Abstract
KRAS mutations are associated with tumor resistance to EGFR TKIs (erlotinib, gefitinib) and to monoclonal antibody against EGFR (cetuximab). Targeted treatment of mutated RAS patients is still considered as a challenge. Inhibitors of c-Met (onartuzumab or tiwantinib) and MEK (selumetinib—a dual inhibitor of MEK1 and MEK2) signaling pathways showed activity in patients with mutations in KRAS that can became an effective approach in carriers of such disorders. BRAF mutation is very rare in patients with NSCLC, and its presence is associated with sensitivity of tumor cells to BRAF inhibitors (vemurafenib, dabrafenib). In the present study, the frequency and type of KRAS and BRAF mutation were assessed in 145 FFPE tissue samples from CNS metastases of NSCLC. In 30 patients, material from the primary tumor was simultaneously available. Real-time PCR technique with allele-specific molecular probe (KRAS/BRAF Mutation Analysis Kit, Entrogen, USA) was used for molecular tests. KRAS mutations were detected in 21.4 % of CNS metastatic lesions and in 23.3 % of corresponding primary tumors. Five mutations were identified both in primary and in metastatic lesions, while one mutation only in primary tumor and one mutation only in the metastatic tumor. Most of mutations were observed in codon 12 of KRAS; however, an individual patient had diagnosed a rare G13D and Q61R substitutions. KRAS mutations were significantly more frequent in adenocarcinoma patients and smokers. Additional analysis indicated one patient with rare coexistence of KRAS and DDR2 mutations. BRAF mutation was not detected in the examined materials. KRAS frequency appears to be similar in primary and CNS.
Keywords: NSCLC, Central nervous system metastases, KRAS mutations, BRAF mutations
Introduction
Activating mutations in the mitogen-activated protein kinase (MAPK) pathway, which incorporates the enzymes RAS (rat sarcoma, encoded by HRAS, NRAS and KRAS genes), RAF (rapidly accelerated fibrosarcoma, encoded by ARAF, BRAF and CRAF genes), MEK (MAPK/extracellular-signal-regulated kinase—ERK, encoded by MEK1 and MEK2 genes), result in constitutive signaling that leads to oncogenic cell proliferation and cells escape from apoptosis [1, 2].
Kirsten rat sarcoma viral oncogene (KRAS) is involved in proper stimulation of MAPK and PI3-K signaling cascades [1–4]. It was previously described that the KRAS gene mutations lead to uncontrolled activation of RAS protein by accumulation of mediators in GTP-binding site [2, 4, 5]. Majority of the KRAS gene abnormalities has a missense character located at codons 12, 13 or 61. Occasionally, substitutions in codons 59, 117 and 146 are also reported. The KRAS gene mutations have been found above in 40 % of colorectal cancers and in 15–25 % of non-small cell lung cancer (NSCLC)—predominantly in patients with adenocarcinoma and smoking history [2, 4–6].
Clinical trials indicated that the KRAS gene mutations are associated with both resistance for reversible EGFR TKIs (epidermal growth factor receptor tyrosine kinase inhibitors: gefitinib, erlotinib) and in reduction of overall survival (OS) in NSCLC patients. For these reasons, the KRAS gene mutations are considered as a negative prognostic biomarker in NSCLC patients. Moreover, KRAS and NRAS genes mutation limits effectiveness of monoclonal antibodies against EGFR (cetuximab, panitumumab) in colorectal cancer patients. [3, 4, 6–8]. Taking into account that RAS protein can activate several signaling pathways, the direct treatment of patients with KRAS mutation has proved to be a challenge. However, effectiveness of inhibitors targeted to c-Met (onartuzumab, tiwantinib), MAPK (vemurafenib, dabrafenib) or MEK (trametinib, selumetinib) cascades is promising [4, 6–10].
BRAF serine/threonine protein kinase is involved in sending signals from HER family receptors through RAS protein to transcription factors, which are involved in cell proliferation. About 40–50 % of melanoma patients and a few percent of colorectal cancer patients harbor a mutation in BRAF gene, mostly substitution in codon 600. BRAF kinase inhibitors: Vemurafenib and dabrafenib are approved for treatment of late-stage melanoma with BRAF mutation. Moreover, in advanced colorectal cancer, BRAF mutations are associated with a poor prognosis and possibly resistance to treatment with monoclonal antibodies against EGFR (cetuximab and panitumumab). However, BRAF gene mutation is very rare in patients with NSCLC (1–2 %)—mostly in non-smokers with adenocarcinoma histology [1, 11].
To date, the majority of published data evaluated the KRAS gene mutations in primary tumors of NSCLC; however, studies assessing these disorders in metastatic lesions are considerably less frequent. For this reason, the main aims of the study were estimation of the incidence of the most common KRAS mutations in codons 12, 13 and 61 and BRAF V600E substitution in the central nervous system (CNS) metastases in Caucasian patients with advanced NSCLC. Moreover, we performed analysis of differences between molecular profile of metastatic lesions and corresponding primary tumors.
Materials and methods
Patients and material
Formalin-fixed, paraffin-embedded (FFPE) tissue samples were enrolled from 145 Caucasian patients with CNS metastases of advanced NSCLC. The corresponding primary NSCLC tumors were simultaneously available from 30 patients. The patients underwent routine neurosurgical procedures with a palliative aim. The median survival time from neurosurgical treatment to death was 9.1 months (information available from 119 patients). All of studied patients were chemotherapy, radiotherapy or molecularly targeted therapies naive. According to number of smoked cigarettes, patients were qualified as heavy smokers (≥15 pack-years), light smokers (<15 pack-years) and non-smokers. Detailed characteristic of studied group has been presented in Table 1.
Table 1.
Characteristic of studied group
| Gender | |
| Male [n (%)] | 100 (69) |
| Female [n (%)] | 45 (31) |
| Age | |
| Median age ± SD (years) | 60 ± 8.8 |
| ≥60 years [n (%)] | 72 (49.7) |
| <60 years [n (%)] | 73 (50.3) |
| Histopathology | |
| Adenocarcinoma [n (%)] | 80 (55.2) |
| Squamous-cell carcinoma [n (%)] | 29 (20) |
| Large-cell carcinoma [n (%)] | 22 (15.1) |
| NSCLC–NOS [n (%)] | 14 (9.7) |
| Smoking status | |
| Current smokers [n (%)] | 73 (50.4) |
| Former smokers [n (%)] | 21 (14.5) |
| Non-smokers [n (%)] | 36 (24.8) |
| Lack of data [n (%)] | 15 (10.3) |
| Performance status (PS) | |
| 0 [n (%)] | 22 (15.2) |
| 1 [n (%)] | 76 (52.4) |
| 2 [n (%)] | 31 (21.4) |
| 3 [n (%)] | 16 (11) |
The study was approved by the ethics committee of the Medical University of Lublin, Poland (No. KE-0254/86/2013).
Mutation analysis
DNA was isolated from FFPE metastatic tissue samples using QIAamp DNA FFPE Tissue Kit (Qiagen, USA) according to a manufacturer’s protocol. Analysis of the KRAS and BRAF genes mutation was conducted using real-time PCR equipment (m2000rt, Abbott, USA) with allele-specific, fluorescent and hydrolysis molecular probes (Entrogen, USA). Each probe contains a fluorophore (FAM or VIC) at the 5′-terminus and a quencher at a 3′-terminus. Entrogene KRAS/BRAF Mutations Analysis Kit is able to identify the presence of G12 V, G12C, G12A, G12R, G12D, G12S, G13D, G13S, G13R, G13A, G13C, Q61 K, Q61L, Q61R and Q61H substitutions in KRAS gene and V600E substitution in BRAF gene. Most samples contain a mixture of wild type (wt) and mutant variants of KRAS and BRAF genes. The assay is designed to preferentially amplify mutant DNA even in samples with advantage of wt DNA. The assay also amplifies an internal control gene in order to ensure that sufficient amount of DNA is available for amplification. The internal control gene is amplified in all samples, regardless of the presence of a mutation in mentioned genes. Moreover, this Entrogene’s real-time PCR assay is certificated for in vitro diagnosis (CE-IVD), and results obtained in this analysis do not require confirmation using other techniques.
The mutations in KRAS and BRAF genes were analyzed in total volume of PCR mixture (25 μl) contained: 12.5 μl of Master Mix, 5.9 μl of allele-specific probe, 1 μl of purified genomic DNA (20 ng/μl) and 5.9 μl of nuclease-free water. The amplification of examined region was performed in 96-well plates in following steps: pre-denaturation 95 °C-10 min and 40 cycles in conditions: 95 °C-15 s and 60 °C-40 s. The negative control was determined with DNA isolated from peripheral blood leukocytes of healthy individuals, and the positive control of the analysis was the reaction with control DNA supplied with the assay by the manufacturer.
In our previous published studies, the incidence of mutation in EGFR (deletions in exon 19 and substitutions: L858R, T790 M, L861Q, S768I, G719X), HER2 (A775YVMA or M774AYMVM insertion) and DDR2 (S768R substitution) genes was assessed in the analyzed material [12–14]. The co-occurrence of these mutations with KRAS and BRAF genes was also presented in this study.
Statistical analysis
Statistical analysis was performed using Statistica version 9.0 (Statsoft, USA) and MedCalc 10 (MedCalc software, Belgium). Associations between the occurrence of KRAS gene mutations and patient clinical factors were examined using the Chi-square test. The Kaplan–Meier method was used to compare the probability of OS in patients with distinct KRAS gene status. Cox regression model with a stepwise selection with minimum AIC factor (Akaike information criterion) was used to assess which of the clinical and genetic factors affect survival. p values <0.05 were considered as statistically significant.
Results
The KRAS gene mutations were detected in 21.4 % (31/145) of CNS metastatic lesions of NSCLC. The mutations were frequently (93.5 %, 29/31) observed in codon 12 (15-G12C; 5-G12 V; 3-G12D; 2-G12A; 2-G12S; 2-G12R); however, 2 rare mutations in codons 13 (G13D) and 61 (Q61R) were also detected. In analyzed metastatic samples, we have not detected any V600E substitution in BRAF gene.
The KRAS gene mutations were significantly more frequent in adenocarcinoma patients than in other types of NSCLC (30 % adenocarcinoma, 6.9 % squamous-cell carcinoma, 13.6 % large-cell carcinoma, 14.3 % not otherwise specified (NOS) NSCLC; p = 0.0391; χ2 = 8.36), in current smokers than in non-smokers and former smokers (22.2 % non-smokers, 42.9 % former smokers, 19.2 % current smokers; p = 0.037; χ2 = 6.567) and in light smokers then heavy smokers (58.8 vs. 16.9 %; p = 0.00027; χ2 = 13.254). On the other hand, there were no differences in the incidence of KRAS gene mutations related to gender (20 % women vs. 22.2 % men; p = 0.786; χ2 = 0.074), performance status and age. Clinical characteristics of all patients with detected mutation in the KRAS gene have been summarized in Table 2.
Table 2.
Clinical characteristics of patients with KRAS gene mutations
| Gender | Diagnosis | Mutation in CNS metastases | Smoking history | Pack-years | OS (mo.) | Age over 60 | PS | Primary tumor |
|---|---|---|---|---|---|---|---|---|
| Male | Adenocarcinoma | G12C | Never smoker | 0 | 10.2 | No | 0 | mt in CNS tu. and primary tu. |
| Female | NSCLC–NOS | G12C | Current smoker | 60 | 43 | Yes | 0 | Unavailable |
| Male | Adenocarcinoma | G12A | Never smoker | 0 | 3.5 | Yes | 1 | Unavailable |
| Female | Adenocarcinoma | G12C | Current smoker | 14 | No data | Yes | 0 | mt in CNS tu., wt in primary tu. |
| Male | Adenocarcinoma | G12C | Current smoker | 40 | 19.8 | Yes | 1 | Unavailable |
| Female | NSCLC–NOS | G12C | Former smoker | 10 | 3.8 | No | 0 | Unavailable |
| Female | NSCLC–NOS | G12A | Current smoker | 35 | No data | Yes | – | Unavailable |
| Male | NSCLC–NOS | Q61R | Current smoker | 30 | 0.2 | Yes | 3 | Unavailable |
| Male | Adenocarcinoma | G12V | Former smoker | 20 | 52.7 | Yes | 0 | mt in CNS tu. and primary tu. |
| Male | Adenocarcinoma | G12V | Current smoker | 40 | 3.9 | Yes | 1 | Unavailable |
| Female | Adenocarcinoma | G12C | Former smoker | 10 | 17 | No | 1 | Unavailable |
| Male | Adenocarcinoma | G12D | Never smoker | 0 | 6.4 | Yes | 1 | Unavailable |
| Male | NSCLC–NOS | G12C | Former smoker | 10 | 20.4 | No | 3 | Unavailable |
| Male | Squamous-cell carcinoma | G12C | Never smoker | 0 | 5.8 | Yes | 0 | mt in CNS tu. and primary tu. |
| Male | Adenocarcinoma | G12C | Current smoker | 15 | 38.3 | No | 1 | mt CNS tu. and primary tu. |
| Male | NSCLC–NOS | G12S | Never smoker | 0 | 3.8 | No | 0 | Unavailable |
| Male | NSCLC–NOS | G12R | Former smoker | 20 | No data | No | 1 | Unavailable |
| Male | Large-cell carcinoma | G12S | Current smoker | 15 | 11 | No | 2 | Unavailable |
| Female | Adenocarcinoma | G12V | Former smoker | 10 | No data | No | – | Unavailable |
| Male | Adenocarcinoma | G12D | Former smoker | 15 | No data | No | – | Unavailable |
| Male | Adenocarcinoma | G12C | Current smoker | 25 | 36.3 | 2 | 2 | Unavailable |
| Male | Adenocarcinoma | G12V | Never smoker | 0 | 19.6 | No | 1 | Unavailable |
| Female | Squamous-cell carcinoma | G12D | Never smoker | 0 | No data | Yes | – | Unavailable |
| Male | Adenocarcinoma | G12C | Current smoker | 40 | 13.6 | No | 2 | Unavailable |
| Male | Adenocarcinoma | G12C | Current smoker | 25 | 93 | No | 1 | Unavailable |
| Male | Adenocarcinoma | G13D | Current smoker | 50 | 26.8 | Yes | 1 | Unavailable |
| Male | Adenocarcinoma | G12C | Current smoker | 30 | 15 | No | 1 | Unavailable |
| Male | Large-cell carcinoma | G12C | Current smoker | 15 | 18.9 | Yes | 1 | mt in CNS tu. and primary tu. |
| Male | Adenocarcinoma | G12V | Never smoker | 5 | 12.5 | No | 1 | Unavailable |
| Female | Adenocarcinoma | G12R | Former smoker | 20 | 6.1 | Yes | 1 | Unavailable |
| Female | Adenocarcinoma | G12C | Former smoker | 0 | 43.3 | No | 1 | Unavailable |
| Male | Large-cell carcinoma | wt | Never smoker | 0 | 29.4 | Yes | 2 | wt in CNS tu. and G12C in primary tu. |
| Male | Large-cell carcinoma | wt | Former smoker | 20 | 4.4 | No | 2 | wt in CNS tu. and G12C in primary tu. |
wt wild type, mt mutant type, CNS central nervous system, tu tumor
The KRAS gene mutations were detected in 23.3 % (7/30) of corresponding primary tumors. However, comparison of molecular profile in matched primary and metastatic lesions indicated some discrepancies. In 5 patients, the KRAS gene mutations occurred simultaneously in primary and metastatic lesions, but in 2 patients, the KRAS gene mutations (G12C) were detected only in primary tumors. Moreover, in one patient, mutation of the KRAS gene (G12C) was observed in metastatic lesion, whereas the status of the KRAS gene in corresponding primary tumors was estimated as wild type. We did not detect any mutation in the BRAF gene in primary tumors.
In previously study, we found 9 common activating EGFR gene mutations (six L858R substitutions and three deletion in exon 19; 6.29 % of studied group), 3 primary T790 M substitution in EGFR gene (2.1 %of studied group), three S768R substitutions in DDR2 gene (2.1 % of studied group) and one insertion in HER2 gene (0.67 % of studied group) in CNS metastases of NSCLC. The most of CNS metastatic lesions were mutually exclusive. However, in one case we have observed coexistence of S768R mutation in DDR2 gene with G12C substitution in KRAS gene [14].
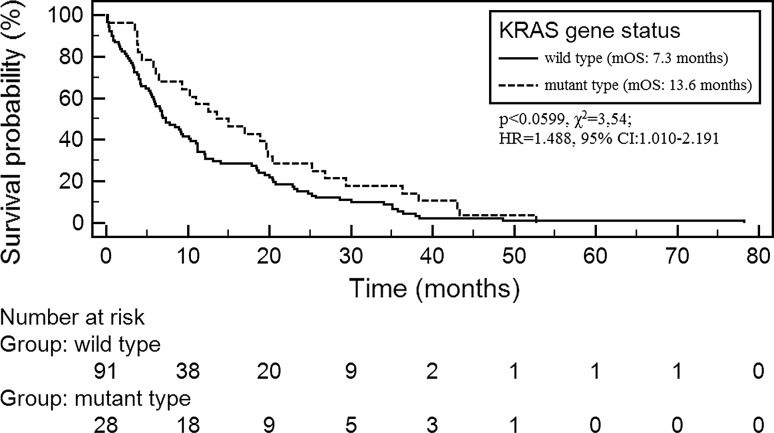
Demographic and clinical factors did not statistically affect on duration of OS in the studied group. There was also no significant association between median OS (mOS) and the occurrence of the KRAS gene mutations. However, patients with the KRAS gene mutations had slightly longer mOS than patients without these mutations (13.6 vs. 7.3 months; p < 0.0599; χ2 = 3.54; HR 1.488, 95 % CI 1.010–2.191; Fig. 1).
Fig. 1.
Overall survival probability in NSCLC patients with different status of KRAS gene
Cox multivariate logistic regression demonstrated the factors that significantly shortened OS in the studied group (overall model fit: χ2 = 6.703, p = 0.035) were as follows: age ≤60 years old (p < 0.0499; HR 0.682, 95 % CI 0.466–0.998) and wild-type status of the KRAS gene (p < 0.0407; HR 0.628, 95 % CI 0.403–0.978).
Discussion
Brain metastases are one of the most common metastatic lesions of NSCLC, which are associated with high mortality of patients. Till date, we have only limited data concerning evaluation of driver mutations incidence (especially EGFR and KRAS genes) in CNS metastases of lung cancer. Descriptions of KRAS gene mutations in CNS metastatic lesions of NSCLC patients occur only in the form of few case reports and in one large study [15]. For this reason, administration of molecularly targeted therapies for such patients is performed only in single cases [15–17].
KRAS and BRAF genes mutation frequency in NSCLC patients
In our analysis, the KRAS gene mutations were detected in 21.4 % of NSCLC CNS metastases using real-time PCR technique. Villalva et al. [15] using pyrosequencing technique detected the KRAS gene mutations in 39 % (30/77) of NSCLC CNS metastases. Moreover, they noticed that pyrosequencing had shown extremely high sensitivity of KRAS gene mutations detection in comparison with techniques used in previous studies. However, such highly sensitive tools are not required to reliably identify KRAS mutations, and real-time technique with CE-IVD molecular probes is recommended as a satisfying in standard diagnostic procedures [18, 19].
In the studies cited below, KRAS gene mutations frequency was analyzed in primary tumors, in metastatic lymph nodes or in available distant metastases. Bauml et al. in group of 374 patients with an informative KRAS mutational analysis found 105 (28.1 %) KRAS gene mutations. Among 366 patients with informative EGFR and KRAS mutational analyses, only 1 (0.3 %) patient exhibited both mutations. The frequency of KRAS mutations was 20.8 % in male patients and 33.2 % in female patients, 8.3 % in never smokers and 32.7 % in ever smokers, 29.9 % in adenocarcinoma tumors and 20.8 % in other NSCLC tumors [20]. Also in the large study of Kris et al., KRAS mutations were the most frequent among other driver mutations in NSCLC patients, and they were found in 182 of 733 analyzed specimens (25 % of patients). Moreover, Kris et al. described 151 EGFR mutations (21 %), 57 ALK gene rearrangements (8 %), 19 HER2 mutations (3 %), 16 BRAF mutations (2 %), 6 PIK3CA mutations (<1 %), 5 NRAS mutations (<1 %) and 1 MEK1 mutation (<1 %) [6]. In European study of Barlesi et al., the 10,000 molecular profiles of NSCLC tumors were characterized. Authors detected 26.9 % tumors with KRAS mutations, 9.4 % tumors with EGFR mutations, 0.9 % tumors with HER2 mutations, 1.6 % tumors with BRAF mutations and 2.6 % PI3KCA mutated tumors as well as 4.0 % tumors with EML4–ALK fusion genes. Double mutations were seen in 0.9 % of the tumors [22]. It was generally reported that the KRAS gene mutations are more frequent in females, smokers and adenocarcinoma subtypes. However, BRAF mutation is extremely rare in NSCLC patients [4, 15, 17, 18]. In our study, we indicated the association between the KRAS gene mutations presence and smoking status as well as adenocarcinoma diagnosis. However, there was no significant association between the presence of KRAS mutations and gender.
Unfortunately, data concerning evaluation of the KRAS gene status simultaneously in corresponding metastatic lesions and primary lung carcinomas are limited. In our study, the corresponding primary tumors were available only in 30 patients; however, it remains a considerable group in comparison with previous reports [13, 18–23]. The KRAS gene mutations were detected in 7 primary tumors (23.4 %) that was in accordance with Kris, Bauml and Barlesi data obtained in higher groups of patients [6, 20, 21]. Additionally, we observed some discrepancies between molecular profile of metastatic and primary lesions. In 71 % of cases (5/7), the same KRAS gene mutations were simultaneously detected in both tumors. However, two mutations were detected only in primary tumors and one only in CNS metastases. Such discrepancies between molecular profile of EGFR and KRAS genes in corresponding primary tumors and various metastatic lesions had also been reported in previous data [17, 24–28].
Manaco et al. detected 11/40 (27.5 %) of the KRAS gene mutations in primary tumors, but the mutations were detected only in 4 (10 %) corresponding metastatic lesions. Moreover, 2 of them had discordant molecular profile of primary and corresponding metastatic lesions [17]. Also Badalian et al. [24] detected three KRAS gene mutations both in primary and in metastatic tumors, but only in one case, the mutations were simultaneously observed in both tumors. Schmid et al. reported that the KRAS gene mutations were more frequent in primary NSCLC than in metastatic lymph nodes (17 vs. 8 %, respectively). However, in one case different type of the KRAS mutation was indicated in primary and metastatic lesions [27]. Similarly, Kalikaki et al. detected the KRAS gene mutations in 5/25 of metastatic and primary samples, but concordance between types of mutation in these lesions was observed only in 2 cases. Moreover, they indicated rare coexistence of the KRAS gene mutations with deletion in EGFR gene that was observed only in primary tumor but not in corresponding metastatic sample [25]. Also Sun et al. [28] described coexistence of the KRAS gene mutation with substitution L858R in EGFR gene. In previous study, we described one coexistence between S768R substitution in DDR2 gene with G12C substitution in KRAS gene in CNS metastases of NSCLC. Unfortunately, in this patient material from corresponding primary tumor was unavailable. The other CNS metastatic lesions were mutually exclusive [14].
The discordance between mutation presence in metastatic and in their corresponding primary NSCLC tumors suggests that molecular status can be changeable during disease progression. Heterogeneity of primary and metastatic tumors indicated that one tissue sample can be considered as representative for this particular lesion but not for all cancer cells [26, 28, 29]. This knowledge can have a potential clinical implication in qualification of patients for molecularly targeted therapies. However, further studies are required to characterize the correlation with the clinical responses to targeted agents in patients with heterogeneous of the driver mutation status between primary and metastatic lesions [18, 26, 28, 29].
KRAS gene mutations as a prognostic factor in NSCLC patients
The KRAS gene mutations were considered as a negative prognostic factor in NSCLC patients. Clinical outcomes were especially poor in patients with KRAS gene mutations after EGFR TKIs therapy [2, 4, 8, 16, 30]. However, clinical trials suggested that the KRAS status has no effect on clinical outcomes to EGFR TKIs therapy in patients without EGFR gene mutations. In this group of patients, the sensitivity of tumors cell on EGFR TKIs therapy is relatively very low [30, 31].
On the other hand, the LACE-bio study suggested that the KRAS gene mutations have no prognostic role in completely resected NSCLC patients treated with adjuvant chemotherapy. However, the overall poor treatment outcomes in the KRAS mutation group treated with adjuvant chemotherapy seemed to be caused by the negative predictive value of codon 13 KRAS gene mutation [32]. In addition, Moran et al. and Garassiono et al. [33, 34] reported that that tumors with KRAS mutations would be more sensitive to pemetrexed. In our study, we observed that patients with mutations in KRAS gene had slightly better prognosis than patients with wt KRAS gene status. However, our study has a significant weakness. Management of patients after neurosurgery was probably very different, and for this reason, the studied group is very heterogeneous. Lack of data about subsequently used therapies excluded possibilities to evaluate KRAS gene status as a reliable prognostic factor.
Therapy strategies
The presence of the KRAS gene mutations is associated with both resistance to EGFR TKIs and reduce of benefits from standard chemotherapy in general group of NSCLC patients [4, 8, 9, 23]. TRIBUTE trial indicated that KRAS gene mutations are associated with worse response rate to standard doubled treatment and erlotinib [35]. The INTEREST trial showed that the KRAS gene mutations were not a predictive factor for a differential survival effect between gefitinib and docetaxel [36]. The BR.21 and the SATURN trials showed that patients with wt of KRAS gene had significant survival benefits from erlotinib in second- or third-line treatment and longer PFS in comparison with patients with KRAS gene mutations [37, 38]. Currently, effective RAS inhibitors are not available. However, selumetinib—an oral, selective, non-ATP competitive inhibitor of MEK1/MEK2 kinases and RAF–MEK–ERK (MAPK) inhibitors—can become a new potential agent in personalized NSCLC therapy [7, 9, 22, 23].
Till date, selumetinib monotherapy had shown any clinical benefits in comparison with standard chemotherapy [39–41]. On the other hand, combination of selumetinib and docetaxel demonstrated significant prolongation of PFS in compared to placebo arm (5.3 vs. 2.1 months, respectively). However, differences in median OS (9.4 vs. 5.2 months, respectively) were statistically insignificant. Moreover, the proportion of serious adverse events (especially neutropenia, diarrhea, nausea and vomiting) was higher in the selumetinib group [41]. Similar, results of preclinical in vivo studies had shown that doublet therapy (selumetinib and docetaxel) leads to more effective inhibition and regression of tumor growth [4, 7, 22]. Moreover, ongoing clinical trials show that combination of targeting agents against different signaling pathways may provide additional benefits in treatment of patients with unregulated MEK, MAPK, RAF and RAS pathways. The presence of BRAF gene mutation is associated with sensitivity of tumor cells to BRAF inhibitors (vemurafenib, dabrafenib). However, before routine application, they need further studies [4, 10, 23, 41].
Conclusions
The KRAS gene mutations (especially in codon 12) are the most frequent genetic abnormalities both in primary and in CNS metastatic lesions of NSCLC. Moreover, the results of this study have indicated discrepancies between molecular profile of some CNS metastases and corresponding primary tumors that may be caused by acquisition of heterogeneity during disease progression. For this reason, secondary tumors or metastatic sites should be retested for molecular abnormalities due to a relatively high rate of possible alterations. Further studies (especially clinical trials) are needed to characterize the correlation between KRAS gene status and clinical outcomes in NSCLC patients.
Conflict of interest
Authors disclosed any conflict of interest.
Abbreviations
- CE-IVD
Certificated for in vitro diagnosis
- CNS
Central nervous system
- KRAS
Kirsten rat sarcoma
- FFPE
Formalin-fixed paraffin-embedded
- NOS
Not otherwise specified
- NSCLC
Non-small cell lung cancer
- OS
Overall survival
- PFS
Progression-free survival
- RR
Response rate
- mt
Mutant type
- wt
Wild type
References
- 1.Jang S, Arkins MB. Treatment of BRAF-mutant melanoma: the role of vemurafenib and other therapies. Clin Pharmacol Ther. 2014;95:24–31. doi: 10.1038/clpt.2013.197. [DOI] [PubMed] [Google Scholar]
- 2.Fiala O, Pesek M, Finek J, et al. The dominant role of G12C over other KRAS mutation types in the negative prediction of efficacy of epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Cancer Genet. 2013;206:26–31. doi: 10.1016/j.cancergen.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Karachaliou N, Mayo C, Costa C, et al. KRAS mutations in lung cancer. Clin Lung Cancer. 2013;14:205–214. doi: 10.1016/j.cllc.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Carpeño J, Belda-Iniesta C. KRAS mutant NSCLC, a new opportunity for the synthetic lethality therapeutic approach. Transl Lung Cancer Res. 2013;2:142–151. doi: 10.3978/j.issn.2218-6751.2013.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011;14:1787–1808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirker R. Novel drugs against non-small-cell lung cancer. Curr Opin Oncol. 2014;26:145–151. doi: 10.1097/CCO.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 8.Guan JL, Zhong WZ, An SJ, et al. KRAS mutation in patients with lung cancer: a predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann Surg Oncol. 2013;20:1381–1388. doi: 10.1245/s10434-012-2754-z. [DOI] [PubMed] [Google Scholar]
- 9.Dienstmann R, Martinez P, Felip E. Personalizing therapy with targeted agents in non-small cell lung cancer. Oncotarget. 2011;2:165–177. doi: 10.18632/oncotarget.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akinleye A, Furqan M, Mukhi N, et al. MEK and the inhibitors: from bench to bedside. J Hematol Oncol. 2013;6:27–32. doi: 10.1186/1756-8722-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2014;116:855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 12.Kamila WK, Michał S, Paweł K, et al. EGFR activating mutations detected by different PCR techniques in Caucasian NSCLC patients with CNS metastases: short report. Clin Exp Metastasis. 2013;30:1063–1071. doi: 10.1007/s10585-013-9603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krawczyk P, Nicoś M, Powrózek T, et al. Sensitive methods for the detection of an insertion in exon 20 of the HER2 gene in the metastasis of non-small cell lung cancer to the central nervous system. Oncol Lett. 2013;6:1063–1067. doi: 10.3892/ol.2013.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicoś M, Powrózek T, Krawczyk P, et al. Sensitive methods for detection of the S768R substitution in exon 18 of the DDR2 gene in patients with central nervous system metastases of non-small cell lung cancer. Med Oncol. 2014;31:176–185. doi: 10.1007/s12032-014-0176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villalva C, Duranton-Tanneur V, Guilloteau K, et al. EGFR, KRAS, BRAF, and HER-2 molecular status in brain metastases from 77 NSCLC patients. Cancer Med. 2013;2:296–304. doi: 10.1002/cam4.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nayak L, Lee EQ, Wen PY. Epidemiology of brain metastases. Curr Oncol Rep. 2012;14:48–54. doi: 10.1007/s11912-011-0203-y. [DOI] [PubMed] [Google Scholar]
- 17.Monaco SE, Nikiforova MN, Cieply K, et al. A comparison of EGFR and KRAS status in primary lung carcinoma and matched metastases. Hum Pathol. 2010;41:94–102. doi: 10.1016/j.humpath.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Alsdorf WH, Clauditz TS, Hoenig T, et al. Intratumoral heterogeneity of KRAS mutation is rare in non-small-cell lung cancer. Exp Mol Pathol. 2013;94:155–159. doi: 10.1016/j.yexmp.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy. Genet Med. 2013;15:517–527. doi: 10.1038/gim.2012.184. [DOI] [PubMed] [Google Scholar]
- 20.Minuti G, D’Incecco A, Cappuzzo F. Targeted therapy for NSCLC with driver mutations. Expert Opin Biol Ther. 2013;13:1401–1412. doi: 10.1517/14712598.2013.827657. [DOI] [PubMed] [Google Scholar]
- 21.Bauml J, Mick R, Zhang Y, et al. Frequency of EGFR and KRAS mutations in patients with non small cell lung cancer by racial background: do disparities exist? Lung Cancer. 2013;81:347–353. doi: 10.1016/j.lungcan.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barlesi F, Blons H, Beau-Faller M et al (2013) Biomarkers (BM) France: Results of routine EGFR, HER2, KRAS, BRAF, PI3KCA mutations detection and EML4-ALK gene fusion assessment on the first 10,000 non-small cell lung cancer (NSCLC) patients (pts). J Clin Oncol. 31 (suppl): abstr 8000.
- 23.Tímár J. The clinical relevance of KRAS gene mutation in non-small-cell lung cancer. Curr Opin Oncol. 2014;26:138–144. doi: 10.1097/CCO.0000000000000051. [DOI] [PubMed] [Google Scholar]
- 24.Badalian G, Barbai T, Rásó E, et al. Phenotype of bone metastases of non-small cell lung cancer: epidermal growth factor receptor expression and K-RAS mutational status. Pathol Oncol Res. 2007;13:99–104. doi: 10.1007/BF02893484. [DOI] [PubMed] [Google Scholar]
- 25.Kalikaki A, Koutsopoulos A, Trypaki M, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–929. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munfus-McCray D, Harada S, Adams C, et al. EGFR and KRAS mutations in metastatic lung adenocarcinomas. Hum Pathol. 2011;42:1447–1453. doi: 10.1016/j.humpath.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Schmid K, Oehl N, Wrba F, et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Zhang Q, Luan H, et al. Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J Exp Clin Cancer Res. 2011;30:30–39. doi: 10.1186/1756-9966-30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu K, Yukawa T, Hirami Y, et al. Heterogeneity of the EGFR mutation status between the primary tumor and metastatic lymph node and the sensitivity to EGFR tyrosine kinase inhibitor in non-small cell lung cancer. Target Oncol. 2013;8:237–242. doi: 10.1007/s11523-012-0241-x. [DOI] [PubMed] [Google Scholar]
- 30.Sun JM, Hwang DW, Ahn JS, et al. Prognostic and predictive value of KRAS mutations in advanced non-small cell lung cancer. PLoS One. 2013;8:64816–64824. doi: 10.1371/journal.pone.0064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackman DM, Miller VA, Cioffredi LA, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15:5267–5273. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shepherd FA, Bourredjem A, Brambilla E, et al. Prognostic and predictive effects of KRAS mutation subtype in completely resected non-small cell lung cancer (NSCLC): a LACE-bio study. J Clin Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran DM, Trusk P, Shell SA et al (2012) KRAS mutation and amplification status predicts sensitivity to anti folate therapies in non-small-cell lung cancer. AACR Cancer Res 72 (Suppl): abstr LB-449.
- 34.Garassino MC, Marabese M, Rusconi P, et al. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann Oncol. 2011;22:235–237. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 35.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2005;23:5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 36.Douillard JY, Shepherd FA, Hirsh V, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol. 2010;28:744–752. doi: 10.1200/JCO.2009.24.3030. [DOI] [PubMed] [Google Scholar]
- 37.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 38.Cappuzzo F, Ciuleanu T, Stelmakh L, et al. Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol. 2010;11:521–529. doi: 10.1016/S1470-2045(10)70112-1. [DOI] [PubMed] [Google Scholar]
- 39.Bodoky G, Timcheva C, Spigel DR, et al. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest New Drugs. 2012;30:1216–1223. doi: 10.1007/s10637-011-9687-4. [DOI] [PubMed] [Google Scholar]
- 40.Bennouna J, Lang I, Valladares-Ayerbes M, et al. A Phase II, open-label, randomised study to assess the efficacy and safety of the MEK1/2 inhibitor AZD6244 (ARRY-142886) versus capecitabine monotherapy in patients with colorectal cancer who have failed one or two prior chemotherapeutic regimens. Invest New Drugs. 2011;29:1021–1028. doi: 10.1007/s10637-010-9392-8. [DOI] [PubMed] [Google Scholar]
- 41.Jänne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]