenotype (SASP) Cellular signalling. 2012;24(4):835–45. doi: 10.1016/j.cellsig.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nature reviews Cancer. 2009;9(2):81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 7.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annual review of pathology. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodier F, Campisi J. Four faces of cellular senescence. The Journal of cell biology. 2011;192(4):547–56. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120(4):513–22. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(21):12072–7. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS biology. 2008;6(12):2853–68. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams PD. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene. 2007;397(1–2):84–93. doi: 10.1016/j.gene.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, Adams PD. Heterochromatin and its relationship to cell senescence and cancer therapy. Cell cycle (Georgetown, Tex. 2007;6(7):784–9. doi: 10.4161/cc.6.7.4079. [DOI] [PubMed] [Google Scholar]
- 14.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–94. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes & development. 2009;23(10):1177–82. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Garcia E, Licht JD. Deregulation of H3K27 methylation in cancer. Nature genetics. 2010;42(2):100–1. doi: 10.1038/ng0210-100. [DOI] [PubMed] [Google Scholar]
- 17.Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Molecular carcinogenesis. 2008;47(9):701–6. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter KR, McCarthy BJ, Freels S, Kim Y, Davis FG. Prevalence estimates for primary brain tumors in the United States by age, gender, behavior, and histology. Neuro-oncology. 2010;12(6):520–7. doi: 10.1093/neuonc/nop066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal S, Hartz AM, Elmquist WF, Bauer B. Breast cancer resistance protein and P-glycoprotein in brain cancer: two gatekeepers team up. Current pharmaceutical design. 2011;17(26):2793–802. doi: 10.2174/138161211797440186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aboody K, Capela A, Niazi N, Stern JH, Temple S. Translating stem cell studies to the clinic for CNS repair: current state of the art and the need for a Rosetta stone. Neuron. 2011;70(4):597–613. doi: 10.1016/j.neuron.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Hui A-M, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer research. 2009;69(23):9065–72. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 24.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer research. 2004;64(18):6503–10. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 26.Cakouros D, Raices RM, Gronthos S, Glackin CA. Twist-ing cell fate: mechanistic insights into the role of twist in lineage specification/differentiation and tumorigenesis. Journal of cellular biochemistry. 2010;110(6):1288–98. doi: 10.1002/jcb.22651. [DOI] [PubMed] [Google Scholar]
- 27.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutova M, Najbauer J, Frank RT, Kendall SE, Gevorgyan A, Metz MZ, et al. Urokinase plasminogen activator and urokinase plasminogen activator receptor mediate human stem cell tropism to malignant solid tumors. Stem cells (Dayton, Ohio) 2008;26(6):1406–13. doi: 10.1634/stemcells.2008-0141. [DOI] [PubMed] [Google Scholar]
- 29.Graham L, Orenstein JM. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nature protocols. 2007;2(10):2439–50. doi: 10.1038/nprot.2007.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–8. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 33.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic acids research. 2012;40(Database issue):D130–5. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing Y, Yu T, Wu YN, Roy M, Kim J, Lee C. An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic acids research. 2006;34(10):3150–60. doi: 10.1093/nar/gkl396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warden CD, Yuan Y-C, Wu X. Optimal Calculation of RNA-Seq Fold-Change Values. Int J Comput Bioinfo In Silico Model. 2014;2(6):285–92. [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 37.Zhao D, Najbauer J, Annala AJ, Garcia E, Metz MZ, Gutova M, et al. Human neural stem cell tropism to metastatic breast cancer. Stem cells (Dayton, Ohio) 2012;30(2):314–25. doi: 10.1002/stem.784. [DOI] [PubMed] [Google Scholar]
- 38.Sun JY, Huang Q, Wang AD, Dong J, Shao NY, Lan Q. Bioinformatic analysis of glioma development relative genes. Ai zheng = Aizheng = Chinese journal of cancer. 2003;22(3):225–9. [PubMed] [Google Scholar]
- 39.Deng X, Warden C, Liu Z, Zhang I, Yuan Y-C. BRAVO: biomarkers recognition and validation online. in preparation. [Google Scholar]
- 40.Carnero A. Markers of cellular senescence. Methods Mol Biol. 2013;965:63–81. doi: 10.1007/978-1-62703-239-1_4. [DOI] [PubMed] [Google Scholar]
- 41.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Molecular cell. 2010;40(4):594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–8. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 43.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & development. 2007;21(5):525–30. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PloS one. 2009;4(5):e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nature reviews Cancer. 2004;4(9):677–87. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 46.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harbor perspectives in biology. 2010;2(12):a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, et al. Lysosome-mediated processing of chromatin in senescence. The Journal of cell biology. 2013;202(1):129–43. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(20):9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Funayama R, Ishikawa F. Cellular senescence and chromatin structure. Chromosoma. 2007;116(5):431–40. doi: 10.1007/s00412-007-0115-7. [DOI] [PubMed] [Google Scholar]
- 50.Mah LJ, El-Osta A, Karagiannis TC. GammaH2AX as a molecular marker of aging and disease. Epigenetics. 2010;5(2):129–36. doi: 10.4161/epi.5.2.11080. [DOI] [PubMed] [Google Scholar]
- 51.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nature cell biology. 2009;11(8):973–9. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kortlever RM, Bernards R. Senescence, wound healing and cancer: the PAI-1 connection. Cell cycle (Georgetown, Tex. 2006;5(23):2697–703. doi: 10.4161/cc.5.23.3510. [DOI] [PubMed] [Google Scholar]
- 53.Samuel JM, Kelberman D, Smith AJ, Humphries SE, Woo P. Identification of a novel regulatory region in the interleukin-6 gene promoter. Cytokine. 2008;42(2):256–64. doi: 10.1016/j.cyto.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Angrisano T, Pero R, Peluso S, Keller S, Sacchetti S, Bruni CB, et al. LPS-induced IL-8 activation in human intestinal epithelial cells is accompanied by specific histone H3 acetylation and methylation changes. BMC microbiology. 2010;10:172. doi: 10.1186/1471-2180-10-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coppe JP, Rodier F, Patil CK, Freund A, Desprez PY, Campisi J. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. The Journal of biological chemistry. 2011;286(42):36396–403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kendall SE, Najbauer J, Johnston HF, Metz MZ, Li S, Bowers M, et al. Neural stem cell targeting of glioma is dependent on phosphoinositide 3-kinase signaling. Stem cells (Dayton, Ohio) 2008;26(6):1575–86. doi: 10.1634/stemcells.2007-0887. [DOI] [PubMed] [Google Scholar]
- 57.Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. The Journal of biological chemistry. 2012;287(53):44508–17. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, et al. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. The Journal of biological chemistry. 1997;272(34):21096–103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 59.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer metastasis reviews. 2010;29(2):273–83. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes & development. 2008;22(7):832–53. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science (New York, NY. 2006;312(5776):1059–63. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(23):8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burtner CR, Kennedy BK. Progeria syndromes and ageing: what is the connection? Nature reviews Molecular cell biology. 2010;11(8):567–78. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- 64.Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, et al. Genomic instability in laminopathy-based premature aging. Nature medicine. 2005;11(7):780–5. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 65.Duesberg P. Chromosomal chaos and cancer. Scientific American. 2007;296(5):52–9. doi: 10.1038/scientificamerican0507-52. [DOI] [PubMed] [Google Scholar]
- 66.Pazolli E, Alspach E, Milczarek A, Prior J, Piwnica-Worms D, Stewart SA. Chromatin remodeling underlies the senescence-associated secretory phenotype of tumor stromal fibroblasts that supports cancer progression. Cancer research. 2012;72(9):2251–61. doi: 10.1158/0008-5472.CAN-11-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, Bartek J. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a) Cell cycle (Georgetown, Tex. 2011;10(3):457–68. doi: 10.4161/cc.10.3.14707. [DOI] [PubMed] [Google Scholar]
- 68.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO reports. 2009;10(8):881–6. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stewart S, Tsun ZY, Izpisua Belmonte JC. A histone demethylase is necessary for regeneration in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(47):19889–94. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. The New England journal of medicine. 1986;315(26):1650–9. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 71.Anders HJ, Romagnani P, Mantovani A. Pathomechanisms: homeostatic chemokines in health, tissue regeneration, and progressive diseases. Trends in molecular medicine. 2014;20(3):154–65. doi: 10.1016/j.molmed.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–54. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–6. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 75.Najbauer J, Huszthy PC, Barish ME, Garcia E, Metz MZ, Myers SM, et al. Cellular host responses to gliomas. PloS one. 2012;7(4):e35150. doi: 10.1371/journal.pone.0035150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt NO, Przylecki W, Yang W, Ziu M, Teng Y, Kim SU, et al. Brain tumor tropism of transplanted human neural stem cells is induced by vascular endothelial growth factor. Neoplasia (New York, NY. 2005;7(6):623–9. doi: 10.1593/neo.04781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao D, Najbauer J, Garcia E, Metz MZ, Gutova M, Glackin CA, et al. Neural stem cell tropism to glioma: critical role of tumor hypoxia. Mol Cancer Res. 2008;6(12):1819–29. doi: 10.1158/1541-7786.MCR-08-0146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.