Abstract
This multicenter study analyzed Nocardia spp., including extraction, spectral acquisition, Bruker matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) identification, and score interpretation, using three Nocardia libraries, the Bruker, National Institutes of Health (NIH), and The Ohio State University (OSU) libraries, and compared the results obtained by each center. A standardized study protocol, 150 Nocardia isolates, and NIH and OSU Nocardia MALDI-TOF MS libraries were distributed to three centers. Following standardized culture, extraction, and MALDI-TOF MS analysis, isolates were identified using score cutoffs of ≥2.0 for species/species complex-level identification and ≥1.8 for genus-level identification. Isolates yielding a score of <2.0 underwent a single repeat extraction and analysis. The overall score range for all centers was 1.3 to 2.7 (average, 2.2 ± 0.3), with common species generally producing higher average scores than less common ones. Score categorization and isolate identification demonstrated 86% agreement between centers; 118 of 150 isolates were correctly identified to the species/species complex level by all centers. Nine strains (6.0%) were not identified by any center, and six (4.0%) of these were uncommon species with limited library representation. A categorical score discrepancy among centers occurred for 21 isolates (14.0%). There was an overall benefit of 21.2% from repeat extraction of low-scoring isolates and a center-dependent benefit for duplicate spotting (range, 2 to 8.7%). Finally, supplementation of the Bruker Nocardia MALDI-TOF MS library with both the OSU and NIH libraries increased the genus-level and species-level identification by 18.2% and 36.9%, respectively. Overall, this study demonstrates the ability of diverse clinical microbiology laboratories to utilize MALDI-TOF MS for the rapid identification of clinically relevant Nocardia spp. and to implement MALDI-TOF MS libraries developed by single laboratories across institutions.
INTRODUCTION
Nocardia spp. are Gram-positive, beaded, branching, and partially acid-fast bacilli belonging to the Corynebacterineae suborder. Nocardiae are environmentally ubiquitous and are recognized to be human pathogens causing both cutaneous and soft tissue infections in immunocompetent patients and opportunistic infections (e.g., lung and brain infections) in immunocompromised hosts. Determination of the Nocardia species causing an infection is important because different species vary in their epidemiology, virulence, and antibiotic susceptibility (1–3). Traditional methods for the determination of Nocardia species include biochemical tests, susceptibility profiling, and sequencing methods (1–5). Recently, matrix-assisted laser desorption ionization (MALDI)–time of flight (TOF) mass spectrometry (MS) has been identified to be a rapid, accurate method for the identification of Nocardia spp. in the clinical laboratory (6–10). Although MALDI-TOF MS promises to aid in the identification of these challenging organisms, several issues related to its universal implementation remain, including the limited representation of rare species in existing databases, novel species detection, and a lack of interchangeable protocols and databases for use in both research and clinical laboratories.
This multicenter study compared Nocardia MALDI-TOF MS extraction, spectral acquisition, and interpretation with the Bruker Biotyper program to determine the accuracy and reproducibility of Nocardia isolate identification by three separate Biotyper libraries: the MALDI-TOF MS Biotyper reference library (Bruker Daltonics Inc., Billerica, MA), the National Institutes of Health (NIH) supplemental Nocardia library (National Institutes of Health, Bethesda, MD), and The Ohio State University (OSU) supplemental Nocardia library (The Ohio State University, Columbus, OH). One hundred fifty isolates representing 30 species or species complexes were exchanged between the centers. Using standardized extraction and run parameters, Nocardia isolate identification and analysis were objectively examined to establish a transferrable platform of the MALDI-TOF MS technology for the identification of Nocardia spp. in the clinical setting.
This study analyzed the ability of individual clinical microbiology laboratories to utilize MALDI-TOF MS for the routine identification of clinically relevant Nocardia spp. and to incorporate MALDI-TOF MS libraries developed by single laboratories into the institutional work flow. The combined use of three Nocardia MALDI-TOF MS libraries provided the most benefit to the centers involved in this study, with 84.2% of isolates being identified to the species or species complex level by all three centers (range, 80 to 89.3% per center) using score cutoffs of ≥2.0 for species/species complex-level identification. Examination of score accuracy thresholds, however, suggested that species- or species complex-level identification could be achieved with MALDI-TOF MS scores of >1.8 with no change in accuracy. Several laboratories have previously published descriptions of the use of this standard for the identification of Mycobacterium spp. (11, 12). Further supplementation of existing libraries will be necessary to improve the identification of select species.
MATERIALS AND METHODS
Isolate exchange.
Fifty isolates previously identified at each center were exchanged. Each isolate was identified on the basis of colony morphology, modified acid-fast stain, and sequencing results. The sequencing target and method varied by center: center 1 performed pyrosequencing of the 16S rRNA gene (13, 14), center 2 performed Sanger sequencing of secA alone or full-length 16S rRNA sequencing (2, 15), and center 3 performed 500-bp Sanger sequencing of the 16S rRNA gene (15). The isolates used for library development were not eligible for isolate exchange and testing.
Exchanged isolates were cultured on Sabouraud dextrose agar (SAB) slants and were assigned a code prior to shipping. Upon receipt, the slants were maintained at 4°C until subculture. A key for the postanalysis identification of these isolates was submitted to A. M. Zelazny (NIH) and was not available to the MALDI-TOF MS operator at each location until the spectral analyses had been submitted, in order to keep the investigators in the study blind to the identity of the isolates and unbiased.
Subculture and growth conditions.
A loop or sterile wooden stick was applied to the SAB slant and used to streak a new SAB plate for organism isolation. The plates were incubated at 35°C in 5% CO2 for 3 to 4 days and examined for purity. Slowly growing isolates were grown for a maximum of 7 days prior to extraction.
MALDI-TOF MS reagents and controls.
Prior to each MALDI-TOF MS run, each facility calibrated its Bruker Biotyper instrument using either a bacterial test standard (BTS; Bruker Daltonics Inc.) or a MALDI-TOF MS calibration standard. To prepare the MALDI-TOF MS calibration standard, 125 μl of 50% acetonitrile–0.1% trifluoroacetic acid was added to a tube of the protein calibration I standard (Bruker Daltonics, Inc.). The components in this solution were briefly mixed, and then the solution was combined with the peptide calibration II standard (Bruker Daltonics, Inc.) to produce the final MALDI-TOF MS calibration standard solution. Standards were stored at −20°C until use. To prepare the working matrix solution, a 400-μl aliquot of stock matrix solution (50% acetonitrile, 2.5% trifluoroacetic acid) was removed from a −20°C freezer and placed on a plate warmer that had been preheated to 44°C for 15 min. Once heated, a saturated α-cyano-4-hydroxycinnamic acid solution was created by adding an excess of α-cyano-4-hydroxycinnamic acid crystals (Bruker Daltonics Inc.) to the aliquot of stock matrix solution to leave 1 to 2 mm of undissolved crystals in the bottom of the tube. The solution was then vortexed and placed on the plate warmer for 15 min.
Two positive controls (Aspergillus ustus strain CBS 261.67T and Nocardia brasiliensis strain ATCC 19296) were included in each extraction, and MALDI-TOF MS spectra were acquired to ensure extraction efficiency and calibration accuracy and to detect potential contamination of the microScout plate (Bruker Daltonics Inc.). The N. brasiliensis control was grown under conditions identical to those described under “Subculture and growth conditions” above. The A. ustus strain was maintained under standard fungal growth conditions (SAB slant, 30°C, no CO2) (16). A negative control was included in each microScout plate run.
MALDI extraction.
Using a loop or a wooden stick, several colonies of each Nocardia isolate were suspended in Eppendorf tubes containing 50 μl of 0.1-mm-diameter zirconia-silica beads and 500 μl of 70% ethanol, vortexed at maximum speed for 15 min, and then centrifuged at 13,000 × g for 2 min to pellet the organism. All ethanol was removed from the tube using a fine-tip pipette. Fifty microliters of 70% formic acid was added to the resulting pellet, and the tubes were vortexed at maximum speed for 5 min. Following this step, the tubes were briefly centrifuged to remove any debris from the lid. Next, 50 μl of 100% acetonitrile was added, and the tube was vortexed for an additional 5 min. Finally, the tubes were centrifuged for 2 min at maximum speed to pellet the Nocardia and beads. The supernatant was immediately spotted in technical duplicate on the clean microScout plate. Following extraction and spotting of the microScout plate, extracts were frozen at −20°C.
Spotting of the microScout plate.
One microliter from each Nocardia sample or control was spotted in duplicate on a 96-well microScout MALDI-TOF MS BigAnchor target plate and allowed to dry on a heat block (44°C). Two microliters of the working matrix solution was added onto each spotted well and allowed to dry. Finally, the microScout plate was allowed to cool briefly on the benchtop before it was loaded into the Microflex instrument.
MALDI-TOF MS method parameters.
Spectra were collected on Bruker MicroFlex instruments using the standard method supplied by Bruker, with the exception that 1,000 shots were acquired in 50-shot steps. The instrument was calibrated prior to each run using the routine calibration standards, either BTS or the MALDI-TOF MS calibration standard, per the routine practice of each lab. Matching was performed using Bruker's recommended parameters with the Biotyper program (v3.1.66; Bruker Daltonics Inc.).
Nocardia libraries.
Three Bruker MALDI-TOF MS Nocardia libraries were examined in this study: the MALDI-TOF MS Biotyper reference library (v4.0.0.1, 5,627 entries total, 72 Nocardia entries; Bruker Daltonics Inc.), the NIH supplemental Nocardia library (v2, 64 entries; National Institutes of Health, Bethesda, MD), and the OSU supplemental Nocardia library (v1.1, 26 entries; The Ohio State University, Columbus, OH). The Bruker library was supplied by Bruker, the NIH library was constructed as previously described for molds with a minimum of six spectra per library submission (16), and the OSU library was constructed as described in the Materials and Methods in the supplemental material. With the exception of the analysis for which the results are reported in Table 7, all analyses described in this report were performed using the top score regardless of library.
TABLE 7.
Contribution of Nocardia libraries to genus- and species-level isolate identification
| Identification level and center | No. (%) of isolates correctly identified by use of the following library(ies): |
|||
|---|---|---|---|---|
| Bruker | Bruker + NIH | Bruker + OSU | Bruker + NIH + OSU | |
| Species level | ||||
| All centers | 213 (47.3) | 309 (68.7) | 326 (72.4) | 379 (84.2) |
| Center 1 | 78 (52.0) | 104 (69.3) | 112 (74.7) | 125 (83.3) |
| Center 2 | 75 (50.0) | 128 (85.3) | 101 (67.3) | 134 (89.3) |
| Center 3 | 60 (40.0) | 77 (51.3) | 113 (75.3) | 120 (80.0) |
| Genus level | ||||
| All centers | 111 (24.7) | 80 (17.8) | 33 (7.3) | 27 (6.0) |
| Center 1 | 28 (18.7) | 25 (16.7) | 7 (4.7) | 10 (6.7) |
| Center 2 | 40 (26.7) | 10 (6.7) | 21 (14.0) | 6 (4.0) |
| Center 3 | 43 (28.7) | 45 (30.0) | 5 (3.3) | 11 (7.3) |
| No identification | ||||
| All centers | 126 (28.0) | 61 (13.6) | 91 (20.2) | 44 (9.8) |
| Center 1 | 44 (29.3) | 21 (14.0) | 31 (20.7) | 15 (10.0) |
| Center 2 | 35 (23.3) | 12 (8.0) | 28 (18.7) | 10 (6.7) |
| Center 3 | 47 (31.3) | 28 (18.7) | 32 (21.3) | 19 (12.7) |
Spectral analysis.
The Bruker, NIH, and OSU Nocardia libraries were challenged against 150 clinical isolates run by each center on duplicate spots. Scores between 1.8 and 1.99 were considered appropriate for genus-level identification, and those ≥2.0 were considered candidates for species-level identification. A degree of score separation of ≥10% from the next-highest putative species- or species complex-level identification was required. Any isolate scoring <2.0 was reextracted and rerun once, in order to mirror clinical practice.
Categorical agreement was defined as the accurate assessment of species- or species complex-level identification within the appropriate MALDI-TOF MS score range. The following species were included in each species complex: the N. nova complex (N. nova, N. elegans, N. kruczakiae, N. veterana), the N. abscessus complex and related species (NARS) (N. abscessus, N. arthritidis, N. asiatica, N. beijingensis, N. pneumoniae), the N. testacea complex (N. testacea, N. sienata), the N. transvalensis complex (N. transvalensis, N. wallacei), and the N. brevicatena-N. paucivorans complex (N. brevicatena, N. paucivorans). With the exception of the scores in Table 6, all identifications were assigned using score cutoffs of 1.8 for the genus level and 2.0 for the species/species complex level.
TABLE 6.
Evaluation of score threshold and margin for identification of Nocardia
| Center | % isolates with the following score threshold/margina: |
|
|---|---|---|
| ≥2.0 cutoff/10% rule | ≥1.8 cutoff/10% rule | |
| 1 | 83.3 | 89.3 |
| 2 | 89.3 | 92.6 |
| 3 | 80.0 | 87.3 |
| Combined | 84.2 | 89.8 |
The cutoff specifies the score required for species- or species complex-level identification; the 10% rule indicates that species- or species complex-level identification is consistent within the top 10% of the highest score.
Calculations.
The average isolate score and standard deviation were calculated using the top scores for the isolate provided by each of the three centers. The average and standard deviation were calculated using Microsoft Excel software.
The average species/species complex score was analyzed by adding the three center scores for each of the isolates assigned to the species/species complex, dividing by the total number of isolates, and multiplying by 3 (as three centers each contributed scores).
The benefit to repeat extraction and the benefit to spotting of duplicates were measured as the number of isolates where reextraction and duplicate spotting improved the overall score above the categorical threshold (<1.8, 1.8 to 1.99, ≥2.0) divided by the total number of isolates for which reextraction or spotting, respectively, was performed. The average of these values was calculated by adding the number of all isolates that exhibited a categorical score change, regardless of center, and dividing by the total number of isolates from any center for which the repeat analysis was performed.
Score agreement among the study centers was assigned by evaluating the score category of each isolate by each center with respect to species/species complex-level identification (i.e., <1.8, 1.8 to 1.99, ≥2.0). A discordant result was one in which the score category of an individual isolate was different for all three centers.
RESULTS
Isolate selection and exchange between three clinical microbiology laboratories.
Fifty isolates from three centers were exchanged in this study (total n = 150; Table 1). Isolates comprised Nocardia spp. commonly encountered in each facility as well as rarer or difficult-to-identify species. All isolates were previously identified by genomic sequencing of the 16S rRNA and/or secA gene prior to inclusion in this study. MALDI-TOF MS operators at each location were kept blind to the identity of the isolates during the study and until all testing was completed and the results were submitted and analyzed.
TABLE 1.
Isolates submitted for multicenter study
| Organism identification | No. of isolates submitted by center: |
||
|---|---|---|---|
| 1 | 2 | 3 | |
| N. abscessus complex and related species (NARS) (n = 10)a | 9 | 1 | |
| N. abscessus (n = 2) | 2 | ||
| N. arthritidis (n = 2) | 2 | ||
| N. asiatica (n = 2) | 2 | ||
| N. beijingensis (n = 3) | 3 | ||
| N. pneumoniae (n = 4) | 4 | ||
| N. brasiliensis (n = 13) | 8 | 1 | 4 |
| N. brevicatena-N. paucivorans complex | |||
| N. brevicatena (n = 1) | 1 | ||
| N. paucivorans (n = 2) | 1 | 1 | |
| N. cyriacigeorgica (n = 17) | 8 | 1 | 8 |
| N. farcinica (n = 16) | 8 | 1 | 7 |
| N. nova complex (n = 21)a | 16 | 5 | |
| N. elegans (n = 2) | 2 | ||
| N. kruczakiae (n = 3) | 2 | 1 | |
| N. nova (n = 22) | 1 | 21 | |
| N. veterana (n = 2) | 2 | ||
| N. transvalensis complex | |||
| N. transvalensis (n = 1) | 1 | ||
| N. wallacei (n = 2) | 2 | ||
| N. anaemiae (n = 2) | 2 | ||
| N. asteroides sensu stricto (n = 1) | 1 | ||
| N. blacklockiae (n = 2) | 2 | ||
| N. exalbida (n = 1) | 1 | ||
| N. mexicana (n = 3) | 3 | ||
| N. niigatensis (n = 1) | 1 | ||
| N. pseudobrasiliensis (n = 3) | 3 | ||
| N. puris (n = 3) | 3 | ||
| N. otitidiscaviarum (n = 4) | 1 | 3 | |
| N. testacea complex | |||
| N. testacea (n = 3) | 3 | ||
| N. sienata (n = 0) | |||
| N. thailandica (n = 1) | 1 | ||
| Nocardia sp. (n = 1)b | 1 | ||
| Total | 50 | 50 | 50 |
The isolates were identified only to the species complex level.
The species of the isolate could not be determined by 16S rRNA or secA sequence analysis.
Species- and species complex-level identification of Nocardia by MALDI-TOF MS.
Isolates were extracted, spotted, and run using a Bruker MicroFlex LT mass spectrometer (Bruker Daltonics Inc.), and the spectra were analyzed against those for the three MALDI-TOF MS Nocardia libraries (the Bruker, NIH, and OSU libraries; Table 2) using Biotyper software (version 3.0; Bruker Daltonics, Inc.) The top 10 results for each isolate were submitted to a single facility for data analysis. The identity of each isolate was compared with that obtained from the genomic sequencing results, and the top score was used to calculate the average score for each species or species complex (Fig. 1). Overall scores for all isolates ranged from 1.3 to 2.7, and the average was 2.2 ± 0.3. Common species, such as N. cyriacigeorgica, generally produced higher average scores than less common ones, such as N. pseudobrasiliensis (Fig. 1).
TABLE 2.
Nocardia isolate contribution, per library
| Organism name | No. of isolates in the following library: |
||
|---|---|---|---|
| Bruker | NIH | OSU | |
| N. abscessus | 2 | 3 | 3 |
| N. africana | 1 | 1 | |
| N. anaemiae | 1 | 1 | |
| N. aobensis | 1 | 1 | |
| N. araoensis | 1 | 1 | |
| N. arthritidis | 1 | 2 | |
| N. asiatica | 1 | 1 | |
| N. asteroides | 3 | 1 | |
| N. asteroides drug pattern II | 1 | ||
| N. beijingensis | 4 | ||
| N. blacklockiae | 1 | ||
| N. brasiliensis | 1 | 2 | 3 |
| N. brevicatena | 1 | ||
| N. carnea | 1 | 1 | |
| N. concava | 1 | 1 | |
| N. cyriacigeorgica | 16 | 4 | 4 |
| N. elegans | 1 | 1 | 1 |
| N. exalbida | 1 | 1 | |
| N. farcinica | 12 | 2 | 4 |
| N. higoensis | 1 | 1 | |
| N. ignorata | 1 | 1 | |
| N. inohanensis | 1 | ||
| N. kruczakiae | 1 | 3 | |
| N. mexicana | 1 | ||
| N. neocaledoniensis | 1 | ||
| N. niigatensis | 1 | 1 | |
| N. ninae | 1 | ||
| N. nova | 2 | 2 | 6 |
| N. otitidiscaviarum | 6 | 2 | |
| N. paucivorans | 2 | 1 | 1 |
| N. pneumoniae | 1 | 1 | |
| N. pseudobrasiliensis | 2 | ||
| N. puris | 1 | ||
| N. salmonicida | 1 | ||
| N. seriolae | 1 | ||
| N. shimofusensis | 1 | ||
| N. sienata | 1 | 1 | |
| Nocardia sp. | 4 | ||
| N. takedensis | 1 | ||
| N. terpenica | 1 | ||
| N. testacea | 1 | 2 | |
| N. thailandica | 1 | 1 | |
| N. transvalensis | 1 | 1 | 1 |
| N. vermiculata | 1 | 1 | |
| N. veterana | 1 | 2 | 3 |
| N. vinacea | 1 | ||
| N. wallacei | 2 | ||
| N. yamanashiensis | 1 | 1 | |
| Total | 72 | 64 | 26 |
FIG 1.
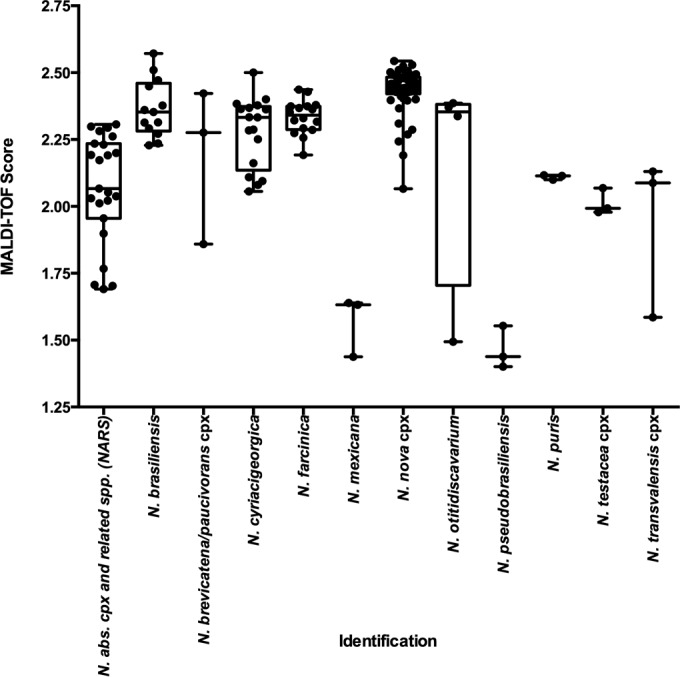
Average MALDI-TOF MS score per species- or species complex-level identification. The average MALDI-TOF MS score per isolate is represented for those species or species complexes with greater than three isolates represented within the study. Horizontal bars within the box-and-whisker plots represent the average for all species or species complex (cpx) isolates in that category.
Isolates with scores of <2.0 were reextracted and rerun once in order to mirror clinical practice. The benefit of reextraction was measured as the number of isolates where reextraction improved the overall score above a specified threshold divided by the total number of isolates for which reextraction was performed. Categorical score improvement occurred with 21.2% of repeat extractions (range, 20.0 to 22.6%; Table 3), demonstrating a clear benefit to this process.
TABLE 3.
Benefit to repeat extractiona
| Center | No. of repeat extractions performed | No. (%) of isolates with improved identification to: |
Overall benefit (%) | |
|---|---|---|---|---|
| Genus level | Species level | |||
| 1 | 31 | 1 (6) | 6 (19) | 22.6 |
| 2 | 20 | 0 (0) | 4 (20) | 20.0 |
| 3 | 34 | 2 (6) | 5 (15) | 20.6 |
Score categories with respect to genus- or species/species complex-level identification were as follows: genus, 1.8 to 1.99; species/species complex, ≥2.0.
The benefit of technical respotting was also examined. An average categorical score improvement of 4.7% was achieved for duplicate spotting (Table 4), but the values differed significantly among the centers (range, 2.0 to 8.7%), highlighting the benefit of a center-specific analysis of this variable.
TABLE 4.
Benefit of spotting of duplicatesa
| Center | No. (%) of isolates with improved identification to: |
Overall benefit (%) | |
|---|---|---|---|
| Genus level | Species level | ||
| 1 | 1 (1) | 2 (1) | 2.0 |
| 2 | 0 (0) | 5 (3) | 3.3 |
| 3 | 8 (5) | 5 (3) | 8.7 |
A total of 150 repeat spottings were performed per center. Score categories with respect to genus- or species/species complex-level identification were as follows: genus, 1.8 to 1.99; species/species complex, ≥2.0.
Score agreement among study centers.
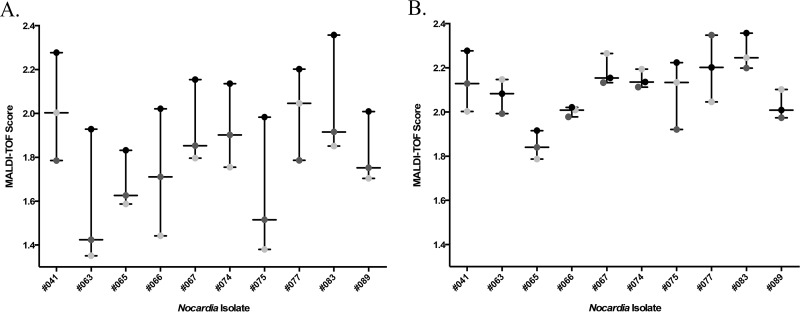
The score agreement among the study centers was analyzed for each score category (≥2.0, 1.8 to 1.99, and <1.8). Isolates were categorized on the basis of their known identity and the isolate MALDI-TOF MS score reported for each isolate per center. Score categorization and isolate identification demonstrated 86.0% overall agreement among all participating centers (Table 5). Categorical score discrepancies among centers occurred for 21 isolates and may have been due to extraction efficiency or variable instrument-specific issues (Table 5, columns 6 to 9). With 11 of these isolates, including two isolates with discordant results, a score of ≥2.0 was produced by at least one center, indicating that identification of the individual isolate was possible with the libraries examined (Fig. 2A; Table 5, columns 6 and 9). To troubleshoot these particular score discrepancies, the extracts from these isolates were shared and reanalyzed by one of the participating institutions. Interestingly, this resolved a majority of the score discrepancies (Fig. 2B). A number of variables were considered to explain the differences in scores between centers when the same set of libraries was used: the instrument used for library construction, microScout plate cleanliness, the length of culture incubation, storage of extracts, the amount of bacteria extracted, laser intensity, source cleanliness, and other instrument settings. Percent source cleanliness is automatically reported by the Biotyper instrument software and indicates the level of ionization debris buildup on the ion source. Figure 2B represents the results of the reanalysis of the extracts at 100% source cleanliness, as the machine had recently been serviced. Interestingly, the effect of source cleanliness on the score varied among the extracts/strains. Studies to further evaluate this and other parameters are under way.
TABLE 5.
Agreement of scores among the three centers, per species complex-level identification
| Isolate or species complex submitted for multicenter study | No. of isolates | No. of isolates with the indicated MALDI-TOF MS score or resulta: |
||||||
|---|---|---|---|---|---|---|---|---|
| All three centers |
Two of three centers |
Discordant | ||||||
| >2.0 | 1.8–1.99 | <1.8 | >2.0 | 1.8–1.99 | <1.8 | |||
| N. nova complex | 50 | 50 | ||||||
| N. abscessus complex and related species (NARS) | 23 | 12 | 1 | 2 | 4 | 2 | 2 | |
| N. cyriacigeorgica | 17 | 16 | 1 | |||||
| N. farcinica | 16 | 16 | ||||||
| N. brasiliensis | 13 | 13 | ||||||
| N. otitidiscaviarum | 4 | 3 | 1 | |||||
| N. mexicana | 3 | 1 | 2 | |||||
| N. puris | 3 | 2 | 1 | |||||
| N. testacea complex | 3 | 1 | 1 | 1 | ||||
| N. pseudobrasiliensis | 3 | 3 | ||||||
| N. transvalensis complex | 3 | 2 | 1 | |||||
| N. niigatensis | 1 | 1 | ||||||
| N. blacklockiae | 2 | 1 | 1 | |||||
| N. anaemiae | 2 | 2 | ||||||
| N. brevicatena-N. paucivorans complex | 3 | 2 | 1 | |||||
| N. exalbida | 1 | 1 | ||||||
| N. thailandica | 1 | 1 | ||||||
| N. asteroides sensu stricto | 1 | 1 | ||||||
| Nocardia sp. | 1 | 1 | ||||||
| Total | 150 | 118 | 2 | 9 | 9 | 4 | 6 | 2 |
The agreement for all three centers was 86.0%, and the agreement between any two centers was 98.7%. A discordant result was one in which the score category of an individual isolate was different for all three centers. (i.e., <1.8, 1.8 to 1.99, ≥2.0).
FIG 2.
Discrepant score agreement among centers. (A) Discrepancies in original MALDI-TOF MS scores submitted for each center for selected isolates; (B) discrepancies in MALDI-TOF MS scores from submitted protein extracts analyzed side by side at a single center. Light gray circles, center 1; black circles, center 2; dark gray circles, center 3.
Score interpretation, threshold, and margins for identification of Nocardia.
Many clinical laboratories use score thresholds of 2.0 for species-level identification and 1.7 for genus-level identification of bacteria with the Bruker Biotyper instrument. We examined whether a lower score threshold of 1.8 for the species/species complex-level identification of Nocardia spp. would decrease that accuracy compared with that achieved with the traditional 2.0 cutoff with this isolate cohort. For this analysis, species/species complex-level identifications that scored ≥1.8 were accepted as definitive, provided that the score of the next-highest match differed by ≥10%. Each center observed an increase in the number of isolates that could be reported with the decreased score threshold (average, 5.6%; range, 3.3 to 7.3%) but did not observe a corresponding decrease in accuracy (Table 6). These results suggest that Nocardia spp. could potentially be reported to the species/species complex level with a score cutoff of 1.8, a finding similar to that obtained by use of the approach taken with Mycobacterium identification at some centers (11, 12). Two entries within the Bruker library complicated this analysis. Nocardia sp. strain MB_9090_05 THL and Nocardia sp. strain N394 IBS consistently grouped with N. brasiliensis and N. cyriacigeorgica, respectively. For the purposes of this study, these entries were ignored, as there was no species information provided.
Analysis of library contribution to Nocardia MALDI-TOF MS identification.
All results presented thus far were based on combined analysis of three MALDI-TOF MS Nocardia libraries; next, we analyzed the contribution of individual libraries. Use of the Bruker, NIH, and OSU libraries provided an overall 36.9% benefit for species-level identification compared to the use of the Bruker library alone (Table 7; all centers, n = 450). Twenty isolates were considered difficult to identify, as the highest MALDI-TOF MS score for at least one center was <1.8. Nine strains (two N. pneumoniae strains, one N. otitidiscaviarum strain, one N. mexicana strain, three N. pseudobrasiliensis strains, one N. wallacei strain, one Nocardia sp. strain) failed to be identified by all centers; six of these were uncommon species with a limited representation in the libraries, and one was a potentially novel species which was not anticipated to be identifiable by the existing libraries. In all, supplementation of the Bruker standard library with either or both of the supplemental libraries greatly improved the scores and identification of Nocardia isolates.
Interestingly, not all library spectra were representative of the diversity of the isolates tested in this study (Fig. 3), as some spectral alignments differed between the library and isolate profiles. Figure 3 highlights these misalignments as several prominent peaks differed between the N. mexicana study isolate profile and a single entry of the reference strain used to build the NIH supplemental Nocardia library. These changes in peak presence and position are consistent with the isolate being a different strain of the same species. Alignment of the study isolate 16S rRNA and secA gene sequences with the N. mexicana reference strain 16S rRNA and secA gene sequences confirmed the identity of this isolate. These data highlight the need for multiple library entries for each species and suggest that additional library entries may be beneficial for less common species.
FIG 3.
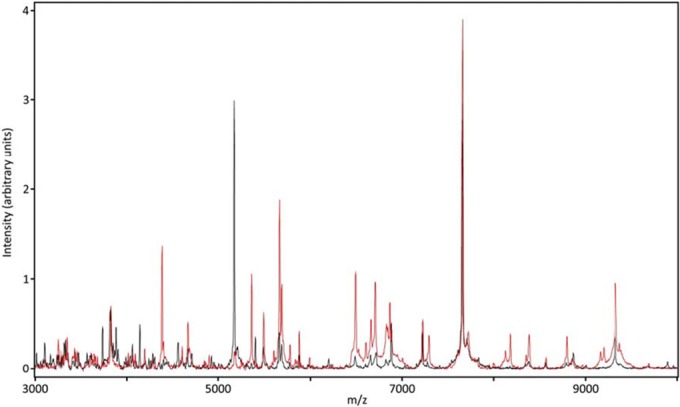
Representative Nocardia mexicana spectral overlay. Library (black) and isolate (red) spectra display peak mismatches.
DISCUSSION
This study demonstrates the ability of diverse clinical microbiology laboratories to utilize MALDI-TOF MS for the rapid, reliable identification of clinically relevant Nocardia spp. and to implement MALDI-TOF MS libraries developed by single laboratories across institutions. The combined use of three Nocardia MALDI-TOF MS libraries provided the most benefit, with 78.7% of isolates being identified to the species or species complex level by all centers.
Previous studies on the ability of supplemental libraries to augment MALDI-TOF MS Nocardia identification demonstrated, on average, an 39% (range, 30 to 75%) improvement in species-level identification with inclusion of an in-house-developed library to the diagnostic algorithm (6, 7, 10). In this study, an overall benefit of 36.9% in species- or species complex-level identification was achieved with supplemental library use; however, isolates of several uncommon species were purposefully included in this study to challenge these commercial and laboratory-created databases and were not represented in previous studies (N. anaemiae, N. blacklockiae, N. kruczakiae, N. mexicana, N. pneumoniae, N. thailandica). As isolates of these species were among those considered difficult to identify in this study, their inclusion led to a negative skew in the calculated benefit for the use of supplemental libraries.
MALDI-TOF MS emerges within the evolving field of Nocardia identification as an identification method that is rapid but perhaps less discriminatory than the widely used sequencing method. As demonstrated in this study, species complex-level identification was faithfully demonstrated for the major species complexes (the N. abscessus complex and related species [NARS], N. brevicatena-N. paucivorans complex, N. nova complex, N. transvalensis complex), but species complex deconvolution was limited. Nevertheless, because of the speed and accuracy of MALDI-TOF MS for rapid genus-level identification and assignment of an isolate to clinically relevant species or species complexes, its use will greatly improve the diagnosis of nocardiosis in the clinical laboratory, with sequencing being available as a secondary method when needed. Several studies have also cited limitations in species complex deconvolution using partial 16S rRNA gene sequencing (5, 10). The limitation of MALDI-TOF MS for the deconvolution of species complexes could potentially impact patient care when a choice between alternative antimicrobial regimens is necessary for effective treatment. Under these circumstances, species-level identification by sequencing may be warranted, especially with organisms grouped by MALDI-TOF MS to be members of the N. abscessus complex and related species (NARS) (N. abscessus, N. arthritidis, N. asiatica, N. beijingensis, N. pneumoniae), which include species with different antibiotic susceptibility patterns (5, 17). Of note, enhanced species complex deconvolution may be achieved with secA-targeted sequencing (2).
The nomenclature for the nocardiae has changed substantially with the transition from biochemical and antimicrobial susceptibility profile methods of identification to sequence-based methods of characterization (4). The discovery of novel species and the use of 16S rRNA gene sequencing in the clinical laboratory have highlighted the limitations of biochemical and susceptibility profiles in providing definitive Nocardia identification (5). The use of additional sequencing targets, such as secA, may provide further resolution from the species complex- to the species-level identification.
A striking difference between this work and previous Nocardia MALDI-TOF MS studies was the isolate extraction method. One notable distinction of our method from other studies was the inclusion of a zirconium bead-beating step in lieu of boiling (6, 8). The direct spotting and standard bacterial extraction methods developed for MALDI-TOF MS are suboptimal for Nocardia spp., due to the hardiness and composition of the cell wall. Verroken et al. demonstrated that a 30-min boiling step followed by formic acid extraction assisted with the acquisition of spectra for Nocardia spp. (6). However, boiling has been shown to complicate protein identification through the addition of formyl groups (18). In this study, the mold extraction protocol published by Lau et al., which uses zirconium beads to mechanically disrupt the cell wall, was modified and shown to enhance the extraction efficiency without producing modified proteins (16). This protocol has been successfully used since 2014 at NIH for the presumptive identification of Nocardia isolates.
In an effort to optimize the methodology of Nocardia MALDI-TOF MS identification across centers, several technical aspects, such as duplicate spotting and repeat isolate extraction, were examined. As demonstrated in Table 4, although two of the three centers found no substantial benefit to duplicate spotting, a third center demonstrated a benefit for 8.7% of isolates examined. In contrast, all centers found a substantial benefit of repeat extraction for isolates that originally scored <2.0 (Table 3). For laboratories in the process of implementing Nocardia MALDI-TOF MS identification, it may be beneficial to include duplicate spotting in the validation phase of method implementation, in order to determine the overall benefit to the institution. Future studies will examine variables that may impact Nocardia extraction and spectral acquisition, including instrument parameters, such as source cleanliness, as well as methods for the deconvolution of species complex-level identification when clinically indicated, which were limitations in this study.
Continued supplementation of the Nocardia MALDI-TOF MS libraries is necessary, as several species present in each Nocardia library are represented only by a single isolate. Even with supplementation by both the NIH and OSU MALDI-TOF MS libraries, many isolates did not achieve >3 representative MALDI-TOF MS spectrum profile (MSPs) per species. For species with a limited representation in the libraries, it may be prudent to report such results as preliminary, pending confirmation by sequencing.
A diagnostic algorithm is proposed on the basis of the results of this study. Suspected Nocardia spp. are grown for 3 to 4 days in SAB plates and identified via MALDI-TOF MS. A score of >1.8 can be assigned a definitive species/species complex-level identification if a ≥10% separation from the next-closest species/species complex is achieved. Additional sequencing may be necessary for species-level identification (i) for the treatment of infections caused by organisms requiring an alternative antibiotic regimen for empirical therapy, i.e., treatment of infections caused by members of the N. abscessus complex and related species (NARS) with amoxicillin-clavulanic acid, clarithromycin, or imipenem, and/or (ii) for epidemiologic purposes. Isolates achieving MALDI-TOF MS scores of <1.8 or those with scores of ≥1.8 not separated from their nearest species-level identification by 10% and requiring species-level identification should be reextracted and could also be subjected to 16S rRNA and/or secA sequencing.
In summary, MALDI-TOF MS was proven to be a quick and reliable method for the identification of Nocardia spp. Although the development of additional spectral databases is necessary to fully optimize this rapid detection method, the centers involved in this study demonstrated the adaptability of MALDI-TOF MS as a system for the detection of Nocardia in the clinical microbiology laboratory. Supplemental Nocardia MALDI-TOF MS libraries beyond commercial libraries that include more species and more strains of each species (particularly those with limited library representation), such as the institutional libraries examined in this study, will continue to greatly assist clinical laboratories worldwide and bridge the gaps of currently existing commercial databases.
Supplementary Material
ACKNOWLEDGMENTS
We thank the medical technologists from the NIH Clinical Center, Department of Laboratory Medicine, Microbiology Service, for their assistance in this study.
The Intramural Research Program of the NIH Clinical Center, Department of Laboratory Medicine, supported this research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02942-15.
REFERENCES
- 1.Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev 19:259–282. doi: 10.1128/CMR.19.2.259-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conville PS, Zelazny AM, Witebsky FG. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J Clin Microbiol 44:2760–2766. doi: 10.1128/JCM.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conville PS, Witebsky FB. 2007. Nocardia, Rhodococcus, Gordonia, Actinomadura, Streptomyces, and other aerobic actinomycetes, p 515–542. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (ed), Manual of clinical microbiology, 9th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 4.Wallace RJ Jr, Steele LC, Sumter G, Smith JM. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob Agents Chemother 32:1776–1779. doi: 10.1128/AAC.32.12.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlaberg R, Fisher MA, Hanson KE. 2014. Susceptibility profiles of Nocardia isolates based on current taxonomy. Antimicrob Agents Chemother 58:795–800. doi: 10.1128/AAC.01531-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verroken A, Janssens M, Berhin C, Bogaerts P, Huang TD, Wauters G, Glupczynski Y. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Nocardia species. J Clin Microbiol 48:4015–4021. doi: 10.1128/JCM.01234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segawa S, Nishimura M, Sogawa K, Tsuchida S, Murata S, Watanabe M, Matsushita K, Kamei K, Nomura F. 2015. Identification of Nocardia species using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Clin Proteomics 12:6. doi: 10.1186/s12014-015-9078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poisnel E, Roseau JB, Landais C, Rodriguez-Nava V, Bussy E, Gaillard T. 2015. Nocardia veterana: disseminated infection with urinary tract infection. Braz J Infect Dis 19:216–219. doi: 10.1016/j.bjid.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McElvania Tekippe E, Shuey S, Winkler DW, Butler MA, Burnham CA. 2013. Optimizing identification of clinically relevant Gram-positive organisms by use of the Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry system. J Clin Microbiol 51:1421–1427. doi: 10.1128/JCM.02680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khot PD, Bird BA, Durrant RJ, Fisher MA. 2015. Identification of Nocardia species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53:3366–3369. doi: 10.1128/JCM.00780-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 49:1790–1794. doi: 10.1128/JCM.02135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilen CB, McMullen AR, Burnham CA. 2015. Comparison of sample preparation methods, instrumentation platforms, and contemporary commercial databases for identification of clinically relevant mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53:2308–2315. doi: 10.1128/JCM.00567-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrell JJ, Tuohy MJ, Brown-Elliott BA, Patel JB, Wallace RJ Jr, Hall GS, Procop GW. 2003. Rapid identification of Nocardia species by pyrosequencing, abstr 338 Abstr Annu Meet Infect Dis Soc Am, San Diego, CA. [Google Scholar]
- 14.Tuohy MJ, Farrell JJ, Brown-Elliott BA, Mann L, Wilson RW, Wallace RJ Jr, Hall GS, Procop GW. 2004. Evaluation of pyrosequencing for the identification of nocardia species., abstr 2578 Abstr 104th Gen Meet Am Soc Microbiol, New Orleans, LA American Society for Microbiology, Washington, DC. [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Lau AF, Drake SK, Calhoun LB, Henderson CM, Zelazny AM. 2013. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:828–834. doi: 10.1128/JCM.02852-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McTaggart LR, Doucet J, Witkowska M, Richardson SE. 2015. Antimicrobial susceptibility among clinical Nocardia species identified by multilocus sequence analysis. Antimicrob Agents Chemother 59:269–275. doi: 10.1128/AAC.02770-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake RR, Boggs SR, Drake SK. 2011. Pathogen identification using mass spectrometry in the clinical microbiology laboratory. J Mass Spectrom 46:1. doi: 10.1002/jms.1854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.