Abstract
The presence of inherited chromosomally integrated human herpesvirus 6 (ciHHV-6) in hematopoietic cell transplant (HCT) donors or recipients confounds molecular testing for HHV-6 reactivation, which occurs in 30 to 50% of transplants. Here we describe a multiplex droplet digital PCR clinical diagnostic assay that concurrently distinguishes between HHV-6 species (A or B) and identifies inherited ciHHV-6. By applying this assay to recipient post-HCT plasma and serum samples, we demonstrated reactivation of HHV-6B in 25% (4/16 recipients) of HCT recipients with donor- or recipient-derived inherited ciHHV-6A, underscoring the need for diagnostic testing for HHV-6 infection even in the presence of ciHHV-6.
INTRODUCTION
Human herpesvirus 6A and -B (HHV-6A and -B) are known to be able to integrate within human chromosomal subtelomeric regions through homologous recombination at HHV-6 direct repeat regions (1). If this integration occurs within germ line cells, the offspring produced from those cells carry a copy of HHV-6A or -B in every nucleated cell; this condition is called inherited chromosomally integrated HHV-6A or -B (ciHHV-6A or -B) (2). The presence of ciHHV-6A or -B in every cell of the body has unknown effects (3), but two recent studies suggested a link between inherited ciHHV-6A or -B and angina pectoris (4, 5). Also, several reports have demonstrated HHV-6 reactivation and associated disease from inherited ciHHV-6 (6, 7).
HHV-6B reactivation (but not HHV-6A reactivation) occurs in 30 to 50% of hematopoietic cell transplant (HCT) recipients, typically within the first 2 to 6 weeks posttransplantation. This has been associated with complications, such as central nervous system dysfunction, fever and rash, myelosuppression, graft rejection, and acute graft-versus-host disease (GVHD), some of which have also been reported after solid organ transplantation (8). While further research is needed to determine the clinical significance of inherited ciHHV-6A or -B after transplantation, it is well recognized that inherited ciHHV-6A or -B complicates molecular diagnostic testing for HHV-6B reactivation (9, 10). Specifically, tissue and blood samples from patients with inherited ciHHV-6A or -B will have high levels of HHV-6A or -B detected by routine quantitative PCR (qPCR), obscuring detection of HHV-6B reactivation.
To aid in the recognition of inherited ciHHV-6 in the transplant setting, we recently developed a clinical droplet digital PCR (ddPCR) assay to identify inherited ciHHV-6 in cellular patient specimens (11). A subsequent study demonstrated the utility of ddPCR in distinguishing HHV-6A and -B (12). Here we describe an improved assay that identifies inherited ciHHV-6 and determines which species, HHV-6A or HHV-6B, is responsible for the integration by using a single reaction mixture. We also show that this assay can aid in the diagnosis of reactivated HHV-6B in HCT recipients with inherited ciHHV-6A, which further highlights the need for improved diagnostics for HHV-6 reactivation, particularly in immunosuppressed patients (3, 9).
(Part of this work was presented previously at the 9th Annual International Conference on HHV-6 and -7, November 2015, Boston, MA [13].)
MATERIALS AND METHODS
Specimens and nucleic acid extraction.
Samples used for test concordance and validation studies were deidentified residual whole-blood specimens, left over from routine clinical testing, which previously tested positive for inherited ciHHV-6. For determining the sensitivity and specificity of the ciHHV-6A/B assay for detecting HHV-6B, a serum control which tested negative for HHV-6A and -B by ddPCR (Bio-Rad Laboratories) was spiked with a constant concentration of ciHHV-6A control cell line DNA (104 copies/ml) and a 10-fold dilution series of HHV-6B particles (HHV-6B strain Z29 quantitated viral load control; Advanced Biotechnologies) to simulate a serum sample from an inherited ciHHV-6A patient with HHV-6B reactivation. Serum samples were extracted on a MagnaPure liquid chromatograph (LC) (Roche, Basel, Switzerland) by utilizing a DNA isolation kit I with 200 μl plasma, giving 100 μl extracted DNA. Each dilution was run in triplicate ddPCRs.
Patients or their donors with inherited ciHHV-6A were identified from a cohort of HCT recipients at the Fred Hutchinson Cancer Research Center (Fred Hutch) from 1992 to 2014. Samples used to identify affected individuals were obtained from the Fred Hutch Research Cell Bank, which collects peripheral blood mononuclear cells (PBMCs) left over from human leukocyte antigen (HLA) typing of donors and recipients for HCT. Sixteen individuals with donor- or recipient-derived inherited ciHHV-6A and available blood specimens were identified by screening samples by qPCR and confirming inherited ciHHV-6 by ddPCR, using previously reported methods (11). Archived plasma and serum samples collected weekly (one pre-HCT and for up to 8 weeks post-HCT) were obtained, and DNAs from these samples were extracted on a MagnaPure LC (Roche, Basel, Switzerland) by utilizing a DNA isolation kit I with 200 μl plasma, giving 100 μl extracted DNA. After extraction, the purified nucleic acids were stored at −20°C. DNAs from patient specimens were run in triplicate ddPCRs. Collection of all patient samples was approved by the University of Washington Institutional Review Board.
ddPCR.
Our original clinical ciHHV-6 ddPCR assay targeted the U67 gene of HHV-6 (14) and used the RPP30 gene as a cellular reference (11). To make the assay more informative, we developed a multiplex ddPCR assay that targets RPP30 and also distinguishes between HHV-6A and HHV-6B by targeting a region of the U67/68 gene that is divergent between the two species (15). This HHV-6A/B assay was annotated as targeting U94 in its original publication (15) due to less sophisticated genome annotation available in prior decades. The assay includes two probes: one is specific for HHV-6A and labeled with a single fluorophore (6-carboxyfluorescein [FAM]), and the other is specific for HHV-6B and labeled with a 2:1 mix of two fluorophores (HEX-FAM). The HHV-6A and HHV-6B probes differ by 5 nucleotides.
ddPCR was performed on a Bio-Rad QX100/200 system as previously described (11), with the following modifications to the master mix and the primer-probe sets. Briefly, for the serum and plasma samples, each ddPCR mixture consisted of 6.25 μl of 4× ddPCR Supermix for Probes (Bio-Rad Laboratories), 1.25 μl each of HHV-6 and RPP30 20× primer-probe mixes, and 16.25 μl of template DNA in a final volume of 25 μl. The 20× primer-probe mix included 5 μM (each) RPP30, HHV-6A FAM, and HHV-6B HEX probes, 2.5 μM HHV-6B FAM probe, and 18 μM (each) RPP30 and HHV-6 primers. The reaction setup was the same for whole-blood samples, with the substitution of 2× ddPCR Supermix for Probes (Bio-Rad) for the 4× ddPCR Supermix. Varying the amounts of the FAM and HEX probes for HHV-6B resulted in droplet plots with distinct patterns that distinguished HHV-6A from HHV-6B for a single multiplex reaction mixture. Data were analyzed with QuantaSoft analysis software (version 1.7.4), and quantification of target molecules is presented as the number of copies per microliter of PCR mix. The number of HHV-6 copies per cell was obtained using the formula HHV-6 copies/(RPP30 copies/2), because there are 2 copies of RPP30 present in a diploid genome. A ratio of close to 1.0 indicates the occurrence of ciHHV-6, while a value of <1 suggests an active HHV-6 infection.
The primer and probe sequences targeting HHV-6 U67/68 (15) and RPP30 (11), which is an RNase reference gene used for cell counts, were as follows: U67/68 forward, TTCCGGTATATGACCTTCGTAAGC; U67/68 reverse, GATGTCTCACCTCCAAATCTTTAGAAAT; U67/68 probe B FAM, FAM-CATTATATATCGAATCTGACGCTACCTTCCG-BHQ1; U67/68 probe B HEX, HEX-CATTATATATCGAATCTGACGCTACCTTCCG-BHQ1; U67/68 probe A FAM, FAM-ACATTATATGTCGAACTTGACACTACCTTCCG-BHQ1; RPP30 forward, GATTTGGACCTGCGAGCG; RPP30 reverse, GCGGCTGTCTCCACAAGT; and RPP30 probe, HEX-TCTGACCTGAAGGCTCTGCGCG-BHQ1.
Confirmatory real-time HHV-6B-specific PCR.
To confirm the observation of HHV-6B reactivation in the background of ciHHV-6A, a real-time HHV-6B-specific PCR targeting U86 (primers and probe, GCAGGATCTTCACTGTTTTTATATCGA, FAM-TTCCAGATCAAAATCCTGGCGCAAAA-6-carboxytetramethylrhodamine [TAMRA], and TCATCAGTCTCTTCATCATGTTCCA) was performed on all samples for each HCT recipient.
Each 30-μl reaction mixture contained 10 μl of extracted serum DNA, 15 μl of 2× Quantitect multiplex PCR mix (Qiagen), 830 nM (each) primers, and 100 μM probe. The thermocycling conditions were as follows: 50°C for 2 min, 95°C for 15 min, and 45 cycles of 94°C for 1 min and 60°C for 1 min. U1102 (type A) DNA and Z29 (type B) DNA were included in each PCR run as controls.
RESULTS
Clinical validation of a multiplex ddPCR assay for detection of inherited ciHHV-6A/B.
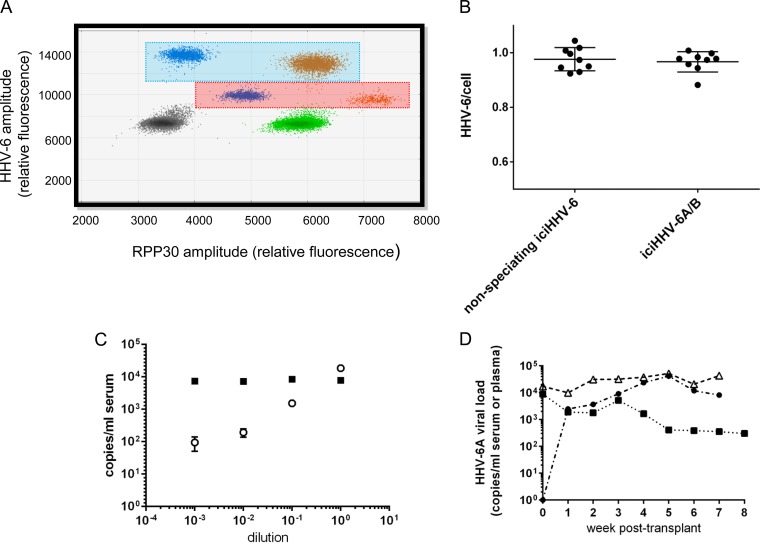
Specimens positive for HHV-6A, HHV-6B, and RPP30 result in a droplet plot that looks like the one shown in Fig. 1A; HHV-6A targets are in droplets within the blue shaded box, while HHV-6B targets are in droplets within the red shaded box. Therefore, for samples with either ciHHV-6A or -B, the distinction between species is clearly evident from the droplet amplitudes (Fig. 1A). The droplet amplitude shifts in the HHV-6B-positive droplets result directly from the use of a probe labeled with a 2:1 HEX-FAM fluorophore mixture.
FIG 1.
A ddPCR multiplex assay for HHV-6A, HHV-6B, and the cell reference gene RPP30 identifies inherited ciHHV-6 (iciHHV-6) and distinguishes HHV-6A from HHV-6B in human whole-blood, serum, and plasma samples. (A) ddPCR droplet plot showing HHV-6A (blue box)- and HHV-6B (red box)-positive droplet populations at different x and y amplitudes. Gray droplets are negative for template DNA. Green droplets are positive for RPP30. Blue droplets are positive for HHV-6A or -B. Orange droplets are positive for both RPP30 and HHV-6A or -B. (B) Concordance between the non-species-discriminating iciHHV-6 ddPCR assay and the iciHHV-6A/B ddPCR assay. (C) Data for a negative-control serum spiked with a ciHHV-6A control cell line (squares) and a 10-fold dilution series of HHV-6B particles (open circles). Bars represent the standard deviations of the means for 3 ddPCRs. (D) Mean HHV-6A loads for three separate classes of iciHHV-6A HCT recipients: recipient integrated (n = 6; filled boxes), donor integrated (n = 7; filled circles), and both donor and recipient integrated (n = 3; open triangles).
As shown in Fig. 1B, the ciHHV-6A/B assay that targets HHV-6 U67/68 is highly concordant with our previous clinical assay for ciHHV-6. Nine residual clinical samples previously positive for inherited ciHHV-6 by ddPCR were available for repeat testing and were run in parallel with both the original clinical assay and the new ciHHV-6A/B assay. The HHV-6/cell ratios for all specimens were very close to 1, allowing identification of inherited ciHHV-6A/B, with no statistically significant difference between the two assays (P = 0.52).
Utility of ciHHV-6A/B ddPCR assay for detection of HHV-6B reactivation in spiked serum samples.
We next determined the sensitivity of the assay for detecting HHV-6B in a background of integrated HHV-6A, with the hypothesis that HHV-6B reactivation events could be identified in serum or plasma samples from HCT recipients with donor- or recipient-derived inherited ciHHV-6A. The assay achieved detection of HHV-6B at levels as low as 95 copies/ml serum in a background of approximately 104 copies/ml ciHHV-6A (Fig. 1C). Additionally, there was no cross talk between HHV-6A and -B: samples spiked with ciHHV-6A were positive only for HHV-6A, and vice versa. This sensitivity and discrimination between species were not feasible with our previous qPCR assay.
Diagnosis of HHV-6B reactivation in HCT recipients with donor- or recipient-derived inherited ciHHV-6A.
Sixteen HCT recipients with donor- or recipient-derived inherited ciHHV-6A were identified from the retrospective cohort and tested for HHV-6B reactivation post-HCT. Up to 8 weekly post-HCT serum or plasma samples, as well as a pre-HCT plasma or serum sample, were available for testing by ciHHV-6A/B ddPCR. Every post-HCT serum and plasma sample (n = 95) from all cases was positive for HHV-6A. HCT recipients harboring inherited ciHHV-6A who received donor cells without inherited ciHHV-6 (n = 6) had decreasing levels of HHV-6A over time as donor cells proliferated (Fig. 1D). Inversely, HCT recipients without inherited ciHHV-6A who received donor cells with inherited ciHHV-6A (n = 7) had increasing levels of HHV-6A over time (Fig. 1D). Finally, there were 3 HCT recipients with inherited ciHHV-6A who received related donor stem cells that also contained inherited ciHHV-6A; these patients had ratios of HHV-6A to genomic equivalents consistent with inherited ciHHV-6A throughout the testing period (mean number of HHV-6 copies/cell = 1.38).
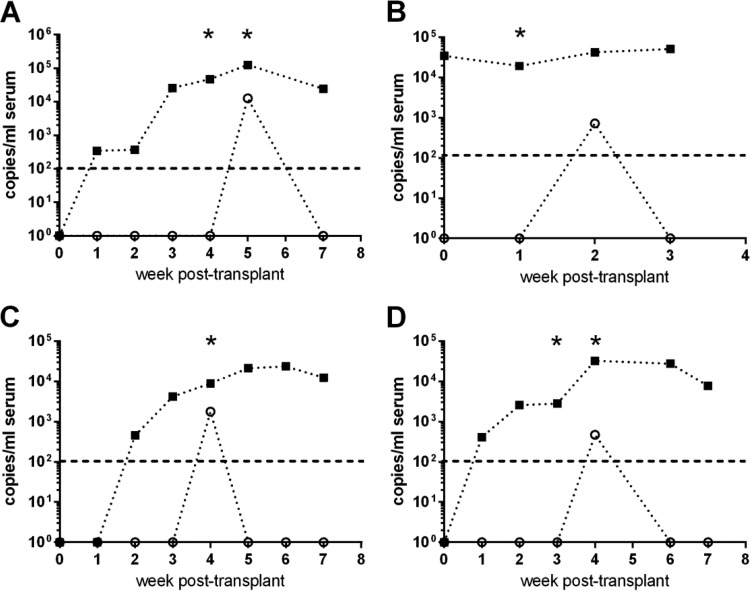
Importantly, the ciHHV-6A/B ddPCR assay identified 4 patients (of 16 total [25%]) with HHV-6B reactivation in a background of ciHHV-6A (Fig. 2). These HCT recipients each had detection of HHV-6B DNA by ddPCR in a single serum sample at week 2, 4, or 5 post-HCT. The ddPCR results for HHV-6B were independently confirmed by HHV-6B-specific real-time PCR for all patients, demonstrating the accuracy of the ddPCR approach (Fig. 2). Of the 7 patients who were inherited ciHHV-6A negative pre-HCT and received ciHHV-6A-positive donor cells, 3 (43%) had reactivated HHV-6B (Fig. 2A, C, and D). Of the 3 HCT recipients who were inherited ciHHV-6A positive and received ciHHV-6A-related donor cells, 1 (33%) had reactivated HHV-6B (Fig. 2B). For this patient, the real-time assay detected HHV-6B in week 1, while the ddPCR assay detected it in week 2, likely because the HHV-6B load was very close to the limit of detection in these samples. Of the 6 HCT recipients who were inherited ciHHV-6A positive and received ciHHV-6-negative donor cells, none had reactivated HHV-6B.
FIG 2.
Multiplex iciHHV-6A/B ddPCR sensitively and simultaneously detects HHV-6B and ciHHV-6A in serum samples from four individual HCT recipients. Sera of ciHHV-6A HCT recipients were tested for reactivation of HHV-6A (squares) and HHV-6B (open circles). Each point represents the mean for 3 ddPCRs. The dashed line at 102 copies/ml represents the limit of detection for HHV-6B. Each asterisk (*) denotes a sample positive for HHV-6B by qualitative real-time PCR. (A, C, and D) Three individual HCT recipients with donor-derived ciHHV-6A; (B) HCT recipient with related donor- and recipient-derived ciHHV-6A.
Clinical findings.
Among the 4 HCT recipients with HHV-6B viremia, 3 were receiving steroids at ≥1 mg/kg of body weight for either graft-versus-host disease (GVHD) of the skin (Fig. 2A and C) or prophylaxis (Fig. 2B). One recipient (Fig. 2D) had skin GVHD shortly before HHV-6B reactivation but was not yet on steroids. The patients whose data are shown in Fig. 2A and D both had generalized seizures and altered mental status within 3 to 6 days of HHV-6B detection, which were attributed to drug toxicity. The patient whose data are shown in Fig. 2B developed bilateral pulmonary infiltrates with diffuse alveolar damage of uncertain etiology within the same time frame, and the patient whose data are shown in Fig. 2C did not have any notable concurrent symptoms.
DISCUSSION
We developed and validated a novel multiplex ddPCR method for identifying inherited ciHHV-6 and determining the HHV-6 species in a single reaction mixture for clinical testing of cellular specimens. We further demonstrated the feasibility of concurrent detection and quantitation of HHV-6A and HHV-6B in the same sample and identified HHV-6B reactivation in 4 of 16 (25%) HCT recipients with donor- or recipient-derived inherited ciHHV-6A. In each of these four HCT recipients, HHV-6B reactivation was detected in only a single serum sample. Infrequent, transient, and low-level HHV-6B reactivation is not atypical post-HCT; in a previous study, patients tested twice weekly for HHV-6B reactivation had a median duration of positivity of only 4 days (16). The absence of reactivation in all inherited ciHHV-6A-positive recipients who received ciHHV-6-negative donor cells (n = 6) generates speculation about whether there may be an immune response in inherited ciHHV-6-positive individuals that reduces the risk of reactivation of latent HHV-6B. Along those lines, recent work showed increased HHV-6-specific functionally active immune responses in individuals with inherited ciHHV-6 (17).
Among the 4 patients with HHV-6B reactivation, 3 were receiving high-dose steroids prior to viral detection, which is a known risk factor for HHV-6B viremia (8). Three of the four patients also had skin GVHD preceding HHV-6B detection. HHV-6B reactivation has been associated with skin GVHD (8), although it is unclear if it played a role in these patients. Interestingly, three patients had significant central nervous system or pulmonary disease within a few days of HHV-6B viremia. Although it is unclear whether HHV-6B contributed to these findings, these are both well-described complications of HHV-6B viremia (8). None of the patients were tested for HHV-6 at the time, although this would have been reasonable to do. Had their blood, cerebrospinal fluid, and bronchoalveolar lavage fluid been tested with traditional qPCR assays, the results would have been very challenging to interpret and might have led to considerable confusion in the therapeutic approach.
While the true characteristics of HHV-6B reactivation (e.g., incidence, duration, and quantitation) in this patient population remain incompletely understood due to the small sample size of this study, these data reveal the need for larger-scale studies. Most importantly, the relatively frequent detection of HHV-6B reactivation underscores the need for advanced testing methodologies to identify viral reactivation in patients with inherited ciHHV-6. Specifically, more advanced diagnostics, such as mRNA detection, are needed to identify reactivation of HHV-6B in ciHHV-6B-positive patients, as this ddPCR assay does not distinguish cellular, integrated HHV-6B from active HHV-6B DNA.
ACKNOWLEDGMENTS
We thank Bio-Rad Laboratories for providing 4× ddPCR Supermix for Probes.
J.A.H. and D.M.Z. received research support from Chimerix Inc. M.B. has served as a consultant and received research support from Chimerix Inc. and Genentech/Roche, in addition to consulting for Clinigen. We have all read the journal's authorship agreement and policy on disclosure of potential conflicts of interest.
This work was supported by a pilot grant from the HHV-6 Foundation (J.A.H.), by a new investigator award from the American Society for Blood and Marrow Transplantation (J.A.H.), and by the National Institutes of Health (grant 1K23AI119133 to J.A.H., grant CA18029 to M.B., and grant HL093294 to M.B.).
REFERENCES
- 1.Morissette G, Flamand L. 2010. Herpesviruses and chromosomal integration. J Virol 84:12100–12109. doi: 10.1128/JVI.01169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daibata M, Taguchi T, Nemoto Y, Taguchi H, Miyoshi I. 1999. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 94:1545–1549. [PubMed] [Google Scholar]
- 3.Flamand L. 2014. Pathogenesis from the reactivation of chromosomally integrated human herpesvirus type 6: facts rather than fiction. Clin Infect Dis 59:549–551. doi: 10.1093/cid/ciu326. [DOI] [PubMed] [Google Scholar]
- 4.Gravel A, Dubuc I, Morissette G, Sedlak RH, Jerome KR, Flamand L. 2015. Inherited chromosomally integrated human herpesvirus 6 as a predisposing risk factor for the development of angina pectoris. Proc Natl Acad Sci U S A 112:8058–8063. doi: 10.1073/pnas.1502741112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarrett R. 2015. iciHHV-6 prevalence and disease associations in the Generation Scotland Study, abstr 8-3 Abstr 9th Int Conf HHV-6 and -7, Boston, MA. [Google Scholar]
- 6.Gravel A, Hall CB, Flamand L. 2013. Sequence analysis of transplacentally acquired human herpesvirus 6 DNA is consistent with transmission of a chromosomally integrated reactivated virus. J Infect Dis 207:1585–1589. doi: 10.1093/infdis/jit060. [DOI] [PubMed] [Google Scholar]
- 7.Endo A, Watanabe K, Ohye T, Suzuki K, Matsubara T, Shimizu N, Kurahashi H, Yoshikawa T, Katano H, Inoue N, Imai K, Takagi M, Morio T, Mizutani S. 2014. Molecular and virological evidence of viral activation from chromosomally integrated human herpesvirus 6A in a patient with X-linked severe combined immunodeficiency. Clin Infect Dis 59:545–548. doi: 10.1093/cid/ciu323. [DOI] [PubMed] [Google Scholar]
- 8.Hill JA, Zerr DM. 2014. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol 9:53–60. doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark DA, Nacheva EP, Leong HN, Brazma D, Li YT, Tsao EH, Buyck HC, Atkinson CE, Lawson HM, Potter MN, Griffiths PD. 2006. Transmission of integrated human herpesvirus 6 through stem cell transplantation: implications for laboratory diagnosis. J Infect Dis 193:912–916. doi: 10.1086/500838. [DOI] [PubMed] [Google Scholar]
- 10.Purev E, Winkler T, Danner RL, Fahle GA, Cook L, Zerr DM, Jerome KR, Childs RW. 2014. Engraftment of donor cells with germ-line integration of HHV6 mimics HHV6 reactivation following cord blood/haplo transplantation. Blood 124:1198–1199. doi: 10.1182/blood-2014-06-577684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sedlak RH, Cook L, Huang ML, Magaret A, Zerr DM, Boeckh M, Jerome KR. 2014. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin Chem 60:765–772. doi: 10.1373/clinchem.2013.217240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leibovitch EC, Brunetto GS, Caruso B, Fenton K, Ohayon J, Reich DS, Jacobson S. 2014. Coinfection of human herpesviruses 6A (HHV-6A) and HHV-6B as demonstrated by novel digital droplet PCR assay. PLoS One 9:e92328. doi: 10.1371/journal.pone.0092328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedlak RH, Hill JA, Nguyen T, Cho M, Levin G, Cook L, Huang M-L, Flamand L, Zerr DM, Boeckh M, Jerome KR. 2015. Abstr 9th Annu Int Conf HHV-6 and -7, November 2015, Boston, MA, abstr 2-4. [Google Scholar]
- 14.Zerr DM, Gooley TA, Yeung L, Huang ML, Carpenter P, Wade JC, Corey L, Anasetti C. 2001. Human herpesvirus 6 reactivation and encephalitis in allogeneic bone marrow transplant recipients. Clin Infect Dis 33:763–771. doi: 10.1086/322642. [DOI] [PubMed] [Google Scholar]
- 15.Zerr DM, Gupta D, Huang ML, Carter R, Corey L. 2002. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin Infect Dis 34:309–317. doi: 10.1086/338044. [DOI] [PubMed] [Google Scholar]
- 16.Zerr DM, Fann JR, Breiger D, Boeckh M, Adler AL, Xie H, Delaney C, Huang ML, Corey L, Leisenring WM. 2011. HHV-6 reactivation and its effect on delirium and cognitive functioning in hematopoietic cell transplantation recipients. Blood 117:5243–5249. doi: 10.1182/blood-2010-10-316083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strenger V, Kayser S, Witte KE, Lassner D, Schwinger W, Jahn G, Urban C, Feuchtinger T. 2016. Individuals with inherited chromosomally integrated HHV-6 (ciHHV-6) have functionally active HHV-6 specific T-cell immunity. Clin Microbiol Infect 22:209.e5–209.e8. doi: 10.1016/j.cmi.2015.10.006. [DOI] [PubMed] [Google Scholar]