Abstract
The emergence of Neisseria gonorrhoeae strains with decreased susceptibility to cephalosporins and azithromycin (AZM) resistance (AZMr) represents a public health threat of untreatable gonorrhea infections. Genomic epidemiology through whole-genome sequencing was used to describe the emergence, dissemination, and spread of AZMr strains. The genomes of 213 AZMr and 23 AZM-susceptible N. gonorrhoeae isolates collected in Canada from 1989 to 2014 were sequenced. Core single nucleotide polymorphism (SNP) phylogenomic analysis resolved 246 isolates into 13 lineages. High-level AZMr (MICs ≥ 256 μg/ml) was found in 5 phylogenetically diverse isolates, all of which possessed the A2059G mutation (Escherichia coli numbering) in all four 23S rRNA alleles. One isolate with high-level AZMr collected in 2009 concurrently had decreased susceptibility to ceftriaxone (MIC = 0.125 μg/ml). An increase in the number of 23S rRNA alleles with the C2611T mutations (E. coli numbering) conferred low to moderate levels of AZMr (MICs = 2 to 4 and 8 to 32 μg/ml, respectively). Low-level AZMr was also associated with mtrR promoter mutations, including the −35A deletion and the presence of Neisseria meningitidis-like sequences. Geographic and temporal phylogenetic clustering indicates that emergent AZMr strains arise independently and can then rapidly expand clonally in a region through local sexual networks.
INTRODUCTION
Gonorrhea is caused by the Gram-negative bacterium Neisseria gonorrhoeae and is the second most commonly reported sexually transmitted bacterial infection in Canada after infections caused by Chlamydia trachomatis. The reported incidence of gonococcal infections in Canada increased from 15 to 39 cases/100,000 population from 1997 to 2013 (1). The public health burden is also high globally, with 111 cases/100,000 population being reported in the United States in 2014 (2), 58 cases/100,000 population being reported in Australia during 2012 (3), and an average of 17 cases/100,000 population being reported in European countries with comprehensive surveillance systems in 2013 (4). Worldwide there are an estimated 106 million new adult cases annually (5).
The ease with which N. gonorrhoeae acquires molecular resistance mechanisms has resulted in the development of resistance to previously recommended antimicrobial therapies (6–9); as a result, first-line empirical treatment in Canada, the United States, Europe, and Australia is now extended-spectrum cephalosporins (ESCs), mainly ceftriaxone (CRO) or cefixime (CFM), and azithromycin (AZM) as a cotherapy (10, 11). In vitro resistance to ESCs, coupled with reports of clinical treatment failures, and the emergence of coresistance to both ESCs and AZM raise concerns over the future treatment of gonorrhea (6, 8, 12–17). Although early studies have reported a low prevalence of azithromycin resistance (AZMr) among N. gonorrhoeae strains (7, 18, 19), recent reports indicate increasing concerns over AZMr (6, 20, 21). From 2003 to 2009, the proportion of AZMr N. gonorrhoeae strains in Canada was low, fluctuating from 0.02% to 0.4%; however, a significant increase was seen in 2010 (1.3%), and then a further increase to 3.3% was seen in 2014 (P < 0.001) (22). Furthermore, the emergence of high-level azithromycin resistance (HL-AZMr; MICs ≥ 256 μg/ml) has been reported in Scotland (23), the United Kingdom (24), Ireland (25), Italy (26), Sweden (27), China (28), Australia (29), Argentina (30), Canada (21), and the United States (31). Increasing AZMr in N. gonorrhoeae threatens the long-term sustainability of current dual-antimicrobial-therapy regimens, which are the last remaining options for first-line empirical treatment in many countries.
AZMr in N. gonorrhoeae has primarily been attributed to two mechanisms: 23S rRNA point mutations, including A2059G (Escherichia coli numbering), conferring high-level resistance (MICs ≥ 256 μg/ml) when present in 3 or 4 of the 4 alleles of the 23S rRNA gene (32), or C2611T (E. coli numbering), conferring low to moderate levels of resistance (MICs = 2 to 32 μg/ml) (33); and the overexpression of the MtrCDE efflux pump, mostly caused by a −35A deletion in the promoter region of the mtrR repressor (34). MtrR A39T and G45D mutations make smaller contributions to AZMr (35, 36). Other macrolide resistance mechanisms, which remain rare, include the mef-encoded and MacAB efflux pumps; the presence of ermA, ermB, ermC, and ermF, encoding 23S rRNA methylases; and mutations in the ribosomal genes rplD and rplV (6).
N. gonorrhoeae multiantigen sequence typing (NG-MAST) and multilocus sequence typing (MLST) have been broadly used to investigate the dynamics of gonococcal infections and antimicrobial-resistant gonococcal strains. More recently, the use of the large-scale next-generation whole-genome sequencing (WGS) technology for genomic epidemiology has been shown to provide a better resolution of strains for comparative studies to identify lineages, monitor the spread of strains, and identify short-term transmission chains (18, 37). In the absence of new antimicrobials for the treatment of gonorrhea, an increased understanding of the emergence and dynamics of antimicrobial-resistant N. gonorrhoeae strains on a national and global basis is crucial.
MATERIALS AND METHODS
Isolates and antimicrobial susceptibility testing.
This study included 236 N. gonorrhoeae isolates recovered from January 1989 to July 2014 across Canada (see Tables S1 and S2 in the supplemental material), which included all viable AZMr isolates in our collection and 23 AZM-susceptible (ASMs) gonococcal isolates. The first AZMr N. gonorrhoeae isolate observed in Canada was isolated in 1997. AZMs isolates and an additional 10 international reference strains, including strains WHO-F, WHO-G, WHO-K, WHO-L, WHO-M, WHO-N, WHO-O, WHO-P, ATCC 49226, and France F89 (14, 38), were included to provide a range of NG-MAST and MLST sequence types (STs), a geographical and temporal distribution of isolates, and isolates for which AZM MICs are 0.5 to 1 μg/ml (close to the AZMr breakpoint of ≥2 μg/ml).
A subset of 23 AZMs isolates was selected from a pool of 249 previously sequenced AZMs isolates to provide an even temporal and geographical distribution (see Tables S1 and S2 in the supplemental material) in order to improve the resolution and clarity of the relationships among AZMr strains presented in Fig. 1. A phylogenetic tree consisting of 459 N. gonorrhoeae isolates that included all 249 AZMs isolates previously sequenced (see Fig. S1 in the supplemental material) was prepared to ensure that the analysis with the smaller number of strains maintained an accurate contextual relationship between AZMs and AZMr strains.
FIG 1.
Whole-genome core SNP maximum likelihood phylogenetic tree of 236 Neisseria gonorrhoeae strains collected from 1989 to 2014 in Canada, including 10 international reference strains. The length of the scale bar in the maximum likelihood tree represents the estimated evolutionary divergence between isolates on the basis of the average pairwise distance between strains (estimated number of substitutions in the sample/total number of high-quality SNPs). NG-MAST and MLST relatedness groups determined by goeBURST analysis and the year and region of collection are indicated. Susceptibility to AZM, CRO, and CFM is shown. Green segments in the molecular marker columns indicate the absence of the particular marker (wild type). The colors of the segments in columns 23S 2059 and 23S 2611 represent the number of alleles with the A2059G or C2611T mutation (E. coli numbering), respectively, as indicated in the key. The mtrR p column depicts disruptions to the mtrR promoter region, including the −35A deletion (red), N. meningitidis-like sequences (light red), and WHO-P-like disruptions (orange).
Susceptibilities to AZM, CRO, and CFM (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) were determined using the agar dilution method and quality control strains as previously described (37, 39). MIC interpretations were based on established criteria: CFM decreased susceptibility was an MIC of ≥0.25 μg/ml, CRO decreased susceptibility was an MIC of ≥0.125 μg/ml (5), and AZMr was an MIC of ≥2.0 μg/ml (2). Low-level AZMr was defined as an MIC of 2 to 4 μg/ml, moderate AZMr was defined as an MIC of 8 to 32 μg/ml, and high-level AZMr was defined as an MIC of ≥256 μg/ml.
Whole-genome sequencing and assembly.
DNA samples were extracted from cultures following a standard protocol with an Epicentre MasterPure complete DNA and RNA extraction kit (Mandel Scientific, Guelph, ON). Multiplexed libraries were created with Nextera XT sample preparation kits (Illumina, San Diego, CA). Paired-end, 300-bp indexed reads were generated on the Illumina MiSeq platform (Illumina, San Diego, CA), yielding an average of 1,211,673 reads per genome and an average genome coverage of 150 times.
De novo assembly.
The quality of the reads was assessed using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and reads were merged using the FLASH program (40), assembled with the SPAdes program (41), and annotated with the Prokka program (42). The average contig length generated was 34,053 bp, and the average N50 contig length was 78,661 bp.
Phylogenomic analysis based on core SNPs.
FASTQ files for forward and reverse reads were concatenated into one FASTQ file per isolate for further analysis. Read ends were trimmed, and poor-quality reads were filtered to improve assembly quality using the script run_assembly_trimClean.pl tool from the CG-Pipeline software program (43) with the following options: −min_quality 25 −bases_to_trim 10 −min_avg_quality 25 −min_length 36 −p 1. Read qualities were assessed using the FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The high-quality reads were then mapped to the publically available reference genome of strain NCCP11945 (GenBank accession no. NC_011035; G. T. Chung; PMID accession no. 18586945) with the SMALT tool (version 0.7.4; http://www.sanger.ac.uk/resources/software/smalt/) and the following options: smalt_index −k 13 −s 6 and smalt_map −f samsoft −r − 1. Single nucleotide variants were called using the FreeBayes variant detector (Erik Garrison and Garbor Marth, 2012, arXiv:1207.3907[q-bio.GN]) and the following parameters: −pvar 0 −ploidy 1 −left-align-indels −min-mapping-quality 30 −min-base-quality 30 −min-alternate-fraction 0.75 −min-coverage 15. Additional variant confirmation was done using the SAMtools mpileup tool (44). The following parameters were used to run SAMtools: samtools mpileup −BQ0 −d100000000 and bcftools view −cg. Positions where variant calls were not in agreement between the two variant callers were excluded. Variant calls within potential problematic regions, including repetitive regions (determined with the MUMmer, version 3.23, program), predicted phages (determined with the PHAST program), genomic islands (determined with the IslandViewer program), and highly recombinant regions containing >90 single nucleotide polymorphisms (SNPs) per 10,000 bp, were excluded from the analysis to minimize the impact of genetic recombination on the phylogeny. All remaining variant calls were merged into a single meta-alignment file. The total number of core SNP positions for the population was 7,614. The meta-alignment of informative core SNP positions was used to create a maximum likelihood phylogenetic tree using the PhyML program with the generalized time-reversible model (45) using the parameters −quiet −b −4 −m GTR −s BEST. The phylogenetic tree was visualized using FigTree software (http://tree.bio.ed.ac.uk/software/figtree/), and phylogenetic clades were determined by cluster analysis using the ClusterPicker program (46) with the following settings: initial and main support thresholds of 0.9, a genetic distance threshold of 4.5, and a large cluster threshold of 10.
Molecular typing.
The WGS data were used for in silico NG-MAST and MLST ST determinations and detection of the presence of molecular antimicrobial resistance markers, including the mtrR −35A deletion, the erm and mef genes, MtrR A39T and G45D, the MacAB efflux pump, the 23S rRNA A2059G and C2611T mutations (E. coli numbering; A2059G and C2611T are A2045G and C2597T, respectively, in N. gonorrhoeae NCCP11945; see Fig. S2 in the supplemental material), and mutations in rplD (L4) and rplV (L22). Typing sequences were submitted to the NG-MAST website (http://www.ng-mast.net/) and the Neisseria MLST website (http://pubmlst.org/neisseria/) to determine STs. Concatenated and aligned NG-MAST porB and tbpB sequences were used to compute a goeBURST full minimum spanning tree (47), and relatedness groups were defined by a maximum difference of 5 SNPs. The allelic profiles of 7 MLST housekeeping genes were used to compute a goeBURST full minimum spanning tree, and profiles with a single locus variation from a founding ST profile were assigned to the same group. The number of 23S rRNA allele mutations was determined by using the core SNP pipeline (37) with an allele of N. gonorrhoeae NCCP11945 (locus tag NGK_RS10650) as a mapping reference and interrogating the allele counts at nucleotide positions 2045 and 2597 (N. gonorrhoeae NCCP11945 numbering; these are A2059G and C2611T in E. coli, respectively; see Fig. S2 in the supplemental material) from the variant call files (.vcf).
Statistical comparisons.
Multiple linear regression analysis was performed, using Microsoft Excel 2010 software (version 14.0.7151.5001; Microsoft Corp.), to determine the relationship of AZM MIC intervals as the dependent variable to the number of mutated 23S rRNA alleles and the presence of mtrR promoter mutations (including the −35A deletion and N. meningitidis-like or WHO-P-like mutations), MtrR A39T and G45D mutations, macAB, and 23S rRNA methylases (ermB, ermC) as independent variables (48). Azithromycin MICs ranging from ≤0.5 to ≥512 μg/ml corresponded to incremental MIC values of 1 to 11, respectively. 23S rRNA A2059G and C2611T (E. coli numbering) variables corresponded to the number of alleles with a respective mutation, whereas other molecular markers were represented by the presence or absence of the mutation, which was given a value of 1 and 0, respectively. The regression model was built from a preliminary analysis that included all independent variables (model 2 in Table S5 in the supplemental material), followed by stepwise removal of variables with nonsignificant individual P values and those causing little change in the adjusted coefficient of determination (R2) value (model 1 in Table S5 in the supplemental material). Adjusted R2 values at a confidence interval of 95% were categorized as follows: values of 0.0 to 0.1 were considered no correlation to a very weak correlation, values of 0.2 to 0.4 were considered a weak correlation, values of 0.5 to 0.7 were considered a moderate correlation, values of 0.8 to 0.9 were considered a strong correlation, and a value of >0.9 was considered a very strong correlation. For the regression significance F test, an overall 2-tailed P value for the combined independent variables of <0.05 at a confidence interval of 95% was considered significant. The measure of the association between two relative proportions was determined using the χ2 or Fisher exact test, and two-tailed P values of <0.05 at 95% confidence were considered significant.
Nucleotide sequence accession numbers.
The 23S rRNA sequences for isolates 35410 (A2059G) and 32380 (C2611T) and unique mtrR DNA sequences were submitted to GenBank under accession numbers KT954109, KT954110, and KT954111 to KT954126, respectively; and WGS read data were submitted to the NCBI Sequence Read Archive under study accession number SRP065041.
RESULTS
N. gonorrhoeae strain distribution.
Of the 236 Canadian isolates, 46 (19.5%) were from females, 188 (79.7%) were from males, and no gender was provided for 2 (0.8%). The patient ages were available for 217 (91.9%) isolates and ranged from 15 to 72 years with a median of 27 years. Isolation sites included the cervix (n = 20), urethra (n = 116), rectum (n = 34), pharynx (n = 36), vagina (n = 1), and unknown (n = 29). Enhanced patient information, such as sexual orientation/partnerships and treatment history/outcome, was not available.
Phylogenomic analysis.
Cluster analysis grouped 189 isolates into 13 clades, and 57 heterogeneous isolates were outside these lineages (Fig. 1). The international reference strains were distributed throughout the phylogeny outside the identified clades, except for France F89 (strain 34842), which clustered into clade H. Clades B (n = 4), E (n = 5), and M (n = 14) were the most homogeneous, with maximum differences of 1, 2, and 6 SNPs, respectively, among the isolates in those clades, whereas clade G (n = 11) was the most diverse, with a maximum difference of 339 SNPs among the isolates in that clade (see Table S3 in the supplemental material).
No phylogenetic association with the clinical isolation site was observed; however, despite a higher proportion of isolates from men overall, isolates in clades D, I, and M were predominantly from women (68.9%, 20/29; P < 0.001). Clades A, B, C, D, F, and J were dominated by isolates from Québec (95.3%, 61/64; P < 0.001), and clades E and M were dominated by strains from Alberta (78.9%, 15/19; P < 0.001). Temporally, the oldest strains appeared in clades K (1997 to 2004) and G (1998 to 2007) and the heterogeneous isolates surrounding these clades of the phyogenetic tree (1989 to 1996). More recent isolates clustered in clade M (2010 to 2011); clades A and B (2010 to 2012); clade L (2012 to 2014); and clades C, D, E, and F (2014). Isolates of clade H spanned the longest time period, with isolation dates ranging from 2005 to 2014.
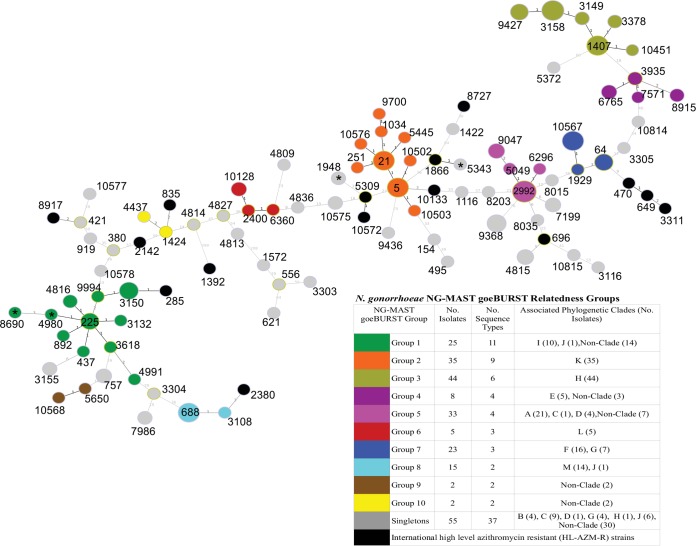
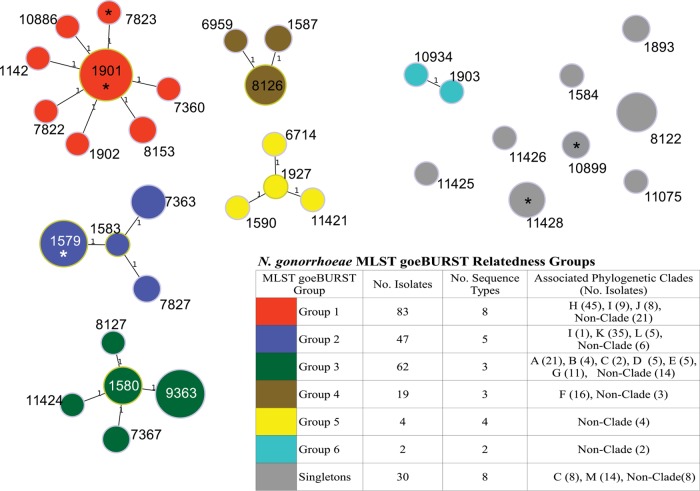
The earliest Canadian AZMr strains were isolated in 1997 in Québec, and these formed a subclade of clade K and were closely related to neighboring subclades, consisting of isolates collected from 1998 to 2002 in British Columbia and Ontario and to more recent isolates of clade L collected in 2013 and 2014 in Ontario and Alberta. A similar relationship was seen with ancestral isolates collected in 1998 from Nova Scotia (strains 20869 and 20870) in clade G surrounded by more recent lineages in clades A to F. Another 1998 Alberta isolate (strain 19328) associated with later lineages in clades J, I, and H (Fig. 1) was also observed. NG-MAST and MLST relatedness groups (Fig. 2 and 3) correlated with the phylogenetic clades, with individual NG-MAST STs being associated with specific subclades, whereas MLST groups associated more broadly in the phylogeny (Fig. 1).
FIG 2.
Genetic relatedness of NG-MAST STs determined by goeBURST minimum spanning tree analysis of Neisseria gonorrhoeae concatenated and aligned NG-MAST porB and tbpB sequences. The number labels on the branches indicate the number of SNP differences between sequence types; branch lengths are not to scale. Node sizes are proportional to the number of isolates in the node. Colored nodes indicate relatedness groups defined by ≤5 SNPs. An asterisk on a node indicates a sequence type of Canadian isolates (ST1948 [n = 2], ST4980, ST5343, ST8690) with HL-AZMr (MIC ≥ 256 μg/ml). Black nodes correspond to HL-AZMr sequence types reported in China (ST1866); Scotland (ST470, ST649); England, Wales, and the United States (ST649); Ireland (ST649, ST3311); Italy (ST835, ST1392, ST2142, ST2386); Sweden (ST285, ST8727); Australia (ST649, ST5309, ST8917, ST10133, ST10572); and Argentina (ST696). ST285 also represents one low-level AZMr Canadian strain grouping into NG-MAST relatedness group 1. Further details on the associated phylogenetic clades are presented in Fig. 1.
FIG 3.
Genetic relatedness of MLSTs determined by goeBURST minimum spanning tree analysis using the Neisseria gonorrhoeae MLST allelic profiles of seven MLST housekeeping genes. Number labels on the branches indicate the number of allelic variations between sequence types; branch lengths are not to scale. Colored nodes indicate relatedness groups defined by a single-locus variation from a founding sequence type. An asterisk on a node indicates a sequence type of Canadian isolates (ST1579, ST1901, ST7823, ST10899, ST11426) with HL-AZMr (MIC ≥ 256 μg/ml). Further details on the associated phylogenetic clades are presented in Fig. 1.
Antimicrobial resistance.
AZM MICs and the molecular AZMr determinants of the N. gonorrhoeae isolates analyzed in this study are summarized in Table 1. Of the 246 N. gonorrhoeae isolates sequenced, 32 were AZMs with MICs of ≤0.5 μg/ml (n = 21) or MICs of 1 μg/ml (n = 11) and 214 were AZMr, where 105 had low-level resistance, 104 had moderate resistance, and 5 had HL-AZMr (Table 1). Similar temporal and regional relationships among lineages were observed when the phylogeny with a smaller number of isolates (Fig. 1) was compared to a phylogeny with a larger number of isolates consisting of 459 N. gonorrhoeae isolates, which included 249 AZMs isolates (see Fig. S1 in the supplemental material), indicating that the phylogenetic analysis with a smaller number of isolates maintained an accurate context of AZMs and AZMr strains, while it increased the clarity of the relationships among the AZMr strains.
TABLE 1.
Molecular markers associated with azithromycin resistance among Canadian and international Neisseria gonorrhoeae isolates collected from 1989 to 2014
| No. of 23S rRNA alleles with a mutation/total no. of alleles in genomea: |
Presence ofb: |
No. of isolates for which the MIC (μg/ml) was as follows: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
mtrR |
erm | |||||||||||||
| A2059G | C2611T | −35Ac | Mend | WHO-Pe | ≤0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥512 | Total | |
| 4/4 | 0/4 | + | − | − | − | 5 | 5 | |||||||
| 0/4 | 4/4 | + | − | − | − | 2 | 28 | 15 | 1 | 46 | ||||
| 0/4 | 4/4 | − | − | − | − | 3 | 18 | 43 | 9 | 73 | ||||
| 0/4 | 3/4 | + | − | − | − | 1 | 5 | 6 | ||||||
| 0/4 | 3/4 | − | − | − | − | 4 | 1 | 5 | ||||||
| 0/4 | 2/4 | + | − | − | − | 5 | 1 | 1 | 7 | |||||
| 0/4 | 2/4 | − | − | − | − | 3 | 3 | |||||||
| 0/4 | 1/4 | + | − | − | − | 2 | 2 | |||||||
| 0/4 | 0/4 | + | − | − | ermB | 1 | 1 | 2 | ||||||
| 0/4 | 0/4 | + | − | − | − | 13 | 7 | 49 | 1 | 70 | ||||
| 0/4 | 0/4 | − | + | − | − | 1 | 11 | 3 | 15 | |||||
| 0/4 | 0/4 | − | − | + | − | 2 | 2 | |||||||
| 0/4 | 0/4 | − | − | − | − | 6 | 3 | 9 | ||||||
| 0/4 | 0/4 | − | − | − | ermC | 1 | 1 | |||||||
| Total | 21 | 11 | 74 | 31 | 78 | 25 | 1 | 5 | 246 | |||||
The data represent the number of alleles with the mutation/total number alleles in the genome. Escherichia coli V00331 (GenBank accession no. V00331.1) numbering (A2059G and C2611T are A2045G and C2599T, respectively, in Neisseria gonorrhoeae NCCP11945 [GenBank accession no. NC_011035.1]).
Symbols: +, present; −, absent.
−35A, a −35A deletion in the mtrR promoter.
Men, N. meningitidis-like sequences.
WHO-P, WHO-P-like sequences.
Among the AZMr isolates, the proportion of pharyngeal isolates with moderate-level resistance (71%, 25/35) was higher than that for isolates from cervical (37%, 7/19; P = 0.018), urethral (48%, 51/107; P = 0.015), and rectal (53%, 16/30; P = 0.144) sources. Phylogenetically, AZMr strains generally clustered clonally into distinct lineages, whereas AZMs strains were generally located outside the clades and among the heterogeneous strains of the phylogenetic tree (Fig. 1; see also Fig. S2 in the supplemental material). Moderate-level AZMr was associated with clades A, B, D, F, H, and L (81%, 78/96), whereas clades C, E, G, I, J, K, and M (11%, 10/93; P < 0.001) were associated with low-level resistance (82%, 76/93; P < 0.001). Coresistance to AZM and an ESC was observed in 8% (18/214) of the AZMr isolates, 16 of which clustered into clade H, and 2 other isolates were distantly separated phylogenetically. These coresistant isolates were isolated over a long time frame (2002 to 2014) from four provinces and were isolated from a variety of clinical isolation sites.
All five HL-AZMr (MIC ≥256 μg/ml) isolates had the A2059G mutation in all four 23S rRNA alleles (Table 1; see also Table S4 in the supplemental material) and were located outside the identified clades of the phylogeny (Fig. 1). Three of these isolates were from Ontario and were collected in 2004 (n = 1) and 2010 (n = 2), and one isolate each was from Québec and British Columbia and were collected in 2009 and 2012, respectively. HL-AZMr was seen in 1 cervical isolate, 2 urethral isolates, 1 rectal isolate, and 1 isolate with an unknown clinical isolation site. High-level coresistance was observed in one HL-AZMr isolate (isolate 31623), collected in 2009 from Québec (NG-MAST ST1948, MLST ST11426), that concurrently had decreased susceptibility to CRO (MIC = 0.125 μg/ml) and an elevated CFM MIC (0.125 μg/ml).
Multiple regression analysis indicated that the number of 23S rRNA alleles with the A2059G or C2611T mutation (E. coli numbering; see Table S4 in the supplemental material) and the presence of mtrR promoter mutations and ermC were strong contributors to increasing AZM MIC increments (adjusted R2 = 0.820, significance F < 0.0001), while the presence of ermB and the MtrR G45D or A39T mutation had little additional effect (adjusted R2 = 0.834, significance F < 0.0001) (see Table S5 in the supplemental material). Increasing AZM MIC increments were strongly associated with an increase in the number of 23S rRNA alleles containing the C2611T mutation, where isolates with two or more mutated alleles had low to moderate resistance (Table 1). This association was further demonstrated by three closely related isolates (33997, 34565, and 33904) that had a maximum genomic difference of one SNP; all were NG-MAST ST757, and all had the mtrR −35A deletion. Isolate 33997 had one 23S rRNA allele with a C2611T mutation and had an AZM MIC of 0.5 μg/ml, isolate 34565 had two mutated alleles and an AZI MIC of 4 μg/ml, and isolate 33904 had three mutated alleles and an AZM MIC of 8 μg/ml (see Table S4 in the supplemental material).
Isolates with the −35A mtrR deletion only (and no 23S rRNA mutations or erm genes; n = 70) had a wide range of MIC values (Table 1) and were distributed throughout the phylogenetic tree (Fig. 1). Isolates of clades C and E with low-level AZMr lacked 23S rRNA mutations or the −35A mtrR deletion but had the N. meningitidis-like mtrR promoter sequences (see the sequences of isolates 37624 and 37088 in Fig. 4) reported by Trembizki et al. (49). Other mtrR promoter mutations similar to those in strain WHO-P were observed in another isolate (isolate 36952) with low-level AZMr.
FIG 4.
Alignment of mtrR-mtrC intergenic DNA sequences of Neisseria gonorrhoeae strains and N. meningitidis M01-240355. Sequence position 1 corresponds to a location −86 bp upstream from the start codon of mtrC. The start codon of mtrR is indicated. The sequences of FA1090 and NCCP11945 were derived from N. gonorrhoeae complete genomes (GenBank accession no. NC_002946 and NC_011035, respectively), and the M01-240355 sequence was derived from the complete genome of N. meningitidis M01-240355 (GenBank accession no. CP002422). Other sequences are those of the following isolates (whose phylogenomic clades are indicated in parentheses in the figure and whose GenBank accession numbers appear in parentheses after the isolate designation): 38202 (KT954111), 38155 (KT954112), WHO-N (KT954113), WHO-L (KT954114), 30978 (KT954115), 29360 (KT954116), WHO-K (KT954117), 38132 (KT954118), WHO-F (KT954119), ATCC 49226 (KT954120), 34106 (KT954121), 36952 (KT954122), and WHO-P (KT954123). The locations of mutations associated with macrolide resistance, the −35A deletion in the mtrR promoter region and MtrR A39T and G45D amino acid substitutions, are indicated. The promoter regions of the sequences of isolates 37624, 38197, and 37088 (GenBank accession no. KT954124, KT954125, and KT954126, respectively) most closely align to the promoter region of the sequence of N. meningitidis M01-240355 with sequence identities of 94%, 98%, and 98%, respectively, using the NCBI BLASTN Suite program. The sequence of isolate 37624 is identical to that of an N. meningitidis-like sequence previously reported by Trembizki et al. (49).
An isolate collected in 2006 (isolate 28759) had a moderate level of AZMr, even though it had no 23S rRNA mutations or mtrR mutations but had a chromosomal ermC (Table 1) flanked by plasmid pEP5289 sequences (GenBank accession no. GU479466). Two other closely related isolates with low-level AZMr had ermB sequences flanked by insertion elements; however, due to the presence of an mtrR −35A deletion, the contribution of ermB to AZMr is uncertain (Table 1). The variables representing the presence of ermB and MtrR G45D/A39T contributed little to the adjusted R2 value in the multiple regression model and, therefore, MIC increments, suggesting a limited role for these determinants in AZMr (Table 1; see also Table S5 in the supplemental material). The MacAB efflux pump was detected in nine isolates with low to moderate AZMr, and these were widely distributed through the phylogeny, but due to the presence of other more dominant resistance determinants, the contribution to overall AZMr could not be resolved. The resistance determinants ermA, ermF, and mef and mutations in L4 (rplD) or L22 (rplV) were not detected.
DISCUSSION
N. gonorrhoeae has developed resistance to all previous antibiotic treatments over the years, and now the currently recommended ESC and AZM cotherapies are threatened (6, 8, 21). Using whole-genome core SNP phylogenomic techniques, the Canadian AZMr N. gonorrhoeae isolates clustered temporally and geographically into distinct lineages, in contrast to reports of the characteristics of N. gonorrhoeae isolates in the United States (7, 18). It has been hypothesized that a genetic event, such as a spontaneous mutation or horizontal gene transfer, initiates the emergence of an antimicrobial-resistant strain, which then rapidly expands clonally in a region through outbreaks among sexual networks (6, 19). Although a definitive evolutionary timeline could not be established due to the highly recombinant nature of the N. gonorrhoeae genome, this trend seems to be reflected in this study, where originator AZMr strains preceded successive related lineages.
All five isolates with HL-AZMr contained the A2059G mutation in all four 23S rRNA alleles. These isolates had a variety of NG-MAST STs (ST8690, ST4980, ST1948 [n = 2], ST5343) and MLST STs (ST1579, ST1901, ST7823, ST11426, ST10899) and were widely separated phylogenomically, temporally, and geographically. Although NG-MAST ST5343 was a single-locus variant of an ST1866 HL-AZMr isolate reported in China (28) and ST1948 was a double-locus variant of ST5309 reported in Australia (29), all other Canadian HL-AZMr isolates had NG-MAST STs unrelated to those in Scotland (ST470, ST649); England, Wales, and the United States (ST649); Ireland (ST649, ST3311); Italy (ST2142, ST835, ST2386, ST1392); Sweden (ST285, ST8727); Australia (ST649, ST5309, ST8917, ST10133, ST10572); and Argentina (ST696) (23–27, 29–31), suggesting that the Canadian strains arose locally through genetic mutations or transformation events and were not expanding clonally; however, the potential of N. gonorrhoeae to develop HL-AZMr is clearly evident.
The 23S rRNA C2611T mutation (E. coli numbering) has been associated with low to moderate AZMr in N. gonorrhoeae (33), which is supported by the findings of this study, where isolates with two or more mutated alleles were resistant (Table 1). Of the isolates with mutated 23S rRNA alleles, the majority had a full complement of four mutated alleles, supporting the suggestion that after an initial allele acquires a mutation, the acquisition of further multiple mutated alleles within the organism occurs easily and rapidly (6, 32) or, alternatively, that there may be a fitness cost associated with partial complements of mutated alleles.
Of the isolates with the mtrR −35A deletion as the sole detected resistance determinant, 71% had low to moderate AZMr; however, 16 Canadian and 4 WHO reference strains with the mtrR −35A deletion were AZMs, suggesting that additional unidentified factors may be required to fully express resistance. Although the mtrR −35A deletion contributed to statistically significantly increased MICs, an additive effect over the resistance conferred by the C2611T mutation was not seen. Another disruption to the mtrR promoter that correlated with AZMr in this study involved N. meningitidis-like sequences (49). While isolates with the mtrR −35A deletion were broadly distributed throughout the phylogeny, isolates with the N. meningitidis-like promoter sequences clustered clonally into recent clades C and E from Québec and Alberta, respectively. The rRNA methylases ermC and ermB were found in only three isolates collected between 2006 and 2008, reflecting the rare occurrence of this mechanism of resistance in AZMr strains (6).
A great threat to the current treatment of gonorrhea is the emergence of strains with coresistance to ESC and AZM, which was found in 18 temporally and geographically diverse isolates in this study. Of particular concern is an isolate collected in 2009 with concurrent HL-AZMr and decreased ESC susceptibility, which, to the best of our knowledge, is the first such report of high-level coresistance. Although 16 of the 18 coresistant isolates were located within clade H, this clade consisted of the global epidemic clone NG-MAST ST1407 and a more recent, related clone, NG-MAST ST3158, which are associated with decreased susceptibility to ESCs (18, 37); therefore, this clade is expected to contain most of the coresistant isolates. Susceptible isolates were generally located through the older nonclustered regions of the phylogenomic tree (Fig. 1; see also Fig. S1 in the supplemental material), a pattern previously observed in the phylogeny of strains with decreased ESC susceptibility (37).
Limitations of this study include the following: the number of bacterial cultures available for testing was limited due to the use of diagnostic nucleic acid amplification tests; information on patient sexual and treatment histories was lacking; and the sample set was primarily composed of antimicrobial-resistant isolates, which may introduce sampling bias, limit contextual relationships, and result in an overrepresentation of isolates from larger population centers, isolates with recent collection dates, and predominant clones. These limitations may have restricted a full comprehensive coverage of AZMr strains, diminished the ability to draw community-specific inferences, and prevented the identification of selective pressures influencing the dynamics of AZMr strains in Canada.
The independent emergence and subsequent dissemination and spread of antimicrobial-resistant N. gonorrhoeae emphasize the need to develop laboratory and epidemiologically linked surveillance systems that not only closely track the dissemination of known resistant strains but also promptly detect novel genetic resistance mechanisms as they emerge. Furthermore, in a clinical setting, it is important to quickly identify and apply an appropriate therapy to limit the expansion of clones through sexual networks. As WGS techniques become more broadly available, genomic epidemiology can provide enhanced insight into the dynamics of N. gonorrhoeae strains to effectively inform public health interventions and ultimately reduce the burden of disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gary Liu, Pam Sawatzky, Karla Montes, and Ravinder Singh Lidder from the Streptococcus and Sexually Transmitted Diseases Unit at the National Microbiology Laboratory (NML) for their laboratory technical assistance; Franklin Bristow, Aaron Petkau, Philip Mabon, Shane Thiessen, Josh Adam, Thomas Matthews, Adrian Zetner, Cameron Sieffert, Adam Olson, and Natalie Knox from the NML Science Technology Cores and Services Division for their infrastructure, technical support, and guidance; and the NML Genomics Core Facility for its next-generation sequencing and analytical expertise. We also thank Patrice Sednaoui of the Institut Alfred Fournier, Centre National de Référence des Gonocoques, Paris, France, for graciously providing N. gonorrhoeae strain France F89.
This work was supported by funding from the Public Health Agency of Canada.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03195-15.
REFERENCES
- 1.Public Health Agency of Canada. 2014. Notifiable diseases on-line. Public Health Agency of Canada, Ottawa, Ontario, Canada: http://dsol-smed.phac-aspc.gc.ca/dsol-smed/ndis/charts.php?c=pl Accessed 1 September 2015. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2015. 2014 STD surveillance report. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/std/stats14/surv-2014-print.pdf Accessed 11 January 2016. [Google Scholar]
- 3.Lahra MM. 2015. Australian Gonococcal Surveillance Programme annual report, 2013. Commun Dis Intell Q Rep 39:E137–E145. [DOI] [PubMed] [Google Scholar]
- 4.European Center for Disease Prevention and Control. 2015. Sexually transmitted infections in Europe 2013. Surveillance report; European Center for Disease Prevention and Control, Solna, Sweden: http://ecdc.europa.eu/en/publications/Publications/sexual-transmitted-infections-europe-surveillance-report-2013.pdf Accessed 1 September 2015. [Google Scholar]
- 5.World Health Organization. 2012. Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. World Health Organization, Geneva, Switzerland: http://www.who.int/reproductivehealth/publications/rtis/9789241503501/en/ Accessed 12 June 2012. [Google Scholar]
- 6.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirkcaldy RD, Soge O, Papp JR, Hook EW III, Rio CD, Kubin G, Weinstock HS. 2015. Analysis of Neisseria gonorrhoeae azithromycin susceptibility in the United States by the Gonococcal Isolate Surveillance Project, 2005 to 2013. Antimicrob Agents Chemother 59:998–1003. doi: 10.1128/AAC.04337-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, Iwasaku K, Nakayama S, Kitawaki J, Unemo M. 2011. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother 55:3538–3545. doi: 10.1128/AAC.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin I, Sawatzky P, Allen V, Hoang L, Lefebvre B, Mina N, Wong T, Gilmour M. 2012. Emergence and characterization of Neisseria gonorrhoeae isolates with decreased susceptibilities to ceftriaxone and cefixime in Canada: 2001-2010. Sex Transm Dis 39:316–323. doi: 10.1097/OLQ.0b013e3182401b69. [DOI] [PubMed] [Google Scholar]
- 10.Unemo M. 2015. Current and future antimicrobial treatment of gonorrhoea—the rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infect Dis 15:364. doi: 10.1186/s12879-015-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Public Health Agency of Canada, Centre for Communicable Diseases and Infection Control, Community Acquired Infections Division. 2013. Canadian guidelines on sexually transmitted infections, gonococcal infections: revised July 2013. Public Health Agency of Canada, Ottawa, Ontario, Canada: http://www.phac-aspc.gc.ca/std-mts/sti-its/index-eng.php Accessed 1 September 2014. [Google Scholar]
- 12.Allen VG, Mitterni L, Seah C, Rebbapragada A, Martin IE, Lee C, Siebert H, Towns L, Melano RG, Lowe DE. 2013. Neisseria gonorrhoeae treatment failure and susceptibility to cefixime in Toronto, Canada. JAMA 309:163–170. doi: 10.1001/jama.2012.176575. [DOI] [PubMed] [Google Scholar]
- 13.Gratrix J, Bergman J, Egan C, Drews SJ, Read R, Singh AE. 2013. Retrospective review of pharyngeal gonorrhea treatment failures in Alberta, Canada. Sex Transm Dis 40:877–879. doi: 10.1097/OLQ.0000000000000033. [DOI] [PubMed] [Google Scholar]
- 14.Unemo M, Golparian D, Nicholas R, Ohnishi M, Gallay A, Sednaouie P. 2012. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob Agents Chemother 56:1273–1280. doi: 10.1128/AAC.05760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unemo M, Golparian D, Syversen G, Vestrheim DF, Moi H. 2010. Two cases of verified clinical failures using internationally recommended first-line cefixime for gonorrhoea treatment, Norway, 2010. Euro Surveill 15(47):pii=19721 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19721. [DOI] [PubMed] [Google Scholar]
- 16.Ison CA, Hussey J, Sankar KN, Evans J, Alexander S. 2011. Gonorrhoea treatment failures to cefixime and azithromycin in England, 2010. Euro Surveill 16(14):pii=19833 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19833. [PubMed] [Google Scholar]
- 17.Unemo M, Golparian D, Stary A, Eigentler A. 2011. First Neisseria gonorrhoeae strain with resistance to cefixime causing gonorrhoea treatment failure in Austria, 2011. Euro Surveill 16(43):pii=19998 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19998. [PubMed] [Google Scholar]
- 18.Grad YH, Kirkcaldy RD, Trees D, Dordel J, Harris SR, Goldstein E, Weinstock H, Parkhill J, Hanage WP, Bentley S, Lipsitch M. 2014. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 14:220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidovic S, Thakur SD, Horsman GB, Levett PN, Anvari V, Dillon J-R. 2012. Longitudinal analysis of the evolution and dissemination of Neisseria gonorrhoeae strains (Saskatchewan, Canada, 2005 to 2008) reveals three major circulating strains and convergent evolution of ciprofloxacin and azithromycin resistance. J Clin Microbiol 50:3823–3830. doi: 10.1128/JCM.01402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole MJ, Spiteri G, Jacobsson S, Pitt R, Grigorjev V, Unemo M. 2015. Is the tide turning again for cephalosporin resistance in Neisseria gonorrhoeae in Europe? Results from the 2013 European surveillance. BMC Infect Dis 15:321. doi: 10.1186/s12879-015-1013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen VG, Seah C, Martin I, Melano RG. 2014. Azithromycin resistance is coevolving with reduced susceptibility to cephalosporins in Neisseria gonorrhoeae in Ontario, Canada. Antimicrob Agents Chemother 58:2528–2534. doi: 10.1128/AAC.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin I, Sawatzky P, Liu G, Allen V, Lefebvre B, Hoang L, Drews S, Horsman G, Wylie J, Haldane D, Garceau R, Ratnam S, Wong T, Archibald C, Mulvey MR. 2016. Decline in decreased cephalosporin susceptibility and increase in azithromycin resistance in Neisseria gonorrhoeae, Canada. Emerg Infect Dis 22:65–67. doi: 10.3201/eid2201.151247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer HM, Young H, Winter A, Dave J. 2008. Emergence and spread of azithromycin-resistant Neisseria gonorrhoeae in Scotland. J Antimicrob Chemother 62:490–494. doi: 10.1093/jac/dkn235. [DOI] [PubMed] [Google Scholar]
- 24.Chisholm SA, Neal TJ, Alawattegama AB, Birley HDL, Howe RA, Ison CA. 2009. Emergence of high-level azithromycin resistance in Neisseria gonorrhoeae in England and Wales. J Antimicrob Chemother 64:353–358. doi: 10.1093/jac/dkp188. [DOI] [PubMed] [Google Scholar]
- 25.Lynagh Y, Mac Aogáin M, Walsh A, Rogers TR, Unemo M, Crowley B. 2015. Detailed characterization of the first high-level azithromycin-resistant Neisseria gonorrhoeae cases in Ireland. J Antimicrob Chemother 70:2411–2413. doi: 10.1093/jac/dkv106. [DOI] [PubMed] [Google Scholar]
- 26.Starnino S, Stefanelli P. 2009. Azithromycin-resistant Neisseria gonorrhoeae strains recently isolated in Italy. J Antimicrob Chemother 63:1200–1204. doi: 10.1093/jac/dkp118. [DOI] [PubMed] [Google Scholar]
- 27.Unemo M, Golparian D, Hellmark B. 2014. First three Neisseria gonorrhoeae isolates with high-level resistance to azithromycin in Sweden: a threat to currently available dual-antimicrobial regimens for treatment of gonorrhea? Antimicrob Agents Chemother 58:624–625. doi: 10.1128/AAC.02093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan L-F, Yin Y-P, Dai X-Q, Pearline RV, Xiang Z, Unemo M, Chen X-S. 2011. Resistance to azithromycin of Neisseria gonorrhoeae isolates from 2 cities in China. Sex Transm Dis 38:764–768. doi: 10.1097/OLQ.0b013e318219cdb5. [DOI] [PubMed] [Google Scholar]
- 29.Stevens K, Zaia A, Tawil S, Bates J, Hicks V, Whiley D, Limnios A, Lahra MM, Howden BP. 2014. Neisseria gonorrhoeae isolates with high-level resistance to azithromycin in Australia. J Antimicrob Chemother 70:1267–1268. doi: 10.1093/jac/dku490. [DOI] [PubMed] [Google Scholar]
- 30.Galarza PG, Alcalá B, Salcedo C, Canigia LF, Buscemi L, Pagano I, Oviedo C, Vázquez JA. 2009. Emergence of high level azithromycin-resistant Neisseria gonorrhoeae strain isolated in Argentina. Sex Transm Dis 36:787–788. doi: 10.1097/OLQ.0b013e3181b61bb1. [DOI] [PubMed] [Google Scholar]
- 31.Katz AR, Komeya AY, Soge OO, Kiaha MI, Lee MV, Wasserman GM, Maningas EV, Whelen AC, Kirkcaldy RD, Shapiro SJ, Bolan GA, Holmes KK. 2012. Neisseria gonorrhoeae with high-level resistance to azithromycin: case report of the first isolate identified in the United States. Clin Infect Dis 54:841–843. doi: 10.1093/cid/cir929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chisholm SA, Dave J, Ison CA. 2010. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother 54:3812–3816. doi: 10.1128/AAC.00309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng L-K, Martin I, Liu G, Bryden L. 2002. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 46:3020–3025. doi: 10.1128/AAC.46.9.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cousin SL Jr, Whittington WLH, Roberts MC. 2003. Acquired macrolide resistance genes and the 1 bp deletion in the mtrR promoter in Neisseria gonorrhoeae. J Antimicrob Chemother 51:131–133. doi: 10.1093/jac/dkg040. [DOI] [PubMed] [Google Scholar]
- 35.Warner DM, Shafer WM, Jerse AE. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol 70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarantonelli L, Borthagaray G, Lee E-H, Shafer WM. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob Agents Chemother 43:2468–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demczuk W, Lynch T, Martin I, Van Domselaar G, Graham M, Bharat A, Allen V, Hoang L, Lefebvre B, Tyrrell G, Horsman G, Haldane D, Garceau R, Wylie J, Wong T, Mulvey MR. 2015. Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J Clin Microbiol 53:191–200. doi: 10.1128/JCM.02589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unemo M, Fasth O, Fredlund H, Limnios A, Tapsall J. 2009. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J Antimicrob Chemother 63:1142–1151. doi: 10.1093/jac/dkp098. [DOI] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement. CLSI document, approved standard M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 43.Kislyuk AO, Katz LS, Agrawal S, Hagen MS, Conley AB, Jayaraman P, Nelakuditi V, Humphrey JC, Sammons SA, Govil D, Mair RD, Tatti KM, Tondella ML, Harcourt BH, Mayer LW, Jordan IK. 2010. A computational genomics pipeline for prokaryotic sequencing projects. Bioinformatics 26:1819–1826. doi: 10.1093/bioinformatics/btq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 46.Ragonnet-Cronin M, Hodcroft E, Hué S, Fearnhill E, Delpech V, Brown AJL, Lycett S. 2013. Automated analysis of phylogenetic clusters. BMC Bioinformatics 14:317. doi: 10.1186/1471-2105-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bremer M, Doerge RW. 2010. Statistics at the bench. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Trembizki E, Doyle C, Jennison A, Smith H, Bates J, Lahra M, Whiley D. 2014. A Neisseria gonorrhoeae strain with a meningococcal mtrR sequence. J Med Microbiol 63:1113–1115. doi: 10.1099/jmm.0.074286-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.