Abstract
The colistin-containing SuperPolymyxin medium was developed for screening polymyxin-resistant Gram-negative bacteria. It was evaluated with 88 polymyxin-susceptible or polymyxin-resistant cultured Gram-negative isolates. Its sensitivity and specificity of detection were ca. 100%. The SuperPolymyxin medium is the first screening medium that is able to detect intrinsic and acquired polymyxin-resistant bacteria.
TEXT
Emergence of polymyxin resistance (PR) is increasingly observed among Gram-negative bacteria (1). The identification of the plasmid-mediated colistin resistance determinant MCR-1 is an additional source of concern (2). Therefore, an early detection of carriers of PR isolates is becoming important.
Polymyxin-containing culture media are known for screening intrinsic PR bacterial species, such as Serratia marcescens and Burkholderia cepacia (3, 4). Those media containing high concentrations of colistin are not adapted for screening isolates with acquired PR that may be of a low level and may contain deoxycholic acids that may interfere with the growth of PR bacteria (18).
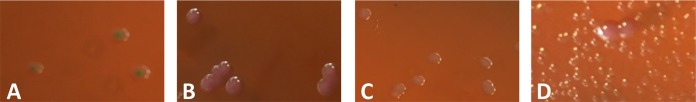
Therefore, our aim was to develop a selective culture medium for screening any type of PR Gram-negative bacteria. The design of this medium took into account the necessity to avoid swarming of Proteus spp. (naturally PR) and to prevent contamination by Gram-positive bacteria and fungi. The optimal screening medium retained was based on the eosin methylene blue (EMB) medium (product no. 70186; EMB Fluka, St. Louis, MO, USA) (5), which is selective for Gram-negative bacteria. This medium may also contribute to species identification by differentiating lactose fermenters (dark blue-brown colonies) from nonfermenters (colorless or light lavender) (Fig. 1). Moreover, differentiation of lactose fermenters was possible with Escherichia coli colonies displaying a characteristic metallic green sheen (Fig. 1A) and Enterobacter spp. and Klebsiella spp. giving brown, dark-centered, and mucoid colonies (Fig. 1B). The optimal colistin (Sigma-Aldrich, St. Louis, MO, USA) concentration was retained at 3.5 μg/ml. Since some Streptococcus and Staphylococcus strains may still grow on EBM medium, daptomycin (Novartis, Horsham, United Kingdom) was added at a concentration of 10 μg/ml. It is noteworthy that daptomycin and not vancomycin was added since vancomycin potentiates the activity of colistin against several Gram-negative bacteria (6, 7). Amphotericin B (Bristol-Myers Squibb, Rueil-Malmaison, France) was also added as an anti-fungi molecule at a concentration of 5 μg/ml.
FIG 1.
Polymyxin-resistant lactose-positive E. coli (A), polymyxin-resistant K. pneumoniae (B), polymyxin-resistant lactose-negative E. coli (C), and a mix of a heavy inoculum of P. mirabilis and a low inoculum of polymyxin-resistant K. pneumoniae (C) growing on the SuperPolymyxin medium.
The stock solutions of colistin, daptomycin, and amphotericin B were prepared as indicated in Table 1 and may be kept at −20°C for 1 year. Notably, glass tubes were used to prepare colistin stock solutions to avoid its binding to polystyrene. Colistimethate sulfate, a therapeutic prodrug of colistin, cannot be used. The diluted powder of EMB was autoclaved at 121°C for 15 min. After cooling the EMB medium for 1 h at 56°C, the antibiotic stock solutions were added. Poured plates were stored at 4°C, protected from direct light exposure for up to 1 week.
TABLE 1.
Preparation of the SuperPolymyxin medium
| Compound | Stock solution (mg/ml) | Quantity or vol to adda | Final concnb |
|---|---|---|---|
| EMB agar powder | 15 g | 3.75% | |
| Distilled water | 400 ml | ||
| Colistin sulfate | 20 In water in glass tubes | 70 μl | 3.5 |
| Daptomycin | 20 In water | 200 μl | 10 |
| Amphotericin B | 20 In D-(+)-glucose 10% | 100 μl | 5 |
The volume of 400 ml of SuperPolymyxin medium was for, i.e., 20 plates.
Concentrations are in micrograms per milliliter unless noted otherwise.
A total of 88 isolates were included in the study to evaluate the performance of the SuperPolymyxin medium that included 52 PR Gram-negative bacteria of worldwide origin (Table 2). Seven strains were intrinsic PR genders (Morganella, Proteus, Providencia, Serratia, and Burkholderia). Forty-five isolates with acquired PR were also included and consisted of 36 enterobacterial strains (E. coli, Klebsiella spp., Enterobacter cloacae, and Hafnia alvei) and 9 nonfermenters (Acinetobacter baumannii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia). Thirty-one polymyxin-susceptible strains were included with 23 enterobacterial strains and 8 nonfermenters (Table 2).
TABLE 2.
MICs of polymyxins for the studied strains and limits of detection of the SuperPolymyxin culture medium
| Straina | Species | Origin | MICb |
Polymyxin resistancec | Mechanism of polymyxin resistance | Lowest detection limit (CFU/ml)d | Lowest detection limit in stools (CFU/ml)d | |
|---|---|---|---|---|---|---|---|---|
| Colistin | Polymyxin B | |||||||
| Yeast isolates | ||||||||
| FR-A | C. albicans | France | NA | NA | NA | NA | >107 | ND |
| Gram-positive cocci isolates | ||||||||
| FR-B | S. aureus | France | NA | NA | NA | NA | >107 | ND |
| FR-C | Staphylococcus epidermidis | France | NA | NA | NA | NA | >107 | ND |
| FR-D | Enterococcus faecium | France | NA | NA | NA | NA | >107 | ND |
| FR-E | Enterococcus faecalis | France | NA | NA | NA | NA | >107 | ND |
| Gram-negative rod isolates naturally resistant to polymyxins | ||||||||
| FR-01 | M. morganii | France | >128 | >128 | R | Intrinsic | 101 | 101 |
| FR-02 | P. mirabilis | France | >128 | >128 | R | Intrinsic | 101 | ND |
| FR-03 | Proteus vulgaris | France | >128 | >128 | R | Intrinsic | 101 | ND |
| FR-04 | Providencia stuartii | France | >128 | >128 | R | Intrinsic | 101 | ND |
| FR-05 | S. marcescens | France | >128 | >128 | R | Intrinsic | 101 | ND |
| FR-201 | B. cepacia | France | >128 | >128 | R | Intrinsic | 101e | ND |
| FR-202 | Burkholderia gladioli | France | >128 | >128 | R | Intrinsic | 101 | ND |
| Gram-negative rod isolates with an acquired mechanism of resistance to polymyxins | ||||||||
| FR-06 | K. pneumoniae | France | 32 | 64 | R | PmrA G53C | 101 | 101 |
| FR-07 | K. pneumoniae | France | 32 | 32 | R | PmrA G53S | 101 | 101 |
| FR-09 | K. pneumoniae | Turkey | 32 | 64 | R | PmrB L17Q | 101 | 101 |
| FR-10 | K. pneumoniae | South Africa | 16 | 8 | R | PmrB T157P | 101 | 101 |
| FR-17 | K. pneumoniae | Turkey | <128 | 128 | R | PhoQ R16C | 101 | 101 |
| FR-21 | K. pneumoniae | France | 32 | 64 | R | MgrB N42Y/K43I | 101 | 101 |
| FR-30 | K. pneumoniae | France | >128 | 64 | R | MgrB truncated | 101 | 101 |
| FR-31 | K. pneumoniae | France | 64 | 32 | R | MgrB truncated | 101 | ND |
| FR-36 | K. pneumoniae | Colombia | 128 | 128 | R | MgrB truncated | 101 | ND |
| FR-40 | K. pneumoniae | France | 64 | 64 | R | MgrB ISEcp1-blaCTX-M-15 | 101 | 101 |
| FR-41 | K. pneumoniae | France | >128 | 128 | R | MgrB IS102-like | 101 | ND |
| FR-47 | K. pneumoniae | Turkey | 64 | 32 | R | MgrB I3903b-like | 101 | ND |
| FR-48 | K. pneumoniae | Spain | 128 | 128 | R | MgrB IS903-like | 101 | ND |
| FR-49 | K. pneumoniae | France | 64 | 32 | R | MgrB IS5-like | 101 | ND |
| FR-54 | K. pneumoniae | Colombia | 128 | 128 | R | MgrB ISKpn13 | 101 | 101 |
| FR-56 | K. pneumoniae | Spain | 64 | 64 | R | MgrB ISKpn26 | 101 | ND |
| FR-68 | K. pneumoniae | Colombia | 64 | 64 | R | MgrB ISKpn14 | 101 | 101 |
| FR-70 | K. pneumoniae | Colombia | 128 | 128 | R | mgrB promoter IS10R | 101 | ND |
| FR-71 | K. pneumoniae | Turkey | 32 | 32 | R | mgrB promoter ISKpn14 | 101 | ND |
| FR-86 | K. pneumoniae | Spain | 64 | 64 | R | mgrB deletion nt 100 | 102 | ND |
| FR-89 | K. pneumoniae | Colombia | >128 | >128 | R | mgrB deletion nt 23 to 33 | 101 | ND |
| FR-92 | K. oxytoca | Colombia | 64 | 64 | R | mgrB promoter ISKpn26-like | 101 | 101 |
| FR-93 | E. coli | France | 4 | 4 | R | Plasmid-mediated mcr-1 gene | 102 | 101 |
| FR-94 | E. coli | South Africa | 16 | 16 | R | Plasmid-mediated mcr-1 gene | 101 | 101 |
| FR-98 | E. coli | South Africa | 8 | 8 | R | Plasmid-mediated mcr-1 gene | 101 | ND |
| FR-203 | A. baumannii | Switzerland | 128 | 128 | R | PmrB G260D | 102 | 101 |
| FR-100 | K. pneumoniae | France | 64 | 64 | R | Unknown | 101 | 101 |
| FR-102 | K. pneumoniae | France | 32 | 32 | R | Unknown | 101 | ND |
| FR-113 | K. pneumoniae | France | >128 | >128 | R | Unknown | 101 | ND |
| FR-114 | K. pneumoniae | Colombia | 64 | 128 | R | Unknown | 101 | ND |
| FR-116 | K. pneumoniae | France | 64 | 32 | R | Unknown | 101 | ND |
| FR-119 | E. coli | France | 8 | 4 | R | Unknown | 101 | ND |
| FR-120 | E. coli | France | 8 | 4 | R | Unknown | 101 | 101 |
| FR-121 | E. coli | France | 4 | 4 | R | Unknown | 101 | ND |
| FR-122 | E. cloacae | Colombia | 32 | 16 | R | Unknown | 101 | 101 |
| FR-126 | E. cloacae | France | >128 | >128 | R | Unknown | 101 | 102 |
| FR-135 | H. alvei | France | 16 | 8 | R | Unknown | 101 | 101 |
| FR-204 | A. baumannii | USA | >128 | >128 | R | Unknown | 102 | 101 |
| FR-205 | A. baumannii | USA | 8 | 8 | R | Unknown | 102 | ND |
| FR-206 | A. baumannii | USA | >128 | >128 | R | Unknown | 102 | ND |
| FR-207 | P. aeruginosa | Colombia | 64 | 64 | R | Unknown | 102f | 101 |
| FR-208 | P. aeruginosa | France | >128 | >128 | R | Unknown | 101e | ND |
| FR-209 | P. aeruginosa | France | >128 | >128 | R | Unknown | 102f | ND |
| FR-210 | S. maltophilia | France | >128 | >128 | R | Unknown | 101e | ND |
| FR-211 | S. maltophilia | France | 32 | 32 | R | Unknown | 5.102e | ND |
| Gram-negative rod isolates susceptible to polymyxins | ||||||||
| FR-136 | E. coli | ATCC 25922 | 0.25 | 0.25 | S | NA | >107 | ND |
| FR-212 | E. coli | USA | 0.12 | 0.12 | S | NA | >107 | ND |
| FR-213 | E. coli | Colombia | 0.12 | 0.12 | S | NA | >107 | ND |
| FR-214 | E. coli | France | 0.12 | 0.12 | S | NA | >107 | ND |
| FR-215 | E. coli | France | 0.25 | 0.12 | S | NA | >107 | ND |
| FR-216 | E. coli | France | 0.25 | 0.12 | S | NA | >107 | ND |
| FR-217 | K. pneumoniae | USA | 0.12 | 0.12 | S | NA | 5 × 106 | ND |
| FR-218 | K. pneumonia | Colombia | 0.12 | 0.5 | S | NA | >107 | ND |
| FR-219 | K. pneumoniae | Colombia | 0.12 | 0.12 | S | NA | >107 | ND |
| FR-220 | K. pneumoniae | Colombia | 0.5 | 0.25 | S | NA | 5 × 106 | ND |
| FR-221 | K. pneumoniae | Colombia | 0.5 | 0.5 | S | NA | >107 | ND |
| FR-222 | K. pneumoniae | France | 0.12 | 0.25 | S | NA | >107 | ND |
| FR-223 | K. pneumoniae | France | 0.12 | 0.12 | S | NA | >106 | ND |
| FR-224 | K. pneumoniae | France | 0.25 | 0.5 | S | NA | >106 | ND |
| FR-225 | K. pneumoniae | France | 0.5 | 0.5 | S | NA | >107 | ND |
| FR-226 | K. pneumoniae | Spain | 0.5 | 0.5 | S | NA | 106 | ND |
| FR-227 | K. pneumoniae | Spain | 0.5 | 0.5 | S | NA | 107 | ND |
| FR-228 | E. cloacae | Colombia | 0.12 | 0.25 | S | NA | >107 | ND |
| FR-229 | E. cloacae | Colombia | 0.12 | 0.12 | S | NA | >107 | ND |
| FR-230 | E. cloacae | France | 0.12 | 0.25 | S | NA | 5 × 106 | ND |
| FR-231 | Enterobacter aerogenes | France | 0.12 | 0.12 | S | NA | >107 | ND |
| FR-232 | Citrobacter freundii | Colombia | 0.25 | 0.25 | S | NA | >107 | ND |
| FR-233 | C. freundii | France | 0.12 | 0.5 | S | NA | 5 × 106 | ND |
| FR-234 | P. aeruginosa | ATCC 27853 | 0.5 | 0.5 | S | NA | 5 × 106 | ND |
| FR-235 | P. aeruginosa | Colombia | 1 | 0.25 | S | NA | >107 | ND |
| FR-236 | P. aeruginosa | Colombia | 2 | 0.5 | S | NA | 106 | ND |
| FR-237 | P. aeruginosa | Colombia | 1 | 0.25 | S | NA | >107 | ND |
| FR-238 | P. aeruginosa | France | 2 | 0.25 | S | NA | 106 | ND |
| FR-239 | A. baumannii | Colombia | 0.5 | 0.12 | S | NA | >107 | ND |
| FR-240 | A. baumannii | France | 0.5 | 0.25 | S | NA | 106 | ND |
| FR-241 | A. baumannii | USA | 0.25 | 0.25 | S | NA | 106 | ND |
The strains FR-01 and FR-136 were used as positive and negative controls, respectively.
MICs of colistin and polymyxin B were determined using the broth microdilution method. NA, not available.
R, resistant; S, susceptible.
Underlined CFU counts are considered to be results below cutoff values set at ≥1 × 103 CFU/ml. ND, not determinate.
Positive culture after 36 h.
Positive culture after 48 h.
MICs of polymyxins (colistin and polymyxin B) were determined using the broth microdilution method in cation-adjusted Mueller-Hinton broth as recommended by the CLSI (8). A final inoculum of 5 × 105 CFU/ml of each strain was distributed in the 96-polystyrene-well tray (Sarstedt, Nümbrecht, Germany). E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as polymyxin-susceptible control strains (9), and all experiments were repeated in triplicate. The results were interpreted according to the CLSI breakpoints (9) for A. baumannii (susceptible [S], ≤2 μg/ml; resistant [R], ≥4 μg/ml) and for Pseudomonas spp. (S, ≤2 μg/ml; R, ≥8 μg/ml) and according to the EUCAST breakpoints for Enterobacteriaceae (i.e., S, ≤2 μg/ml; R, >2 μg/ml) (10). MIC values for colistin and polymyxin B were superimposable, permitting an easy classification between PR and polymyxin-susceptible strains (Table 2).
The known genes possibly involved in chromosomally encoded PR in E. coli, Klebsiella pneumoniae, Klebsiella oxytoca, P. aeruginosa, and A. baumannii were sequenced as described previously (11–15). The PCR detection of the mcr-1 gene was carried out as described previously (2) (Table 2). MIC values of polymyxins for strains with acquired resistance were variable and were lower than those for intrinsically PR strains (Table 2).
Using an inoculum with an optical density of 0.5 McFarland standard (inoculum of ∼108 CFU/ml), serial 10-fold dilutions of the isolates were made in normal saline, and 100-μl portions were plated onto the SuperPolymyxin medium. To quantify the viable bacteria in each dilution, Trypticase soy agar was inoculated concomitantly with 100 μl of suspension and was incubated overnight at 37°C. The number of viable colonies was counted after 24 h of culture at 37°C (and after 36 and 48 h for B. cepacia, P. aeruginosa, and S. maltophilia). PR and polymyxin-susceptible control strains were FR-01 (Morganella morganii) and FR-136 (E. coli ATCC 25922), respectively (Table 2). The lowest limit of detection for the tested strains was determined using the SuperPolymyxin medium. The sensitivity and specificity cutoff values were set at 1 × 103 CFU/ml, i.e., a limit value of 1 × 103 CFU/ml and above was considered not efficiently detected (16). All of the PR strains grew on the SuperPolymyxin medium in 24 h except P. aeruginosa, S. maltophilia, and the intrinsically PR Burkholderia genus that grew in 24 to 48 h (Table 2). The lowest limit of detection was below the cutoff value for all PR strains, whereas the limit of detection of the polymyxin-susceptible strains was above the cutoff value at ≥1 × 106 CFU/ml (Table 1). The sensitivity and specificity of the SuperPolymyxin medium for selecting PR isolates were 100% in both cases. Moreover, this medium was tested with a light growth of a PR K. pneumoniae isolate (FR-10) among a heavier growth of Proteus mirabilis (FR-02) and revealed that it nicely discriminated between the two species (Fig. 1D).
Spiked stools were also tested with a representative collection of 22 PR isolates of various species with various levels and mechanisms of PR (Table 2). Spiked fecal samples were made by adding 100 μl of each strain dilution to 900 μl of fecal suspension that was obtained by suspending 5 g of freshly pooled feces from five healthy volunteers in 50 ml of distilled water as described previously (17). A nonspiked fecal suspension was used as a negative control. The lowest detection limit of the PR isolate was determined by plating 100 μl of each dilution on the medium. The sensitivity and specificity were determined using the same cutoff value set at ≥103 CFU/ml (17). This value may correspond to a low-level carriage of PR bacteria in stools. All of the PR isolates spiked in stools grew with a lowest detection limit ranging from 101 to 102 CFU/ml (Table 2).
Similar results were obtained with 20 colistin-susceptible and polymyxin B-susceptible strains and 20 colistin-resistant and polymyxin B-resistant strains by using polymyxin B at the same concentration instead of colistin sulfate.
Finally, to assess the storage stability of the SuperPolymyxin medium, Candida albicans, Staphylococcus aureus, and colistin-susceptible E. coli ATCC 25955 were subcultured daily onto SuperPolymyxin agar plates from a single batch of medium stored at 4°C. Growth of those isolates were consistently inhibited during at least a 7-day period.
The SuperPolymyxin medium constitutes a screening medium aimed to detect any PR bacteria regardless of its resistance mechanism and of its level. This medium may be used in human medicine for detecting carriers and in veterinary medicine for surveillance surveys.
ACKNOWLEDGMENTS
We are in debt to several colleagues who sent us polymyxin-resistant isolates, i.e., A. Brink, Y. Doi, J. Fernandez-Dominguez, J.-W. Decousser, J.-Y. Madec, P. Plésiat, and M.-V. Villegas.
This work was financed by the University of Fribourg, Switzerland. An international patent form has been filed on behalf of the University of Fribourg (Switzerland) corresponding to this selective culture medium.
REFERENCES
- 1.Bialvaei AZ, Samadi Kafil H. 2015. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin 31:707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 2.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Grasso GM, D'Errico MM, Schioppa F, Romano F, Montanaro D. 1988. Use of colistin and sorbitol for better isolation of Serratia marcescens in clinical samples. Epidemiol Infect 101:315–320. doi: 10.1017/S0950268800054248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilligan PH, Gage PA, Bradshaw LM, Schidlow DV, DeCicco BT. 1985. Isolation medium for the recovery of Pseudomonas cepacia from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol 22:5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine M. 1943. The effect of concentration of dyes on differentiation of enteric bacteria on eosin-methylene-blue agar. J Bacteriol 45:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon NC, Png K, Wareham DW. 2010. Potent synergy and sustained bactericidal activity of a vancomycin-colistin combination versus multidrug-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 54:5316–5322. doi: 10.1128/AAC.00922-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randall CP, Mariner KR, Chopra I, O'Neill AJ. 2013. The target of daptomycin is absent from Escherichia coli and other Gram-negative pathogens. Antimicrob Agents Chemother 57:637–639. doi: 10.1128/AAC.02005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2012. Methods for dilution of antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—9th ed. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.EUCAST. 2014. Breakpoints tables for interpretation of MICs and zone diameters. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf.
- 11.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayol A, Poirel L, Villegas MV, Nordmann P. 2015. Modulation of mgrB gene expression as a source of colistin resistance in Klebsiella oxytoca. Int J Antimicrob Agents 46:108–110. doi: 10.1016/j.ijantimicag.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Lee JY, Ko KS. 2014. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn Microbiol Infect Dis 78:271–276. doi: 10.1016/j.diagmicrobio.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Turkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 15.Quesada A, Porrero MC, Tellez S, Palomo G, Garcia M, Dominguez L. 2015. Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J Antimicrob Chemother 70:71–74. doi: 10.1093/jac/dku320. [DOI] [PubMed] [Google Scholar]
- 16.Nordmann P, Girlich D, Poirel L. 2012. Detection of carbapenemase producers in Enterobacteriaceae by use of a novel screening medium. J Clin Microbiol 50:2761–2766. doi: 10.1128/JCM.06477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naas T, Ergani A, Carrër A, Nordmann P. 2011. Real-time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob Agents Chemother 55:4038–4043. doi: 10.1128/AAC.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froelich JM, Tran K, Wall D. 2006. A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J Bacteriol 188:1180–1183. doi: 10.1128/JB.188.3.1180-1183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]