Abstract
MIC results for 115 Staphylococcus intermedius group isolates are presented. Of these, 33% were methicillin resistant, among which 51.4% were susceptible to doxycycline, 29.7% to clindamycin, and 21.6% to trimethoprim-sulfamethoxazole. All of the isolates were susceptible to ceftaroline, daptomycin, linezolid, nitrofurantoin, quinupristin-dalfopristin, rifampin, tigecycline, and vancomycin. Of all the isolates, 82.6%, 67.8%, and 23.5% were susceptible to ciprofloxacin, erythromycin, and penicillin, respectively. No isolates harbored mupA or qacA/B genes, which suggested a lack of resistance to mupirocin or chlorhexidine.
TEXT
The Staphylococcus intermedius group (SIG) is comprised of Staphylococcus intermedius, Staphylococcus pseudintermedius, and Staphylococcus delphini. These Gram-positive cocci (except for S. intermedius) are positive for tube coagulase and negative for slide coagulase and may be misidentified as Staphylococcus aureus by clinical laboratories that test human specimens (1). A colonizer of the nares and anal mucosa of cats and dogs, S. pseudintermedius is increasingly being recognized in human diagnostic specimens (2). This may be due in part to improved diagnostic technologies, such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), now being used in many clinical laboratories. S. pseudintermedius has been documented to cause invasive infections, including brain abscesses, endocarditis, and bacteremia, in humans (3). Methicillin resistance among S. pseudintermedius isolated from dogs is increasing (4), with rates of up to 47% in some regions of the world (5). This resistance is predominantly due to the dissemination of the ST71 clonal lineage in Europe and the ST68 clonal lineage in North America (4). Methicillin-resistant isolates often display resistance to other classes of antimicrobials used in veterinary medicine, including aminoglycosides, fluoroquinolones, lincosamides, macrolides, and tetracyclines, and to chloramphenicol and trimethoprim-sulfamethoxazole (SXT) (6). However, limited susceptibility data are available for S. pseudintermedius with antimicrobials used for humans. We recently conducted a study to evaluate oxacillin and cefoxitin disk and MIC results as predictors of methicillin resistance (encoded by mecA) in a collection of 115 SIG isolates from human and veterinary specimens associated with clinical infections. This study documented that cefoxitin testing, which is recommended by the Clinical and Laboratories Standards Institute (CLSI) to predict methicillin resistance for other species of staphylococci, is a poor predictor of mecA in SIG, whereas both oxacillin disk and MIC tests accurately detect mecA-mediated oxacillin resistance in these isolates (7). As a result of our study, CLSI published S. pseudintermedius-specific oxacillin breakpoints in the 26th edition of the M100S standard (8). In the present study, we document the results of antimicrobial susceptibility testing (AST) for this collection of 115 SIG isolates, including 111 isolates of S. pseudintermedius (45 from human, 56 from canine, 7 from feline, 2 from avian, and 1 from porcine sources) and 4 isolates of S. delphini (3 from equine and 1 from avian sources).
Bacterial isolates were described in our previous article (7). AST was performed according to the CLSI reference broth microdilution (BMD) MIC method (8), using panels prepared in-house with cation-adjusted Mueller-Hinton broth (MHB). MHB was supplemented with 50 mg/liter CaCl2 for daptomycin testing and 2% NaCl for oxacillin testing (9). Fifteen antimicrobial agents were tested (Table 1). BMD tests were read after 16 to 20 h of incubation at 35°C in ambient air for all of the antimicrobials except oxacillin and vancomycin, where the final reading was done after 24 h of incubation. MIC results were interpreted according to the Staphylococcus spp. breakpoints listed in CLSI M100S, 26th edition, including use of the new oxacillin S. pseudintermedius breakpoints and ceftaroline and vancomycin breakpoints for S. aureus (8). Because there are no CLSI tigecycline breakpoints, the Food and Drug Administration (FDA) breakpoint for S. aureus was used. All isolates with penicillin-susceptible MICs (≤0.12 μg/ml) were tested by penicillin disk diffusion using the standard CLSI method and examined for β-lactamase production using a BBL Cefinase disk (BD, Sparks, MD). In addition to taking zone measurements, the zone edges were evaluated for sharp versus fuzzy borders around the penicillin disks. β-Lactamase testing was performed using growth taken from the zone margin surrounding a penicillin disk test on BBL Mueller-Hinton agar (MHA) (BD) after 16 to 18 h of incubation. mecA PCR and SCCmec typing were performed as described in our previous article (7). Mupirocin resistance was determined by PCR for the mupA gene, and chlorhexidine resistance was determined by PCR for the qacA/B gene, as described elsewhere (10).
TABLE 1.
MIC values of 15 antimicrobial agents for Staphylococcus intermedius group (n = 115) when tested by the CLSI reference broth microdilution MIC method in cation-adjusted Mueller-Hinton broth
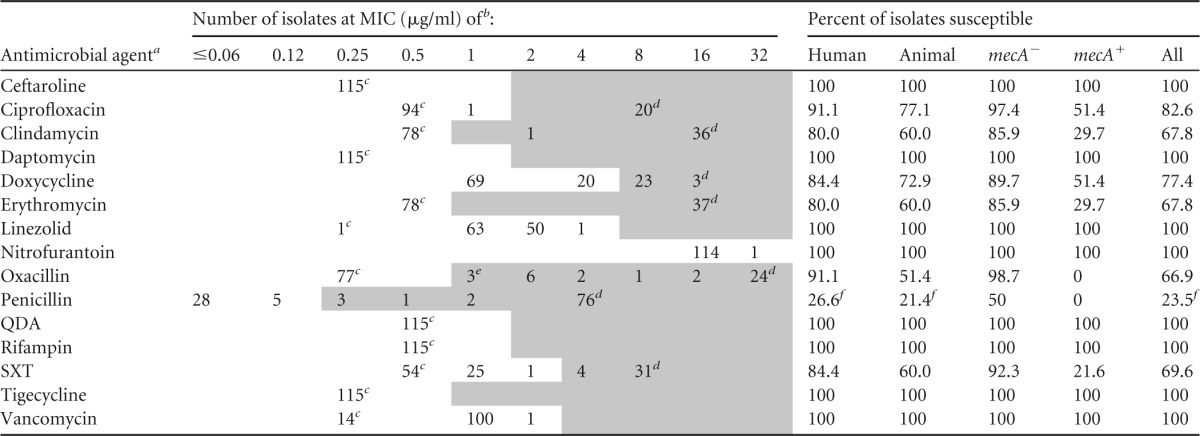
a QDA, quinupristin-dalfopristin; SXT, trimethoprim-sulfamethoxazole.
b MIC values in the unshaded part of the table fall in the susceptible interpretive category; those in the shaded parts of the table are in the intermediate and/or resistant category.
c MIC less than or equal to value in column header.
d Value greater than or equal to value in column header.
e Includes 1 isolate that was mecA negative.
f Includes 5 human isolates and 1 animal isolate that had penicillin-susceptible MICs but were β-lactamase positive.
MIC results obtained for the 115 isolates are shown in Table 1. Thirty-seven isolates (32.2%), 4 of human origin and 33 of veterinary origin, harbored the mecA gene. Using CLSI M100S Staphylococcus spp. interpretive criteria, 33 (42.3%) of the 78 mecA-negative isolates had penicillin-susceptible MICs of ≤0.12 μg/ml (Table 1). For 27 of 33 isolates, MICs were ≤0.06 μg/ml, penicillin zone measurements were susceptible at ≥29 mm, and induced nitrocefin tests were negative. Six (18.2%) of 33 isolates, 5 human and 1 animal isolate, yielded a positive induced nitrocefin test, indicating the presence of a β-lactamase. Six isolates demonstrated penicillin zones of ≤28 mm (resistant), and all had sharp zone edges. Five of these isolates had penicillin MICs of 0.12 μg/ml, and 1 isolate had a penicillin MIC of ≤0.03 μg/ml. Repeat testing in two laboratories confirmed the results. When the nitrocefin tests were performed using uninduced colonies (i.e., not from a penicillin zone margin), variable results were obtained, with 0 to 4 of the 6 isolates yielding a positive result in different laboratories on different days, when testing colonies grown on blood agar plates or on MHA. As such, a test for β-lactamase production should be performed for all penicillin-susceptible S. pseudintermedius isolates, as done for other Staphylococcus spp. Whether a penicillin zone edge test is sufficient for this purpose or an induced nitrocefin-based test is needed remains to be determined. However, in our limited analysis, the penicillin zone edge test was 100% concordant with the nitrocefin results obtained when testing induced colonies. All isolates were susceptible to ceftaroline, the cephalosporin with high-affinity binding to PBP 2a expressed by mecA.
With regard to the non- β-lactam agents, significant differences were noted in the percentage of methicillin-resistant isolates susceptible to doxycycline, SXT, and clindamycin, compared to what has been documented with contemporary isolates of S. aureus (11). This constellation of multidrug resistance is consistent with the multidrug-resistant (MDR) S. pseudintermedius clones ST68 and ST71, which harbor mutations within gyrA and grlA (conferring resistance to fluoroquinolones), as well as a Tn5404-like transposon element that harbors the dfrG (sulfamethoxazole resistance) and ermB (clindamycin and erythromycin resistance) genes (4). Interestingly, differences were noted in our collection based on the SCCmec type. Isolates with SCCmec V were more commonly resistant to erythromycin and clindamycin (10/11 isolates, 90.9%), SXT (10/11 isolates, 90.9%), doxycycline (8/11 isolates, 72.7%), and ciprofloxacin (9/11 isolates, 81.8%) than those with SCCmec types IV or III. For SCCmec type IV, 4 of 8 (50.0%), 8 of 8 (100%), 1 of 8 (12.5%), and 0 of 8 (0.0%) isolates were resistant to these antimicrobials, respectively. For isolates with SCCmec type III, 4 of 9 (44.4%), 2 of 9 (22.2%), 4 of 9 (44.4%), and 0 of 9 (0.0%) were resistant. Isolates of the MDR North American ST68 lineage harbor SCCmec V, similar to the more resistant isolates in our collection (4).
Doxycycline susceptibility was 89.7% among mecA-negative isolates and only 51.4% among mecA-positive isolates (Table 1). This was in striking contrast to doxycycline susceptibility rates among human isolates of methicillin-resistant S. aureus (MRSA), which were 96% among a collection of >4,000 isolates recovered from human diagnostic specimens in 2010 (12). Doxycycline susceptibility rates were similarly high among methicillin-resistant coagulase-negative Staphylococci (CoNS), at 94.1% in one study of 1,473 isolates (13). Our data are consistent with previous studies that documented 31% to 38% doxycycline susceptibility among methicillin-resistant S. pseudintermedius (MRSP) isolates from canine sources (14, 15). No difference was found in susceptibility to doxycycline between human (n = 5, 40.0% susceptible) and veterinary (n = 32, 53.1% susceptible) MRSP isolates in the present study.
Of note, canine-specific breakpoints for doxycycline have been proposed to accommodate the pharmacokinetics of doxycycline doses used for dogs. The canine breakpoints are ≤0.125 μg/ml (susceptible), 0.25 μg/ml (intermediate), and ≥0.5 μg/ml (resistant) (16). The lowest concentration of doxycycline tested in our study was 1 μg/ml, so we cannot estimate the effect that these breakpoints would have on our collection of isolates. However, 35% of mecA-positive and 10.2% of mecA-negative isolates had MICs of 2 to 4 μg/ml, which are considered resistant by the canine breakpoints but susceptible by the human breakpoints. Resistance to the tetracyclines is mediated through acquisition of tetracycline resistance genes (tet genes), four of which have been identified among S. pseudintermedius isolates: tet(M) and tet(O), which mediate ribosomal protection, and tet(K) and tet(L), which encode efflux pumps. The most commonly occurring of these genes are tet(M) and tet(K) in S. pseudintermedius (16, 17). Isolates that harbor none of these genes typically have MICs of ≤0.125 μg/ml to doxycycline, whereas acquisition of the tet(M) gene can be associated with MICs that are elevated but below the susceptible breakpoint of 4 μg/ml given in CLSI document M100S, 26th edition. Clinically, it is unclear whether isolates that are susceptible by the CLSI M100S breakpoint and harbor a tet gene are associated with treatment failures, but these isolates would be considered resistant by the proposed veterinary breakpoint (16). The EUCAST susceptible breakpoint for doxycycline is ≤1 μg/ml for human isolates of Staphylococcus spp. (www.eucast.org), and when applying this breakpoint, only 18.1% of methicillin-resistant and 79.5% of methicillin-susceptible isolates in our study would be considered doxycycline susceptible. Regardless, the tet genes are carried on Tn5801 and Tn916 elements (6), the same as found in human and veterinary isolates of tetracycline-resistant S. aureus (18). The Tn916 tet(M) gene was found in all isolates of the clonal complex (CC) 398 of S. aureus, suggesting this element was integrated into the genome of the clone early and disseminated vertically. This may be the case for the ST71 and ST68 clonal lineages of S. pseudintermedius and may account for the common occurrence of doxycycline resistance in these isolates. Doxycycline resistance may also be selected for through the common use of this agent for the treatment of pyoderma in small-animal veterinary medicine.
SXT susceptibility was only 21.6% among mecA-positive isolates. In contrast, human isolates of MRSA are typically susceptible to this agent; in 2013, 98.0% of isolates in a collection of >9,000 MRSA isolates were susceptible to SXT (19). SXT susceptibility is lower among CoNS. In the same study conducted in 2013, 52.7% of 2,268 methicillin-resistant CoNS isolates were susceptible to SXT (19).
All isolates in this study that were resistant to erythromycin were also resistant to clindamycin, and susceptibility rates for both agents were only 29.7% among MRSP isolates (Table 1). Consequently, no inducible clindamycin resistance was observed, although an inducible erm gene was previously documented in S. pseudintermedius (20).
We documented 51.4% ciprofloxacin susceptibility in MRSP isolates, which was similar to observations in MRSA and MR CoNS isolates (19). However, this susceptibility rate was significantly higher than rates documented in some studies of veterinary SIG isolates, where susceptibility rates as low as 2.7% were reported using the same susceptible breakpoint of 1 μg/ml (21). A single point mutation in topoisomerase II or IV genes confers fluoroquinolone resistance in S. pseudintermedius (22).
All isolates were susceptible to ceftaroline, daptomycin, linezolid, nitrofurantoin, quinupristin-dalfopristin, rifampin, tigecycline, and vancomycin. There are currently no vancomycin breakpoints for SIG, as CLSI only publishes S. aureus and CoNS breakpoints for this antimicrobial agent. However, unlike for CoNS, where the modal MIC for vancomycin is 2.0 μg/ml, we found the vancomycin MIC mode to be 1.0 μg/ml, similar to that documented for S. aureus. As such, it may be reasonable for clinical laboratories to interpret vancomycin MICs by using the more conservative S. aureus susceptible breakpoints of ≤2.0 μg/ml when SIG is encountered, compared to the ≤4-μg/ml breakpoint for CoNS in CLSI M100S or for Staphylococcus spp. in the CLSI VET01 standards. Similar to other studies of SIG (23), we did not find any cases of high-level mupirocin resistance among the isolates in this collection, nor did we detect the presence of the qacA/B gene in any isolates, suggesting the absence of chlorhexidine resistance in these isolates.
In summary, we present in vitro susceptibility results for a large collection of SIG clinical isolates tested by the CLSI reference BMD MIC method. Laboratories should carefully review susceptibility results for all coagulase-positive staphylococci and consider using additional identification procedures, such as MALDI-TOF MS or an automated instrument, for isolates that are doxycycline and/or SXT resistant, a phenotype common to S. pseudintermedius but unusual for S. aureus. This is important, as correct identification of these isolates is critical to accurate testing of SIG with oxacillin to detect methicillin resistance. Clinicians should be cognizant of the dramatic difference in SXT, clindamycin, and doxycycline susceptibility between SIG and S. aureus, as these agents are commonly prescribed as empirical therapy for MRSA in wound and skin structure infections. While susceptibility to these antimicrobials was higher in human than in animal isolates overall (Table 1), this was likely due to the significantly higher proportion of mecA-positive isolates in the veterinary collection, a bias of our data set. A second limitation of the present study was the inclusion of only 4 S. delphini and no S. intermedius isolates; further data will determine if susceptibility rates differ significantly between these isolates and S. pseudintermedius. It is worth noting, however, that S. intermedius is rarely isolated in veterinary or human clinical laboratories but rather is a constituent of the normal nares flora of the wild pigeon (24).
REFERENCES
- 1.Borjesson S, Gomez-Sanz E, Ekstrom K, Torres C, Gronlund U. 2015. Staphylococcus pseudintermedius can be misdiagnosed as Staphylococcus aureus in humans with dog bite wounds. Eur J Clin Microbiol Infect Dis 34:839–844. doi: 10.1007/s10096-014-2300-y. [DOI] [PubMed] [Google Scholar]
- 2.Lee J, Murray A, Bendall R, Gaze W, Zhang L, Vos M. 2015. Improved detection of Staphylococcus intermedius group in a routine diagnostic laboratory. J Clin Microbiol 53:961–963. doi: 10.1128/JCM.02474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelesidis T, Tsiodras S. 2010. Staphylococcus intermedius is not only a zoonotic pathogen, but may also cause skin abscesses in humans after exposure to saliva. Int J Infect Dis 14:e838–e841. doi: 10.1016/j.ijid.2010.02.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perreten V, Kadlec K, Schwarz S, Gronlund Andersson U, Finn M, Greko C, Moodley A, Kania SA, Frank LA, Bemis DA, Franco A, Iurescia M, Battisti A, Dulm B, Wagenaar JA, van Duijkeren E, Weese JS, Fitzgerald JF, Rossano A, Guardabassi L. 2010. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother 65:1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Tian W, Lin D, Luo Q, Zhou Y, Yang T, Deng Y, Liu YH, Liu JH. 2012. Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius in pets from South China. Vet Microbiol 160:517–524. doi: 10.1016/j.vetmic.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy AJ, Harrison EM, Stanczak-Mrozek K, Leggett B, Waller A, Holmes MA, Lloyd DH, Lindsay JA, Loeffler A. 2015. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. J Antimicrob Chemother 70:997–1007. doi: 10.1093/jac/dku496. [DOI] [PubMed] [Google Scholar]
- 7.Wu MT, Burnham CA, Westblade LF, Dien Bard J, Lawhon SD, Wallace MA, Stanley T, Burd E, Hindler J, Humphries RM. 2016. Evaluation of oxacillin and cefoxitin disk and MIC breakpoints for the prediction of methicillin resistance in human and veterinary isolates of Staphylococcus intermedius group. J Clin Microbiol 54:535–542. doi: 10.1128/JCM.02864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; 26th ed CLSI M100S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—9th ed. CLSI M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Fritz SA, Hogan PG, Camins BC, Ainsworth AJ, Patrick C, Martin MS, Krauss MJ, Rodriguez M, Burnham CA. 2013. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother 57:559–568. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter SS, Diekema DJ, Heilmann KP, Dohrn CL, Crispell EK, Riahi F, McDanel JS, Satola SW, Doern GV. 2014. Activities of vancomycin, ceftaroline, and mupirocin against Staphylococcus aureus isolates collected in a 2011 national surveillance study in the United States. Antimicrob Agents Chemother 58:740–745. doi: 10.1128/AAC.01915-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones RN, Stilwell MG, Wilson ML, Mendes RE. 2013. Contemporary tetracycline susceptibility testing: doxycycline MIC methods and interpretive criteria (CLSI and EUCAST) performance when testing Gram-positive pathogens. Diagn Microbiol Infect Dis 76:69–72. doi: 10.1016/j.diagmicrobio.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Jones RN, Farrell DJ, Sader HS, Castanheira M. 2011. Abstr 21st ECCMID, abstr 944. [Google Scholar]
- 14.Weese JS, Sweetman K, Edson H, Rousseau J. 2013. Evaluation of minocycline susceptibility of methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol 162:968–971. doi: 10.1016/j.vetmic.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Hnot ML, Cole LK, Lorch G, Papich MG, Rajala-Schultz PJ, Daniels JB. 2015. Evaluation of canine-specific minocycline and doxycycline susceptibility breakpoints for meticillin-resistant Staphylococcus pseudintermedius isolates from dogs. Vet Dermatol 26:e334–e371. doi: 10.1111/vde.12227. [DOI] [PubMed] [Google Scholar]
- 16.Maaland MG, Papich MG, Turnidge J, Guardabassi L. 2013. Pharmacodynamics of doxycycline and tetracycline against Staphylococcus pseudintermedius: proposal of canine-specific breakpoints for doxycycline. J Clin Microbiol 51:3547–3554. doi: 10.1128/JCM.01498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz S, Roberts MC, Werckenthin C, Pang Y, Lange C. 1998. Tetracycline resistance in Staphylococcus spp. from domestic animals. Vet Microbiol 63:217–227. doi: 10.1016/S0378-1135(98)00234-X. [DOI] [PubMed] [Google Scholar]
- 18.de Vries LE, Christensen H, Skov RL, Aarestrup FM, Agerso Y. 2009. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J Antimicrob Chemother 64:490–500. doi: 10.1093/jac/dkp214. [DOI] [PubMed] [Google Scholar]
- 19.Sader HS, Flamm RK, Jones RN. 2013. Antimicrobial activity of ceftaroline tested against staphylococci with reduced susceptibility to linezolid, daptomycin, or vancomycin from U.S. hospitals, 2008 to 2011. Antimicrob Agents Chemother 57:3178–3181. doi: 10.1128/AAC.00484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gold RM, Lawhon SD. 2013. Incidence of inducible clindamycin resistance in Staphylococcus pseudintermedius from dogs. J Clin Microbiol 51:4196–4199. doi: 10.1128/JCM.02251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruscher C, Lubke-Becker A, Semmler T, Wleklinski CG, Paasch A, Soba A, Stamm I, Kopp P, Wieler LH, Walther B. 2010. Widespread rapid emergence of a distinct methicillin- and multidrug-resistant Staphylococcus pseudintermedius (MRSP) genetic lineage in Europe. Vet Microbiol 144:340–346. doi: 10.1016/j.vetmic.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Descloux S, Rossano A, Perreten V. 2008. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius. J Clin Microbiol 46:1818–1823. doi: 10.1128/JCM.02255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godbeer SM, Gold RM, Lawhon SD. 2014. Prevalence of mupirocin resistance in Staphylococcus pseudintermedius. J Clin Microbiol 52:1250–1252. doi: 10.1128/JCM.03618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K. 2007. Reclassification of phenotypically identified Staphylococcus intermedius strains. J Clin Microbiol 45:2770–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]


