Abstract
Background
The overexpression of the chemokine receptor 4 (CXCR4) in different epithelial, mesenchymal, and hematopoietic cancers makes CXCR4 an attractive diagnostic and therapeutic target. However, targeting the CXCR4 receptor with small cyclic pentapeptide-based radiopharmaceuticals remains challenging because minor structural modifications within the ligand-linker-chelate structure often significantly affect the receptor affinity. Based on the excellent in vivo properties of CXCR4-directed pentapeptide [68Ga]pentixafor (cyclo(-d-Tyr-N-Me-d-Orn(AMB-DOTA)-l-Arg-l-2-Nal-Gly-)), this study aims to broaden the spectrum of applicable (radio)metal-labeled pentixafor analogs.
Methods
Cyclic pentapeptides, based on the pentixafor scaffold, were synthesized by a combined solid- and solution-phase peptide synthesis. The CXCR4 receptor affinities of the cold reference compounds were determined in competitive binding assays using CXCR4-expressing Jurkat T - cell leukemia cells and [125I]FC131 as the radioligand.
Results
Metalated pentixafor derivatives with cyclic and acyclic chelators were synthesized by solid-phase peptide synthesis and evaluated in vitro. The resulting CXCR4 affinities (IC50) were highly dependent on the chelator and metal used. Two pentapeptides, Ga-NOTA and Bi-DOTA conjugates, offer an improved affinity compared to [68Ga]pentixafor.
Conclusions
Based on the pentapeptide [68Ga]pentixafor, a broad range of metal-labeled analogs were investigated. The affinities of the new compounds were found to be strongly dependent on both the chelator and the metal used. Bi-labeled pentixafor showed high receptor affinity and seems to be a promising ligand for further preclinical evaluation and future α-emitter-based endoradiotherapy.
Keywords: GPCR, CXCR4, [68Ga]pentixafor, Pentapeptide, DOTA, Chelator, Radiopharmaceutical, Tracer, Cancer
Background
The chemokine receptor 4 (CXCR4) and its sole known natural ligand stromal cell derived factor-1 (SDF-1, CXCL12) are physiologically involved in leukocyte recruitment, homing, and retention of hematopoietic stem and progenitor cells [1, 2]. CXCR4 also holds a substantial role in various pathological conditions and represents a highly attractive therapeutic target. Overexpression of CXCR4 has been linked to cancer proliferation, cell migration, and tissue-specific homing of cancer cells as well as resistance to conventional and targeted therapies [3–5]. After the identification of CXCR4 as a co-receptor for the entry of the HIV virus [6], great efforts have been made towards the development of CXCR4 antagonists. Recently, our group has developed [68Ga]pentixafor (cyclo(-d-Tyr-N-Me-d-Orn(aminomethylbenzoyl (AMB)-DOTA)-l-Arg-l-2-Nal-Gly-), [68Ga]1, Fig. 1), a cyclic pentapeptide with 5 nM affinity and high selectivity towards hCXCR4 [7, 8]. [68Ga]pentixafor exhibits high uptake and long retention in CXCR4-expressing tissues (6.16 % ± 1.16 % ID/g, 1 h post-injection (p.i.) and 4.63 % ± 1.54 %, 2 h p.i. in OH1 human small cell lung cancer tumor-bearing mice) and is rapidly cleared from non-target tissue (1.08 % ± 0.27 % in the blood, 1 h p.i.) and renally excreted [7]; the corresponding tumor-to-blood and tumor-to-muscle ratios are substantially higher than those compared to other peptidic CXCR4 imaging agents [8]. First, studies in patients suffering from lymphoproliferative diseases [9, 10] demonstrated its excellent properties, including a favorable dosimetry for CXCR4-receptor mapping by means of positron emission tomography (PET) [11].
Fig. 1.
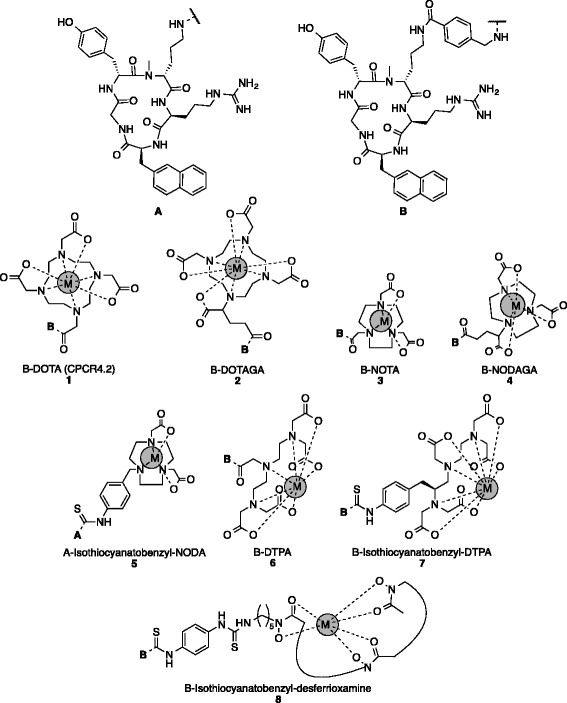
Pentixafor-based analogs with different chelators and metals (M, gray). A constitutes the pentapeptide core cyclo(- d -Tyr-N-Me- d -Orn- l -Arg- l -2-Nal-Gly-) whereas B contains an additional aminomethylbenzoyl (AMB) linker between the peptide and chelator
Based on the success of [68Ga]pentixafor, this study aims to broaden the potential spectrum of radiometal-labeled CXCR4 ligands, both for imaging and therapy, by development and in vitro evaluation of a range of tracers labeled with alternative radionuclides, such as 111In3+ (for single photon emission computed tomography (SPECT)); 18F− and 89Zr4+ (for PET); or 90Y3+, 177Lu3+, and 213Bi3+ (for endoradiotherapy).
However, the experiences gained during the development of pentixafor have shown that, compared with [68Ga]pentixafor, unlabeled pentixafor and other radiometalated pentixafor derivatives exhibit significantly lower CXCR4 receptor affinities. Thus, in contrast to other peptides, such as somatostatin receptor (SSTR), gastrin-releasing peptide receptor (GRPR), or αvβ3 binding peptides, the affinity of [68Ga]pentixafor towards CXCR4 is determined by the entire ligand-spacer-chelator-radiometal construct. Consequently, a more or less independent “bioactive substructure” or “pharmacophor” (e.g., the pentapeptide core A depicted in Fig. 1) cannot be identified. In this study, we investigated pentixafor derivatives with alternative cyclic and acyclic chelators and evaluated these ligands in vitro. With regard to the utilized chelators, the following nuclides relevant for medical purposes have been investigated: Ga3+, AlF2+, Zr4+, Cu2+, In3+, Lu3+, Y3+, and Bi3+ (Fig. 1).
Methods
General
Trityl chloride polystyrene (TCP) resins were purchased from PepChem (Tübingen, Germany) and Sigma-Aldrich (Steinheim, Germany). 9-fluorenylmethyloxycarbonyl (Fmoc) and all other protected amino acid analogs were obtained from Iris Biotech (Marktredwitz, Germany) or Bachem (Bubendorf, Switzerland). Chelators were obtained from CheMatech (Dijon, France, or Macrocyclics (Dallas, USA)) while all other chemicals were bought from Sigma-Aldrich, Fluka, or Merck (Darmstadt, Germany) if not stated otherwise. Solvents and all other organic reagents were purchased from Sigma-Aldrich (Munich, Germany), CLN (Freising, Germany), and VWR (Darmstadt, Deutschland). Water for reversed phase (RP)-HPLC was filtered through a 0.2-μm filter (Thermo Scientific, Barnstead Smart2Pure, Niederelbert, Germany). Analytical RP-HPLC was performed on a Nucleosil 100 C18 (5 μm, 125 × 4.0 mm2) column (CS GmbH, Langerwehe, Germany) using a Sykam gradient HPLC System (Sykam GmbH, Eresing, Germany). For elution, linear gradients of acetonitrile (0.1 % (v/v) trifluoroacetic acid (TFA), solvent B) in water (0.1 % TFA (v/v), solvent A) at a constant flow of 1 mL/min over 15 min were used. UV detection was performed at 220 and 254 nm using a 206 PHD UV-vis detector (LinearTM Instruments Corporation, Reno, USA). Purities were determined at 220 nm using LabSolutions software by Shimadzu Corp. Preparative RP-HPLC was performed on a Sykam gradient HPLC System (Sykam GmbH, Eresing, Germany) equipped with a Multospher 100 RP 18-5 (250 × 20 mm2) column (CS GmbH, Langerwehe, Germany) at a constant flow of 10 mL/min using the same solvents as stated above (duration of gradient, 20 min). Electrospray ionization (ESI)-mass spectra were recorded on a Varian 500-MS IT mass spectrometer (Agilent Technologies, Santa Clara, USA).
General SPPS
Peptides were synthesized manually on solid TCP support using standard Fmoc strategy and an Intelli-Mixer syringe shaker (Neolab, Heidelberg, Germany). As side chain protecting groups, N-[1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl] (Dde) for d-Orn and t-butyl and 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Pbf) groups for d-Tyr and Arg, respectively, were utilized.
O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) and N-hydroxybenzotriazole (HOBt) were used as coupling reagents. N-alkylated amines were acylated using (1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate) (HATU) with 1-hydroxy-7-azabenzotriazole (HOAt) as racemization suppressant.
Cleavage of the peptide from the TCP support with retention of acid-labile protection groups was achieved by treating the peptidyl resin with a solution of dichloromethane (DCM):trifluoroethanol:acetic acid (6:1:3) (v/v/v); for deprotection of acid-labile groups, TFA:triisopropylsilane (TIPS):H2O (95:2.5:2.5) (v/v/v) was used (2× 30 min). Deprotection of Dde was carried out with 2 % hydrazine monohydrate in dimethylformamide (DMF) (v/v).
For Fmoc-deprotection, the peptide was treated with 20 % piperidine in N-methyl-2-pyrrolidone (NMP) (v/v) for 20 min.
For general coupling of amino acids, a solution of Fmoc-Xaa-OH (1.5 equiv), TBTU (1.5 equiv), HOBt (1.5 equiv), and N,N-diisopropylethylamine (DIPEA) (5 equiv) in NMP (1 mL/g resin) was added to the resin-bound peptide and shaken for 90 min at room temperature and washed six times with NMP.
Synthesis of A
The peptide was synthesized according to a previously published procedure [12]. In short, synthesis was carried out using a standard Fmoc strategy using a TCP resin as solid support and HOBt/TBTU as coupling reagents. After selective N-methylation of d-Orn, d-Tyr was coupled to the peptide with HATU/HOAt, cleaved from the resin, and finally cyclized.
Coupling AMB to the d-Orn side chain was carried out using Fmoc- or Boc-protected AMB (1.5 equiv). AMB was preactivated with DIC (N,N′-diisopropyl-carbodiimide) (1.5 equiv), HOAt (1.5 equiv), and DIPEA (4.5 equiv) in 5 mL DMF for 10 min. The d-Orn deprotected peptide precursor was dissolved in DMF, and the preactivated linker was added. After complete reaction (2 h), Fmoc deprotection and semi-preparative HPLC purification, appropriate chelators were coupled to the peptide. HPLC (50 to 100 % B in 15 min): tR = 8.0 min; ESI-mass spectra (MS): calculated for (C61H78N10O10S): 1142.5; found: m/z = 1143.6 [M+H]+.
Coupling of chelators and metal complexation
A molar excess of the activated chelator was added to a free amino group of the peptide analog. Subsequent to successful coupling, the chelator-conjugated peptide was deprotected and purified. Metal complexation was performed in the presence of weakly chelating acetate buffers to reduce the likelihood of hydrolysis. Solutions for metal labeling comprised LuCl3 (20 mM), pH = 6.0; InCl3 (20 mM), pH = 4.5; and YCl3 (20 mM), pH = 5.9, each in ammonium acetate (0.1 M) and Ga(NO3)3 (2 mM) pH = 3.0; Cu(OAc)2 pH = 6.0; and ZrCl4 (20 mM) pH = 1.3, each in water. The chelator-conjugated peptide (250 μL, 2 mM, 1 equiv) was dissolved in H2O and DMSO up to 50 % (v/v), if necessary, and the metal (1–10 equiv) was added, pH adjusted, and heated for 30 min. Final metalated peptides were obtained in a purity ≥ 95 %, and used for in vitro studies without further purification, unless stated otherwise.
Coupling of DOTA or NOTA
For DOTA/NOTA derivatization, 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) or 1,4,7-triazacyclononane-triacetic acid (NOTA) (1 equiv) was preactivated for 20 min using N-hydroxysuccinimide (NHS) (1.25 equiv), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) (1.25 equiv), and DIPEA (2 equiv) dissolved in water (1 mL/0.3 mmol). The peptide B (0.25 or 0.3 equiv, for DOTA or NOTA, respectively) was dissolved in DMF (1 mL per 0.15 mmol of peptide) and slowly added to the reaction mixture according to a previously published protocol [13]. 3: HPLC (30 to 55 % B in 15 min): tR = 10.1 min; ESI-MS: calculated for (C56H73N13O12): 1119.6; found: m/z = 1121.0 [M+H]+, 1143.0 [M+Na]+; 1: HPLC (30 to 55 % B in 15 min): tR = 8.8 min; ESI-MS: calculated for (C60H80N14O14): 1220.6; found: m/z = 1221.6 [M+H]+, 1243.6 [M+Na]+.
Coupling of DOTAGA-anhydride
2,2′,2″-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid (DOTAGA-anhydride) (2 equiv) was dissolved in anhydrous DMF to obtain a white suspension and added to a solution of B (1 equiv) and triethylamine (10 equiv) in anhydrous DMF. 2: HPLC (30 to 55 % B in 15 min): tR = 9.8 min; ESI-MS: calculated for (C63H84N14O16): 1292.6; found: m/z = 1293.5 [M+H]+, 1315.5 [M+Na]+.
Coupling of NODAGA
4-(4,7-bis(2-(tert-butoxy)-2-oxoethyl)-1,4,7-triaza cyclononan-1-yl)-5-(tert-butoxy)-5-oxopentanoic acid ((NODAGA)(tBu)3) (3 equiv) was coupled to B (1 equiv) using a solution of DIC (2 equiv), HOAt (2 equiv), and DIPEA (6 equiv) in anhydrous DMF. The product was fully deprotected and purified. HPLC (30 to 55 % B in 15 min): tR = 10.8 min; ESI-MS: calculated for (C59H77N13O14): 1191.6; found: m/z = 1193.0 [M+H]+, 1215.0 [M+Na]+.
Coupling of DTPA
3,6,9-tris(2-(tert-butoxy)-2-oxoethyl)-13,13-dimethyl-11-oxo-12-oxa-3,6,9-triazatetradecan-1-oic acid (DTPA)(tBu)4 (2 equiv) was coupled to B (1 equiv) using a solution of DIC (1.5 equiv), HOAt (1.5 equiv), and DIPEA (4.5 equiv) in anhydrous DMF. After successful coupling, the product was deprotected and purified. HPLC (30 to 60 % B in 15 min): tR = 8.0 min; ESI-MS: calculated for (C58H75N13O16): 1209.6; found: m/z = 1210.8 [M+H]+, 1232.6 [M+Na]+.
Coupling of p-SCN-Bn-DTPA
2-(4-isothiocyanatobenzyl)diethylenetriaminepentaacetic acid (p-SCN-Bn-DTPA) (21.0 mg, 32.4 μmol, 1.5 equiv) was coupled to B (18.0 mg, 21.6 μmol, 1 equiv) with DIPEA in anhydrous DMF to obtain pH ≈ 9. HPLC (30 to 60 % B in 15 min): tR = 11.2 min; ESI-MS: calculated for (C66H82N14O17S): 1374.6; found: m/z = 1375.8 [M+H]+.
Coupling of p-SCN-Bn-DFO
p-Isothiocyanatobenzyl-desferoxamine (DFO-Bn-NCS) (21.0 mg, 32.4 μmol, 1.5 equiv) was coupled to B (18.0 mg, 21.6 μmol, 1 equiv) in anhydrous DMF and adjusted with NEt3 to pH = 9.5. HPLC (30 to 60 % B in 15 min): tR = 14.0 min; ESI-MS: calculated for (C77H106N18O15S2): 1586.8; found: m/z = 1587.0 [M+H]+.
Coupling of NCS-MP-NODA
2,2′-(7-(4-isothiocyanatobenzyl)-1,4,7-triazonane-1,4-diyl)diacetic acid (NCS-MP-NODA) (1.9 mg, 5.4 μmol, 1.5 equiv) was coupled to A (3 mg, 3.6 μmol, 1 equiv) with DIPEA (1.1 μL, 3.2 μmol, 2 equiv) in anhydrous DMF to obtain pH ≈ 9. HPLC (20 to 50 % B in 15 min): tR = 9.0 min; ESI-MS: calculated for (C54H71N13O10S): 1093.5; found: m/z = 1095.1 [M+H]+, 1116.7 [M+Na]+.
Chelation of Ga3+ with DOTA, DOTAGA, NOTA, NODAGA conjugates
A solution of Ga(NO3)3 (250 μL, 2 mM, 1 equiv) in water, pH = 3.0, was added to the chelator-conjugated peptide, (250 μL, 2 mM, 1 equiv) dissolved in H2O and, if necessary, in DMSO up to 50 % (v/v). The final pH of the mixture was adjusted to 4–6 and heated at 90 °C for 30 min. [natGa]1: HPLC (30 to 55 % B in 15 min): tR = 9.0 min; ESI-MS: calculated for (C60H78GaN14O14): 1287.5; found: m/z = 1287.7 [M+H]+, 1311.7 [M+Na]+; [natGa]2: HPLC (30 to 55 % B in 15 min): tR = 9.8 min (Ga); ESI-MS: calculated for (C63H82GaN14O16): 1359.5; found: m/z = 1361.3 [M+H]+, 1383.2 [M+Na]+; [natGa]3: HPLC (30 to 55 % B in 15 min): tR = 9.3 min; ESI-MS: calculated for (C56H71GaN13O12): 1186.5; found: m/z = 1188.9 [M+H]+, 1209.9 [M+Na]+; [natGa]4: HPLC (30 to 55 % B in 15 min): tR = 10.8 min; ESI-MS: calculated for (C59H74GaN13O14): 1257.5; found: m/z = 1258.8 [M+H]+.
Chelation of In3+, Y3+, and Lu3+ with DOTA, DOTAGA, and DTPA conjugates
A solution of either YCl3, LuCl3, or InCl3 (250 μL, 20 mM, 10 equiv) in ammonium acetate (0.1 M, pH = 6.0) was added to the chelator-conjugated peptide (250 μL, 2 mM, 1 equiv). The pH was adjusted to 4–6, and the mixture heated at 90 °C for 30 min. [natLu]1 and [natY]1: HPLC (10 to 50 % B in 15 min): tR = 10.7 min each; ESI-MS: calculated for (C60H77LuN14O14): 1392.5; found: m/z = 1393.8 [M+H]+; ESI-MS: calculated for (C60H77N14O14Y): 1306.5; found: m/z = 1308.5 [M+H]+, 1329.7 [M+Na]+; [natLu]2 and [natY]2: HPLC (30 to 55 % B in 15 min): tR = 9.8 min each; ESI-MS: calculated for (C63H80LuN14O16): 1463.5; found: m/z = 1465.3 [M+H]+, 1488.1 [M+Na]+; ESI-MS: calculated for (C63H80N14O16Y): 1377.5; found: m/z = 1379.3 [M+H]+, 1402.2 [M+Na]+; [natIn]7: HPLC (30 to 55 % B in 15 min): tR = 10.3 min; ESI-MS: calculated for (C66H77InN14O17S): 1484.4; found: m/z = 1487.4 [M+H]+; [natLu]6, [natIn]6, and [natY]6: HPLC (30 to 60 % B in 15 min): tR = 9.1 min each; ESI-MS: calculated for (C58H71LuN13O16): 1380.5; found: m/z = 1382.8 [M+H]+, for (C58H71N13O16Y): 1294.4; found: m/z = 1296.7 [M+H]+, for (C58H71InN13O16): 1320.4; found: m/z = 1322.9, [M+H]+, 1344.9 [M+Na]+.
Chelation of Zr4+ with 8
To obtain [natZr]8, a solution of ZrCl4 in water (pH = 1.3) (20 mM) was added to the DFO-bearing peptide (2 mM). Quantitative complexation at room temperature occurred within a few minutes without forming any side products. HPLC (30 to 60 % B in 15 min): tR = 14.0 min; ESI-MS: calculated for (C77H103N18O15S2Zr): 1673.6; found: m/z = 1674.5 [M+H]+.
Chelation of Bi3+ with DOTA and DOTAGA conjugates
[natBi]1 and [natBi]2 were prepared by the dropwise addition of natBi(H3CCOO)3 (10 equiv) to a solution of DOTA or DOTAGA-peptide at pH ~ 5. [natBi]1: HPLC (30 to 55 % B in 15 min): tR = 8.8 min; ESI-MS: calculated for (C60H77BiN14O14): 1426.6; found: m/z = 1427.4 [M+H]+, 1451.5 [M+Na]+, 1465.1 [M+K]+; [natBi]2: HPLC (30 to 55 % B in 15 min): tR = 9.8 min; ESI-MS: calculated for (C63H80BiN14O16): 1497.6; found: m/z = 1499.3 [M+H]+, 1521.3 [M+Na]+, 1537.3 [M+K]+.
Chelation of Cu2+ with DOTA, DOTAGA, NOTA, NODAGA, and DTPA conjugates
A solution of Cu(OAc)2 (250 μL, 2 mM, 1 equiv) in water, pH = 6.0, was added to the chelator-conjugated peptide, (250 μL, 2 mM, 1 equiv). The mixture was incubated at room temperature for 30 min. HPLC revealed quantitative complexation. [natCu]1: HPLC (30 to 55 % B in 15 min): tR = 10.8 min; ESI-MS: calculated for (C60H77CuN14O14): 1280.5; found: m/z = 1282.8 [M+H]+, 1306.8 [M+Na]+.
[natCu]2: HPLC (30 to 55 % B in 15 min): tR = 11.0 min; ESI-MS: calculated for (C63H82CuN14O16): 1353.5; found: m/z = 1354.8 [M+H]+, 1377.8 [M+Na]+; [natCu]3: HPLC (30 to 55 % B in 15 min): tR = 10.5 min; ESI-MS: calculated for (C56H71CuN13O12): 1180.5; found: m/z = 1181.8 [M+H]+, 1203.6 [M+Na]+; [natCu]4: HPLC (30 to 55 % B in 15 min): tR = 10.9 min; ESI-MS: calculated for (C59H74CuN13O14): 1251.5; found: m/z = 1253.8 [M+H]+; [natCu]6: HPLC (30 to 55 % B in 15 min): tR = 9.8 min; ESI-MS: calculated for (C58H72CuN13O16): 1269.5; found: m/z = 1271.7 [M+H]+, 1232.8 [M+Na]+.
Chelation of AlF2+ with NOTA and NODA conjugates
The peptide was labeled with [natF]AlF by mixing AlCl3 (1.2 equiv, 0.468 μmol in 0.5 M NaOAc, pH = 4.0), NaF (1.2 equiv 0.468 μmol in 0.5 M sodium acetate, pH = 4.0) with 3 or 5 (1 equiv, 0.39 μmol). The [natF]AlF-labeled peptide was purified using RP-HPLC on a C18 (5 μm, 125 × 4.0 mm). [natF]AlF-3: HPLC (23 % B in 15 min): tR = 10.5 min; ESI-MS: calculated for (C56H71AlFN13O12): 1163.5; found: m/z = 1164.9 [M+H]+; [natF]AlF-5: HPLC (20 to 50 % B in 15 min): tR = 8.2 min; ESI-MS: calculated for (C54H69AlFN13O10S): 1137.5; found: m/z = 1139.0 [M+H]+, 1160.9 [M+Na]+.
Cell culture and determination of CXCR4 receptor affinity (IC50)
For in vitro experiments, the Jurkat T - cell line was used. The cells were maintained in RPMI 1640 medium (Biochrom) containing 10 % fetal calf serum (FCS) (Biochrom). The cell line was cultured at 37 °C in a humidified atmosphere with 5 % CO2 and passaged two to three times a week, depending on the cell count.
CXCR4 affinities were determined in competitive binding assays using Jurkat cells with [125I]FC131 as the radioligand according to a protocol similar to previously published [7]. FC131 (cyclo(-d-Tyr-l-Arg-l-Arg-l-Nal-Gly-)) [14] was synthesized and iodinated as described previously [7]. Jurkat cells (4 × 105 cells per vial) were incubated with the respective peptide of interest at the final concentrations ranging from 10−11 to 10−5 M and app. 0.1 nM of [125I]FC131. The total sample volume was 250 μL. After an incubation time of 120 min, the vials were centrifuged at 1300 rpm (Heraeus Megafuge, Thermo) for 3 min and the supernatant was removed. The cells were washed twice with 200 μL ice-cold Hank’s balanced salt solution (HBSS). After each washing step, the samples were centrifuged and the supernatant removed. Finally, the amount of displaced and bound radioligand in the combined fractions of the supernatant and the cell pellet was quantified. The half maximal inhibitory concentration (IC50) values were determined using GraphPad Prism software.
Results
Based on the promising results of [68Ga]pentixafor [7–11, 15], the influence of different pentixafor-based peptide-linker-chelators on the CXCR4 affinity (IC50) has been evaluated. For this purpose, seven different chelators have been coupled via an AMB moiety to the d-Orn side chain of the CPCR4.2 core peptide A (Fig. 1): (a) DOTA, (b) DOTAGA, (c) NOTA, (d) 1,4,7-triazacyclononane,1-glutaric acid-4,7-acetic acid (NODAGA), (e) DTPA, (f) p-SCN-Bn-DTPA, (g) p-SCN-Bn-Desferrioxamine (DFO), and (h) p-SCN-Bn-NODA, whereby SCN-Bn-NODA was directly coupled to the pentapeptide core without ABS. Depending on the preferred metals for each chelator, reference Ga3+, complexes with AlF2+, Zr4+, Cu2+, In3+, Lu3+, Y3+, and Bi3+ were prepared and evaluated in competitive binding assays on Jurkat T - cells with [125I]FC131 as the radioactive reference (Table 1).
Table 1.
IC50 values of different metal-chelate conjugates consisting of cyclo(-d-Tyr-N-Me-d-Orn(spacer-[M 3+ ]chelator)-l-Arg-l-2-Nal-Gly-), expressed as mean ± SD (n = 3)
| Compound | IC50/nM | Compound | IC50/nM |
|---|---|---|---|
| 1 (DOTA) | 102 ± 17 | 2 (DOTAGA) | 654 ± 263 |
| [natGa3+]1 | 24.8 ± 2.5 | [natGa3+]2 | 380 ± 102 |
| [natBi3+]1 | 22.1 ± 7.0 | [natBi3+]2 | 29.4 ± 7.6 |
| [natCu2+]1 | 131 ± 11 | [natCu2+]2 | 1165 ± 220 |
| [natLu3+]1 | 40.9 ± 12 | [natLu3+]2 | 188 ± 2.8 |
| [natY3+]1 | 40.8 ± 27 | [natY3+]2 | 126 ± 89 |
| 3 (NOTA) | 253 ± 49 | 4 (NODAGA) | 275 ± 106 |
| [natGa3+]3 | 17.8 ± 7.7 | [natGa3+]4 | 342 ± 72 |
| [natCu2+]3 | 46.1 ± 26 | [natCu2+]4 | 343 ± 10 |
| [natF]AlF-3 | 220 ± 57 | ||
| 5 (SCN-Bn-NODA) | 115 ± 24 | 6 (DTPA) | 53 ± 7.9 |
| [natF]AlF-5 | 329 ± 49 | [natY3+]6 | 156 ± 6.7 |
| 7 (SCN-Bn-DTPA) | 200 ± 73 | [natLu3+]6 | 111 ± 77 |
| [natIn3+]7 | 74.1 ± 2.4 | [natIn3+]6 | 90 ± 42 |
| 8 (SCN-Bn-DFO) | 105 ± 35 | [natCu2+]6 | 175 ± 66 |
| [natZr4+]8 | 148 ± 27 |
FC131 as control 9.9 ± 2.4 nM
As already demonstrated [7, 8], non-metalated pentixafor (1) shows low affinity (IC50: 102 ± 17 nM) towards CXCR4, whereas the Lu3+ and Y3+ complexes (IC50: 40.9 ± 12 and 40.8 ± 27 nM, respectively) exhibit affinities quite similar to [natIn]pentixafor (IC50: 44 ± 4 nM) [7]. Surprisingly, the affinity of the Bi3+ complex is even higher (IC50: 22.1 ± 7.0 nM) than the affinity of [natGa]pentixafor (IC50: 24.6 ± 2.5 nM) thus making [213Bi]pentixafor an attractive α-particle emitting therapeutic analog for endoradiotherapy. The affinity of the corresponding Cu2+ complex was low (131 ± 11 nM). With regard to previous reports [7], it is important to notice that absolute IC50 values for [natGa]pentixafor differ because higher cell numbers were used for the IC50 experiments in this study (4 × 105 vs 2 × 105 cells/sample).
When switching to the corresponding DOTAGA derivatives, the IC50 values indicate a similar order within the series of investigated metal complexes with generally lower affinities (Table 1), except the Bi3+-complex, which, again surprisingly, showed only a small decrease of CXCR4 affinity. Thus, with respect to receptor affinity, DOTAGA-pentixafor was found to offer no advantage over DOTA-pentixafor analogs.
A corresponding evaluation of the NOTA and NODAGA derivatives identified [natGa3+]3 as the ligand with the highest affinity in this study (IC50: 17.8 ± 7.7 nM). All other complexes, including the [natF]AlF-NOTA derivative, seem to be unsuitable for further preclinical evaluation or potential clinical application. Similar disappointing results were obtained for NODA-Bn-SCN, DTPA, DTPA-Bn-SCN, and DFO-BN-SCN derivatives.
Discussion
Experiences in the development of CXCR4-targeting peptides showed that affinities to the CXCR4 receptor can be significantly affected by even moderate structural modifications in the pentapeptide core [16–19], the linker unit [20], or the chelate [7, 8]. This is in contrast to previous experiences of GPCR, glycoprotein, or enzyme-targeting peptides, such as SSTRs, GRPRs (bombesin), and integrins (e.g., αvß3, RGD peptides), as well as the prostate-specific membrane antigen (PSMA), to mention only a few. A variety of peptides towards these targets have been developed and—unlike CXCR4—conjugated with a broad spectrum of linker/chelator moieties. For αvß3-binding RGD peptides, the Lys side chain of the typically used c(Arg-Gly-Asp-d-Phe-Lys) does not influence the binding of the peptide in the cleft between the two αv and β3 subunits that forms the heterodimeric transmembrane glycoprotein [21]. Thus, these RGD peptides tolerate the introduction of spacers and chelator or the formation of multimers, such as dimers, tetramers, and octamers [22, 23]. Similar freedom of variation, although not that multifarious, has been found for SST ligands. Tyr3-octreotate for instance was conjugated to both DTPA and DOTA and labeled with natIn, natGa, or natY. All conjugates, including metal-free octreotate bound hSST2 with high affinity (0.2 to 3.9 nM), regardless of the chelator and metal used [24]. This was confirmed by other SST2 binding peptides, coupled to NODAGA, CB-TE2A, or DOTA and labeled with natGa or natCu resulting in affinities in a range from 1.3 to 12.5 nM [25]. Moreover, modification of Tyr3-octreotate by glucose or cellobiose and 4-[18F]fluorobenzaldehyde or 2-fluoropropionic acid resulted in almost unchanged SST2 affinities in the range of 1.3 to 3.1 nM [26]. Hence, compared to CXCR4-binding pentixafor derivatives, where discrepancies varied from 17.6 to 1165 nM (Ga-NOTA and Cu-DOTAGA conjugated to the same highly affine scaffold of CPCR4.2), the effect of chelator and metal exchange was much less pronounced.
Profound investigations on bombesin-receptor mediated imaging agents have shown that bombesin analogs can be conjugated and labeled with a broad range of chelators and metals under retention of their receptor affinity [27]. Smith et al. listed 12 bombesin conjugates, labeled with a variety of metal chelation systems, all of them with an unchanged affinity in the range of 0.5 to 10.5 nM [28, 29].
Regarding PSMA, 14 99mTc-based imaging agents and five copper compounds were investigated with various common chelators of 99mTc and 64Cu, resulting in high affinity for every compound with Ki values ranging from 0.03 to 16.3 nM [30] and 0.19 to 13.26 nM [31] for Tc and Cu compounds, respectively. Free, Ga3+-, and Lu3+-labeled PSMA DOTA and DOTAGA conjugates were shown to be highly specific as well, ranging from 10.2 to 54.7 nM [32]. This list can be prolonged with 18F-tracers developed by Pomper et al. [33, 34] and other further 68Ga-tracers developed by Eder et al. [35, 36].
Although different groups used different models for affinity evaluation, the relative trend shows that, in contrast to the examples above, the CXCR4 affinity is strongly influenced by the entire ligand-spacer-chelator-radiometal construct. During the development of linker-bridged dimers, Demmer et al. could demonstrate that even dimers, consisting of one high affinity and one “non-” CXCR4-binding peptide exhibit higher affinity when compared with the high affinity monomer conjugated with the used linker [20]. The authors conclude a subsite binding of the second peptide unit close to the main binding pocket. Based on the results of this study, we conclude that binding of the AMB-[M3+]chelator moiety of [natGa]pentixafor and [natGa3+]3 significantly contributes to and is a prerequisite for high affinity binding of the entire peptide ligand. Consequently, depending on the chelator, metalation can have a significant effect on the affinity towards CXCR4.
Conclusions
In summary, these studies demonstrated that pentixafor, consisting of the cyclic peptide cyclo(-d-Tyr-N-Me-d-Orn-l-Arg-l-2-Nal-Gly-) and conjugated at the Orn side chain with AMB-[natGa]DOTA, represents a highly optimized ligand. As a result of this study, two further ligands, a Ga-NOTA ([natGa3+]3) and a Bi-DOTA ([natBi3+]1) derivative with slightly higher affinity to hCXCR4, have been developed. Whereas the Ga3+-ligand [natGa3+]3 suffers from a lower hydrophilicity and thus presumably inferior pharmacokinetics compared to [natGa]pentixafor, the Bi3+-complex is expected to be a very promising new ligand for further studies towards α-emitter-based endoradiotherapeutic approaches, including multiple myeloma and other lymphoproliferative disorders.
Acknowledgements
The research leading to these results has received funding from the Deutsche Forschungsgemeinschaft (DFG) under Grant Agreement No. SFB 824 project Z1 and B5. The authors thank V. Felber, S. Hintze, and M. Konrad for synthetic assistance and [natF]AlF-labeling of NOTA- and NODA-ligands and M. Wirtz and J. Notni for supportive discussions.
Abbreviations
- (NODAGA)(tBu)3
4-(4,7-bis(2-(tert-butoxy)-2-oxoethyl)-1,4,7-triazacyclononane-1-yl)-5(tert-butoxy)-5-oxopentanoic acid
- AMB
aminomethylbenzoyl
- CXCR4
chemokine receptor 4
- DCM
dichloromethane
- Dde
N-[1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl]
- DIC
N,N′-diisopropyl-carbodiimide
- DIPEA
N,N-diisopropylethylamine
- DMF
dimethylformamide
- DOTA
1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid
- DOTAGA
1,4,7,10-tetraazacyclododecane,1-(glutaric acid)-4,7,10-triacetic acid
- DOTAGA-anhydride
2,2′,2″-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid
- DTPA
diethylenetriaminepentaacetic acid
- DTPA(tBu)4
3,6,9-tris(2-(tert-butoxy)-2-oxoethyl)-13,13-dimethyl-11-oxo-12-oxa-3,6,9-triazatetradecan-1-oic acid
- EDCI
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- FCS
fetal calf serum
- Fmoc
fluorenylmethyloxycarbonyl
- GRPR
gastrin-releasing peptide receptor
- HATU
1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- HBSS
Hank’s balanced salt solution
- HOAt
1-hydroxy-7-azabenzotriazole
- HOBt
N-hydroxybenzotriazole
- IC50
half maximal inhibitory concentration
- NCS-MP-NODA
2,2′-(7-(4-isothiocyanatobenzyl)-1,4,7-triazonane-1,4-diyl)diacetic acid
- NHS
N-hydroxysuccinimide
- NMP
N-methyl-2-pyrrolidone
- NODAGA
1,4,7-triazacyclononane,1-glutaric acid-4,7-acetic acid
- NOTA
1,4,7-triazacyclononane-triacetic acid
- Pbf
2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl
- Pentixafor
cyclo(-d-Tyr-N-Me-d-Orn(AMB-DOTA)-l-Arg-l-2-Nal-Gly-)
- PET
positron emission tomography
- p-SCN-Bn-DFO
(4-isothiocyanatophenyl)-3-[6,17-dihydroxy-7,10,18,21-tetraoxo-27-(N-acetylhydroxylamino)-6,11,17,22-tetraazaheptaeicosine] thiourea
- p-SCN-Bn-DTPA
2-(4-isothiocyanatobenzyl)-diethylenetriamine pentaacetic acid
- PSMA
prostate-specific membrane antigen
- SDF-1
stromal cell derived factor-1
- SPECT
single photon emission computed tomography
- SPPS
solid-phase peptide synthesis
- SSTR
somatostatin receptors
- TBTU
O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate
- TCP
trityl chloride polystyrene
- TFA
trifluoroacetic acid
- TIPS
triisopropylsilane
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AP planned and carried out the synthesis and in vitro evaluation of the compounds. MS participated in the design of the study, contributed to data interpretation, and revised the manuscript. MS helped with coordination of the experiments, and HJW helped analyzing and interpreting the data and revised the manuscript. HK and HJW initiated and designed the study. All authors approved the final manuscript.
References
- 1.Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, et al. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23(7):879–94. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- 2.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107(5):1761–7. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 3.Kuil J, Buckle T, van Leeuwen FW. Imaging agents for the chemokine receptor 4 (CXCR4) Chem Soc Rev. 2012;41(15):5239–61. doi: 10.1039/c2cs35085h. [DOI] [PubMed] [Google Scholar]
- 4.Weiss ID, Jacobson O. Molecular imaging of chemokine receptor CXCR4. Theranostics. 2013;3(1):76–84. doi: 10.7150/thno.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodard LE, Nimmagadda S. CXCR4-based imaging agents. J Nucl Med. 2011;52(11):1665–9. doi: 10.2967/jnumed.111.097733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–7. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 7.Demmer O, Gourni E, Schumacher U, Kessler H, Wester HJ. PET imaging of CXCR4 receptors in cancer by a new optimized ligand. Chemmedchem. 2011;6(10):1789–91. doi: 10.1002/cmdc.201100320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourni E, Demmer O, Schottelius M, D’Alessandria C, Schulz S, Dijkgraaf I, et al. PET of CXCR4 expression by a (68)Ga-labeled highly specific targeted contrast agent. J Nucl Med. 2011;52(11):1803–10. doi: 10.2967/jnumed.111.098798. [DOI] [PubMed] [Google Scholar]
- 9.Philipp-Abbrederis K, Herrmann K, Knop S, Schottelius M, Eiber M, Luckerath K, et al. In vivo molecular imaging of chemokine receptor CXCR4 expression in patients with advanced multiple myeloma. EMBO Mol Med. 2015;7(4):477–87. doi: 10.15252/emmm.201404698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wester HJ, Keller U, Schottelius M, Beer A, Philipp-Abbrederis K, Hoffmann F, et al. Disclosing the CXCR4 expression in lymphoproliferative diseases by targeted molecular imaging. Theranostics. 2015;5(6):618–30. doi: 10.7150/thno.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrmann K, Lapa C, Wester HJ, Schottelius M, Schiepers C, Eberlein U, et al. Biodistribution and radiation dosimetry for the chemokine receptor CXCR4-targeting probe 68Ga-pentixafor. J Nucl Med. 2015;56(3):410–6. doi: 10.2967/jnumed.114.151647. [DOI] [PubMed] [Google Scholar]
- 12.Demmer O, Dijkgraaf I, Schottelius M, Wester HJ, Kessler H. Introduction of functional groups into peptides via N-alkylation. Org Lett. 2008;10(10):2015–8. doi: 10.1021/ol800654n. [DOI] [PubMed] [Google Scholar]
- 13.Schottelius M, Schwaiger M, Wester HJ. Rapid and high-yield solution-phase synthesis of DOTA-Tyr(3)-octreotide and DOTA-Tyr(3)-octreotate using unprotected DOTA. Tetrahedron Lett. 2003;44(11):2393–6. doi: 10.1016/S0040-4039(03)00221-1. [DOI] [Google Scholar]
- 14.Fujii N, Oishi S, Hiramatsu K, Araki T, Ueda S, Tamamura H, et al. Molecular-size reduction of a potent CXCR4-chemokine antagonist using orthogonal combination of conformation- and sequence-based libraries. Angew Chem. 2003;42(28):3251–3. doi: 10.1002/anie.200351024. [DOI] [PubMed] [Google Scholar]
- 15.Dijkgraaf I, Demmer O, Schumacher U, Feldhaus S, Anton M, Brandau W, et al. CXCR4 receptor targeting for in-vivo imaging of metastases. J Nucl Med Meeting Abstracts. 2008;49:103P-b. [Google Scholar]
- 16.Mungalpara J, Zachariassen ZG, Thiele S, Rosenkilde MM, Vabeno J. Structure-activity relationship studies of the aromatic positions in cyclopentapeptide CXCR4 antagonists. Org Biomol Chem. 2013 doi: 10.1039/c3ob41941j. [DOI] [PubMed] [Google Scholar]
- 17.Mungalpara J, Thiele S, Eriksen O, Eksteen J, Rosenkilde MM, Vabeno J. Rational design of conformationally constrained cyclopentapeptide antagonists for C-X-C chemokine receptor 4 (CXCR4) J Med Chem. 2012;55(22):10287–91. doi: 10.1021/jm300926y. [DOI] [PubMed] [Google Scholar]
- 18.Debnath B, Xu S, Grande F, Garofalo A, Neamati N. Small molecule inhibitors of CXCR4. Theranostics. 2013;3(1):47–75. doi: 10.7150/thno.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demmer O, Frank AO, Hagn F, Schottelius M, Marinelli L, Cosconati S, et al. A conformationally frozen peptoid boosts CXCR4 affinity and anti-HIV activity. Angew Chem. 2012;51(32):8110–3. doi: 10.1002/anie.201202090. [DOI] [PubMed] [Google Scholar]
- 20.Demmer O, Dijkgraaf I, Schumacher U, Marinelli L, Cosconati S, Gourni E, et al. Design, synthesis, and functionalization of dimeric peptides targeting chemokine receptor CXCR4. J Med Chem. 2011;54(21):7648–62. doi: 10.1021/jm2009716. [DOI] [PubMed] [Google Scholar]
- 21.Haubner R, Gratias R, Diefenbach B, Goodman SL, Jonczyk A, Kessler H. Structural and functional aspects of RGD-containing cyclic pentapeptides as highly potent and selective integrin αVβ3 antagonists. J Am Chem Soc. 1996;118(32):7461–72. doi: 10.1021/ja9603721. [DOI] [Google Scholar]
- 22.Poethko T, Schottelius M, Thumshirn G, Herz M, Haubner R, Henriksen G, et al. Chemoselective pre-conjugate radiohalogenation of unprotected mono- and multimeric peptides via oxime formation. Radiochim Acta. 2004;92(4-6):317–27. doi: 10.1524/ract.92.4.317.35591. [DOI] [Google Scholar]
- 23.Schottelius M, Wester HJ. Molecular imaging targeting peptide receptors. Methods. 2009;48(2):161–77. doi: 10.1016/j.ymeth.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Reubi JC, Schar JC, Waser B, Wenger S, Heppeler A, Schmitt JS, et al. Affinity profiles for human somatostatin receptor subtypes SST1-SST5 of somatostatin radiotracers selected for scintigraphic and radiotherapeutic use. Eur J Nucl Med. 2000;27(3):273–82. doi: 10.1007/s002590050034. [DOI] [PubMed] [Google Scholar]
- 25.Fani M, Del Pozzo L, Abiraj K, Mansi R, Tamma ML, Cescato R, et al. PET of somatostatin receptor-positive tumors using 64Cu- and 68Ga-somatostatin antagonists: the chelate makes the difference. J Nucl Med. 2011;52(7):1110–8. doi: 10.2967/jnumed.111.087999. [DOI] [PubMed] [Google Scholar]
- 26.Schottelius M, Poethko T, Herz M, Reubi JC, Kessler H, Schwaiger M, et al. First (18)F-labeled tracer suitable for routine clinical imaging of sst receptor-expressing tumors using positron emission tomography. Clin Cancer Res. 2004;10(11):3593–606. doi: 10.1158/1078-0432.CCR-03-0359. [DOI] [PubMed] [Google Scholar]
- 27.Sancho V, Di Florio A, Moody TW, Jensen RT. Bombesin receptor-mediated imaging and cytotoxicity: review and current status. Curr Drug Deliv. 2011;8(1):79–134. doi: 10.2174/156720111793663624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CJ, Volkert WA, Hoffman TJ. Gastrin releasing peptide (GRP) receptor targeted radiopharmaceuticals: a concise update. Nucl Med Biol. 2003;30(8):861–8. doi: 10.1016/S0969-8051(03)00116-1. [DOI] [PubMed] [Google Scholar]
- 29.Smith CJ, Volkert WA, Hoffman TJ. Radiolabeled peptide conjugates for targeting of the bombesin receptor superfamily subtypes. Nucl Med Biol. 2005;32(7):733–40. doi: 10.1016/j.nucmedbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Ray Banerjee S, Pullambhatla M, Foss CA, Falk A, Byun Y, Nimmagadda S, et al. Effect of chelators on the pharmacokinetics of (99m)Tc-labeled imaging agents for the prostate-specific membrane antigen (PSMA) J Med Chem. 2013;56(15):6108–21. doi: 10.1021/jm400823w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee SR, Pullambhatla M, Foss CA, Nimmagadda S, Ferdani R, Anderson CJ, et al. (6)(4)Cu-labeled inhibitors of prostate-specific membrane antigen for PET imaging of prostate cancer. J Med Chem. 2014;57(6):2657–69. doi: 10.1021/jm401921j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weineisen M, Simecek J, Schottelius M, Schwaiger M, Wester HJ. Synthesis and preclinical evaluation of DOTAGA-conjugated PSMA ligands for functional imaging and endoradiotherapy of prostate cancer. EJNMMI Res. 2014;4(63):1–15. doi: 10.1186/s13550-014-0063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Pullambhatla M, Foss CA, Byun Y, Nimmagadda S, Senthamizhchelvan S, et al. 2-(3-{1-Carboxy-5-[(6-[18F]fluoro-pyridine-3-carbonyl)-amino]-pentyl}-ureido)-pentanedioic acid, [18F]DCFPyL, a PSMA-based PET imaging agent for prostate cancer. Clin Cancer Res. 2011;17(24):7645–53. doi: 10.1158/1078-0432.CCR-11-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mease RC, Dusich CL, Foss CA, Ravert HT, Dannals RF, Seidel J, et al. N-[N-[(S)-1,3-Dicarboxypropyl]carbamoyl]-4-[18F]fluorobenzyl-L-cysteine, [18F]DCFBC: a new imaging probe for prostate cancer. Clin Cancer Res. 2008;14(10):3036–43. doi: 10.1158/1078-0432.CCR-07-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eder M, Neels O, Muller M, Bauder-Wust U, Remde Y, Schafer M, et al. Novel preclinical and radiopharmaceutical aspects of [68Ga]Ga-PSMA-HBED-CC: a new PET tracer for imaging of prostate cancer. Pharmaceuticals (Basel) 2014;7(7):779–96. doi: 10.3390/ph7070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eder M, Schafer M, Bauder-Wust U, Hull WE, Wangler C, Mier W, et al. 68Ga-complex lipophilicity and the targeting property of a urea-based PSMA inhibitor for PET imaging. Bioconjug Chem. 2012;23(4):688–97. doi: 10.1021/bc200279b. [DOI] [PubMed] [Google Scholar]


