Abstract
Group B streptococcus (GBS) capsular serotypes are major determinants of virulence and affect potential vaccine coverage. Here we report a whole-genome-sequencing-based method for GBS serotype assignment. This method shows strong agreement (kappa of 0.92) with conventional methods and increased serotype assignment (100%) to all 10 capsular types.
TEXT
Streptococcus agalactiae, or group B streptococcus (GBS), is an important pathogen in neonates (1–3), with early infections being acquired from the maternal genitourinary tract (4). In addition, GBS is now recognized as an increasingly important pathogen among immunosuppressed and elderly individuals in high-income regions (5, 6).
GBS expresses a capsular polysaccharide that is involved in virulence and immune evasion. Ten different serotype variants (i.e., Ia, Ib, II, III, IV, V VI, VII, VIII, and IX), which differ in their disease-causing abilities, have been described. Conjugate vaccines targeting the most common disease-causing serotypes are currently in development (7). Establishment of vaccine serotype coverage is important, as is postintroduction surveillance to monitor for potential serotype replacement, as has been seen following the introduction of other conjugate vaccines (8).
Current methods for GBS serotype allocation rely on latex agglutination assays or PCR assays (9). Recent advances in whole-genome sequencing (WGS) have enabled the development of approaches that can be used in place of traditional microbiological methods, such as strain typing and antibiotic susceptibility profiling (10–12). A major advantage of this approach is that the cost of sequencing can be mitigated by the ability to use the same data to generate multiple outputs. Given the decreasing cost of WGS (13) and the ongoing increase in WGS data generation, we sought to establish and to validate a WGS-based method for GBS capsular typing.
We developed an algorithm for serotype assignment on the basis of sequence similarity between a given de novo assembly and capsular gene sequences of the 10 GBS serotypes. For nine serotypes, published sequences were used as references (Table 1); for serotype IX, however, only a partial capsular locus sequence has been published (14). A suitable reference for the full capsular locus region was therefore determined by WGS of a serotype IX isolate obtained from the Statens Serum Institute (Copenhagen, Denmark).
TABLE 1.
Reference sequences used for sequence-based serotype allocation
| Serotype | GenBank accession no. | Region (bp) | Reference |
|---|---|---|---|
| Ia | AB028896.2 | 6982–11695 | Yamamoto et al. (20) |
| Ib | AB050723.1 | 2264–6880 | Watanabe et al. (21) |
| II | EF990365.1 | 1915–8221 | Martins et al. (22) |
| III | AF163833.1 | 6592–11193 | Chaffin et al. (23) |
| IV | AF355776.1 | 6417–11656 | Cieslewicz et al. (24) |
| V | AF349539.1 | 6400–12547 | Cieslewicz et al. (24) |
| VI | AF337958.1 | 6437–10913 | Cieslewicz et al. (24) |
| VII | AY376403.1 | 3403–8666 | Cieslewicz et al. (24) |
| VIII | AY375363.1 | 2971–7340 | Cieslewicz et al. (24) |
| IX | NAa | NA | This study |
NA, not applicable.
To assign the serotype for a given isolate, a BLAST database was generated from the de novo assembly and queried with the variable region of the capsular locus sequence for each serotype (cpsG-cpsK for serotypes Ia to VII and IX and cpsR-cpsK for serotype VIII), using BLASTn with an E value threshold of 1e−100 and otherwise default parameters. A serotype was considered correct if it showed ≥95% sequence identity over ≥90% of the sequence length. These thresholds were chosen on the basis of being stringent enough to provide differentiation between the various reference sequences while maximizing serotype allocation for an initial test set of publicly available GBS WGS data, for which serotype information was not available (therefore, we had no way of knowing whether the assigned serotypes were actually correct).
This sequence-based method for serotype allocation was validated using WGS with a set of 223 colonizing or invasive human isolates from Canada, Latin America, Singapore, the United Kingdom, the United States, and Thailand for which serotypes had been determined previously using conventional latex agglutination assays, with PCR assays being used to confirm weak positive or negative results in a subset (15–17). For two rare serotypes (serotypes VIII and IX), one isolate of each was obtained from the Statens Serum Institute. GBS isolates stored at −80°C were subcultured on Columbia blood agar for 24 to 48 h, followed by DNA extraction from a single colony using a commercial kit (QuickGene; Fujifilm, Tokyo, Japan). High-throughput sequencing was performed at the Wellcome Trust Centre for Human Genetics (Oxford University, Oxford, United Kingdom) using the Illumina HiSeq2500 platform, generating 150-base paired-end reads. De novo assembly was performed using Velvet and VelvetOptimiser (18, 19). Agreement between serotype allocations was tested with the kappa statistic.
High-quality sequence data were obtained for all 223 GBS isolates (median read number, 2,975,508 [range, 1,798,744 to 13,073,718]; median contig number, 46 [range, 16 to 106]; median assembly length, 2.05 Mb [range, 1.94 to 2.22 Mb]). Each isolate was allocated to a single serotype using the WGS data (Table 2). Three isolates that did not have a capsular type assigned by latex agglutination methods had serotypes Ib, VI, and VII assigned. For all previously serotyped GBS isolates with a known capsule type, the kappa statistic of 0.92 indicated very strong agreement between WGS-predicted and conventional serotypes. Nine isolates had discordant results. In each case, there was strong support for the sequence-allocated serotype, with >98% sequence identity over 100% of the reference length in all nine cases (Fig. 1). Across all isolates, differences in relatedness between the capsular locus sequences of the different serotypes led to characteristic relationships between the allocated serotype (best match) and the second-best match. For example, all isolates assigned to serotype Ia had serotype III as the second-best match. In all cases, the second-best match was substantially poorer than the best match, demonstrating that there was no ambiguity in the predicted serotype (Fig. 1 and Table 3).
TABLE 2.
Serotype allocation by WGS versus serotype allocation by latex agglutination
| Latex agglutination serotype | No. with WGS serotype of: |
Total no. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VI | VII | VIII | IX | ||
| Ia | 34 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 35 |
| Ib | 0 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| II | 0 | 0 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 |
| III | 3 | 0 | 0 | 111 | 0 | 0 | 0 | 0 | 0 | 1 | 115 |
| IV | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| V | 0 | 0 | 0 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 16 |
| VI | 0 | 0 | 0 | 0 | 0 | 1 | 8 | 0 | 0 | 0 | 9 |
| VII | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 5 |
| VIII | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1a | 0 | 1 |
| IX | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1a | 2 |
| Nontypeable | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 3 |
| Total | 37 | 11 | 26 | 112 | 1 | 17 | 10 | 6 | 1 | 2 | 223 |
Reference GBS isolates from Statens Serum Institute for serotypes VIII and IX.
FIG 1.
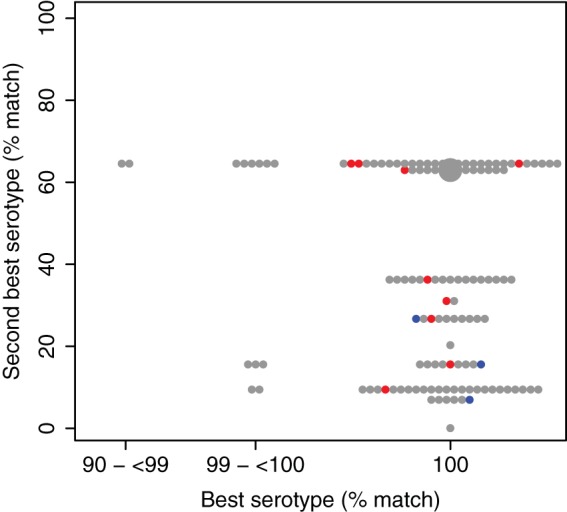
Relationships between allocated serotypes (best matches) and second-best matches. For each isolate, the percentage of the capsular locus region present (≥95% sequence identity) for the assigned serotype is shown on the x axis and that for the serotype showing the next best match is shown on the y axis. Gray circles, isolates showing agreement between sequence-based serotyping and conventional serotyping; blue circles, isolates classified as nontypeable by conventional methods; red circles, isolates with discordant results. Small circles, single isolates; large circle, 100 isolates. For each serotype, the second-best match is identical in all cases, leading to the observed horizontal banding (details in Table 3).
TABLE 3.
Relationships between allocated serotypes and second-best matchesa
| Allocated serotype | Match (%) for allocated serotype | Second-best serotype | Match (%) for second-best serotype |
|---|---|---|---|
| Ia | 93.91–100 | III | 64.56 |
| III | 100 | Ia | 62.98 |
| V | 100 | IX | 36.26 |
| IX | 100 | V | 31.05 |
| VI | 100 | III | 26.68 |
| IV | 100 | Ia | 20.3 |
| Ib | 99.61–100 | VI | 15.55 |
| II | 99.86–100 | IV | 9.45 |
| VII | 100 | Ib | 6.95 |
| VIII | 100 | None | 0 |
See also Fig. 1.
The nine isolates with discordant results and the three nontypeable isolates were retested by latex agglutination assays (Table 4) and were resequenced using the Illumina MiSeq platform, with sequence processing and WGS-based serotype prediction performed as described above. In all cases, resequencing results were consistent with the initial WGS classification. For 6/9 isolates with discordant results, the new latex agglutination results matched the WGS-based prediction, suggesting that the initial discordance might have resulted from incorrect latex agglutination typing or sample mislabeling. The other three isolates with discordant results and the three nontypeable isolates were all classified as nontypeable with retesting.
TABLE 4.
Retyping of isolates with discordant results and nontypeable isolates
| Isolate | Reason for retyping | Latex agglutination serotype |
WGS serotype |
||
|---|---|---|---|---|---|
| Initial | Repeat | Initial | Repeat | ||
| CB466 | Discordant results | III | Ia | Ia | Ia |
| IW8194 | Discordant results | III | IX | IX | IX |
| IW8466 | Discordant results | Ia | III | III | III |
| IW8471 | Discordant results | III | Ia | Ia | Ia |
| IW7157 | Discordant results | Ib | II | II | II |
| SMRU1 | Discordant results | VI | V | V | V |
| SMRU25 | Discordant results | IV | NTa | VI | VI |
| SMRU4 | Discordant results | IX | NT | Ib | Ib |
| SMRU59 | Discordant results | III | NT | Ia | Ia |
| Z41 | NT | NT | NT | Ib | Ib |
| UK22 | NT | NT | NT | VII | VII |
| IW2723 | NT | NT | NT | VI | VI |
| CB454 | Control | III | III | III | III |
| IW4445 | Control | Ia | Ia | Ia | Ia |
| IW4077 | Control | II | II | II | II |
NT, nontypeable.
This WGS-based method for GBS serotyping, which was validated using 223 isolates that had been typed using conventional methods, was therefore highly accurate. Although WGS currently may not be cost-effective for direct replacement of traditional serotyping, costs are likely to decrease further. Furthermore, WGS may already be the cheapest option for combined studies, with possibilities for utilizing the resulting data for additional analyses, such as multilocus sequence typing, analyses of relatedness to other sequenced isolates, and detailed phylogenetic analyses.
ACKNOWLEDGMENTS
We thank the Wellcome Trust Centre for Human Genetics, University of Oxford, where the whole-genome sequencing was performed, and we thank the library and sequencing teams. We thank the Shoklo Malaria Research Unit (Thailand) for providing GBS isolates.
The Modernising Medical Microbiology Informatics Group includes Jim Davies, Charles Crichton, Milind Acharya, and Carlos del Ojo Elias.
Funding Statement
This publication presents independent research supported by the Wellcome Trust (grants 093804 and 098532 to A.C.S. and J.A.B.), the Health Innovation Challenge Fund (grants HICF-T5-358 and WT098615/Z/12/Z), a parallel funding partnership between the Department of Health and the Wellcome Trust (D.W.C., A.E.S., and A.S.W.), the United Kingdom Clinical Research Collaboration (a parallel funding partnership between the Medical Research Council [grant G0800778], the Biotechnology and Biological Sciences Research Council, and the Wellcome Trust [grant 087646/Z/08/Z]), and the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre. D.W.C. is a NIHR senior investigator.
The views expressed in this publication are those of the authors and not necessarily those of the funders.
REFERENCES
- 1.Baker CJ, Barrett FF, Gordon RC, Yow MD. 1973. Suppurative meningitis due to streptococci of Lancefield group B: a study of 33 infants. J Pediatr 82:724–729. doi: 10.1016/S0022-3476(73)80606-7. [DOI] [PubMed] [Google Scholar]
- 2.Barton LL, Feigin RD, Lins R. 1973. Group B beta hemolytic streptococcal meningitis in infants. J Pediatr 82:719–723. doi: 10.1016/S0022-3476(73)80605-5. [DOI] [PubMed] [Google Scholar]
- 3.Communicable Disease Surveillance Centre. 1985. Neonatal meningitis: a review of routine national data 1975–83. Br Med J (Clin Res Ed) 290:778–779. doi: 10.1136/bmj.290.6470.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon HC Jr, Gray E, Pass MA, Gray BM. 1982. Anorectal and vaginal carriage of group B streptococci during pregnancy. J Infect Dis 145:794–799. doi: 10.1093/infdis/145.6.794. [DOI] [PubMed] [Google Scholar]
- 5.Schuchat A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin Microbiol Rev 11:497–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA 299:2056–2065. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 7.Madhi SA, Dangor Z, Heath PT, Schrag S, Izu A, Sobanjo-Ter Meulen A, Dull PM. 2013. Considerations for a phase-III trial to evaluate a group B Streptococcus polysaccharide-protein conjugate vaccine in pregnant women for the prevention of early- and late-onset invasive disease in young-infants. Vaccine 31(Suppl 4):D52–D57. doi: 10.1016/j.vaccine.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 8.Mulholland K, Satzke C. 2012. Serotype replacement after pneumococcal vaccination. Lancet 379:1387. doi: 10.1016/S0140-6736(12)60588-1. [DOI] [PubMed] [Google Scholar]
- 9.Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, Creti R. 2010. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J Microbiol Methods 80:212–214. doi: 10.1016/j.mimet.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker TM, Kohl TA, Omar SV, Hedge J, Del Ojo Elias C, Bradley P, Iqbal Z, Feuerriegel S, Niehaus KE, Wilson DJ, Clifton DA, Kapatai G, Ip CL, Bowden R, Drobniewski FA, Allix-Beguec C, Gaudin C, Parkhill J, Diel R, Supply P, Crook DW, Smith EG, Walker AS, Ismail N, Niemann S, Peto TE, Modernizing Medical Microbiology Informatics Group. 2015. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 15:1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ. 2012. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 14.Slotved HC, Kong F, Lambertsen L, Sauer S, Gilbert GL. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J Clin Microbiol 45:2929–2936. doi: 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisharat N, Crook DW, Leigh J, Harding RM, Ward PN, Coffey TJ, Maiden MC, Peto T, Jones N. 2004. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J Clin Microbiol 42:2161–2167. doi: 10.1128/JCM.42.5.2161-2167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones N, Oliver K, Jones Y, Haines A, Crook D. 2006. Carriage of group B streptococcus in pregnant women from Oxford, UK. J Clin Pathol 59:363–366. doi: 10.1136/jcp.2005.029058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies HD, Jones N, Whittam TS, Elsayed S, Bisharat N, Baker CJ. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J Infect Dis 189:1097–1102. doi: 10.1086/382087. [DOI] [PubMed] [Google Scholar]
- 18.Gladman S, Seeman T. 2012. VelvetOptimiser. Victorian Bioinformatics Consortium, Clayton, Australia: http://bioinformatics.net.au/software.velvetoptimiser.shtml. [Google Scholar]
- 19.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto S, Miyake K, Koike Y, Watanabe M, Machida Y, Ohta M, Iijima S. 1999. Molecular characterization of type-specific capsular polysaccharide biosynthesis genes of Streptococcus agalactiae type Ia. J Bacteriol 181:5176–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe M, Miyake K, Yanae K, Kataoka Y, Koizumi S, Endo T, Ozaki A, Iijima S. 2002. Molecular characterization of a novel β1,3-galactosyltransferase for capsular polysaccharide synthesis by Streptococcus agalactiae type Ib. J Biochem 131:183–191. doi: 10.1093/oxfordjournals.jbchem.a003086. [DOI] [PubMed] [Google Scholar]
- 22.Martins ER, Melo-Cristino J, Ramirez M. 2007. Reevaluating the serotype II capsular locus of Streptococcus agalactiae. J Clin Microbiol 45:3384–3386. doi: 10.1128/JCM.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaffin DO, Beres SB, Yim HH, Rubens CE. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J Bacteriol 182:4466–4477. doi: 10.1128/JB.182.16.4466-4477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cieslewicz MJ, Chaffin D, Glusman G, Kasper D, Madan A, Rodrigues S, Fahey J, Wessels MR, Rubens CE. 2005. Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect Immun 73:3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


