CASE
An 11-year-old, previously healthy boy in north Texas was riding his bicycle along a pond when his bicycle tire became stuck in the mud. The patient fell off his bike into the pond and struck his right knee on a hard object, acquiring a 2-cm-long horseshoe-shaped laceration. The patient's mother brought the patient to the emergency department (ED) due to concern for a foreign body. There was no erythema or swelling, and the site was nontender. The wound was irrigated and was closed with 4 sutures. There was no visible foreign body. Antibiotic ointment was applied, and the patient was discharged home.
Six days later, the patient returned to the ED with complaints of 2 days of mild erythema, pain, and swelling around the wound site. Earlier that day, the patient had bumped his knee on the edge of a table, and 10 to 15 ml of purulent material was discharged from the wound.
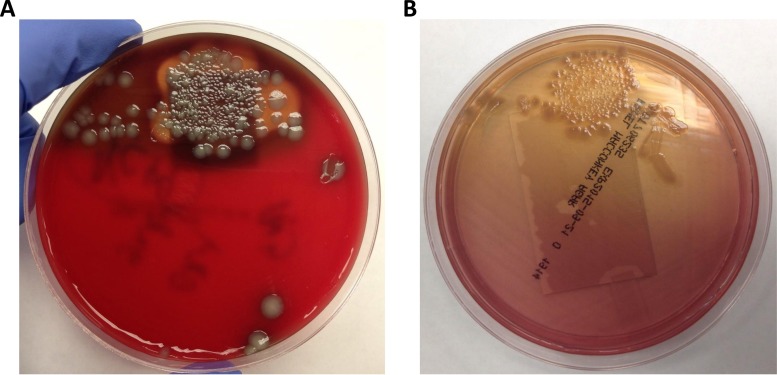
While in the ED, an ultrasound test of the knee was performed, which revealed a 1.1-cm-by-1.1-cm-by-0.3-cm irregular fluid collection incorporating a sinus tract. The ED physician expressed a small amount of “thin, cloudy, yellow-white liquid” from the wound which was sent for culture. The specimen Gram stain showed few white blood cells, no squamous epithelial cells, and no organisms. The patient was prescribed clindamycin and discharged home. The next day, the blood agar plate revealed two colony types, one beta-hemolytic and the other nonhemolytic. Both isolates grew on MacConkey agar but did not ferment lactose (Fig. 1). The different colony morphologies were identified as representing the same species. The isolates were oxidase positive and indole positive. They were also positive for lysine and ornithine carboxylase and positive for arginine dihydrolase.
FIG 1.
Original (A) blood agar and (B) MacConkey agar plates from the patient's wound culture.
The organism was originally identified as P. shigelloides at 99% probability by the use of a Vitek 2 system and a GN card (bioMérieux, Durham, NC). The microbiology technologist realized this was an unusual identification for a wound isolate and set up an additional identification procedure using a MicroScan Negative Combo 44 (NC44) Gram-negative panel. This panel also identified the isolate as P. shigelloides (99.99% probability). The nonhemolytic isolate was later tested by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonics, Inc., Billerica, MA) and was identified as P. shigelloides with a score value of 2.391 using Bruker Biotyper 4.0 software and the RUO 5627 library. As a result of the organism identification, the patient was subsequently prescribed trimethoprim-sulfamethoxazole. The next day, the susceptibility results indicated the isolate was susceptible to all agents tested, which included piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, meropenem, ciprofloxacin, levofloxacin, trimethoprim-sulfamethoxazole, gentamicin, tobramycin, and amikacin. Clindamycin, a lincosamide antibiotic, which the patient was originally prescribed, has no activity against aerobic Gram-negative organisms.
The patient followed up with his primary care physician 4 days later, and the wound showed decreased redness and swelling. The wound had healed by the time the patient was seen 2 weeks later for a sports physical.
DISCUSSION
Plesiomonas shigelloides is the only species member in the Plesiomonas genus, and it is the only oxidase-positive member of the Enterobacteriaceae. Plesiomonas was originally classified with Aeromonas in the Vibrionaceae, but it was found to be more similar to the Enterobacteriaceae. The organism was discovered in 1947 and was originally referred to as strain C27, which was a motile organism that cross-reacted with Shigella sonnei antisera, hence the current species name (1). The designation C27 was later changed to Pseudomonas shigelloides, Pseudomonas michigani, Aeromonas shigelloides, Fergusonia shigelloides, Vibrio shigelloides, and, ultimately, Plesiomonas shigelloides (2).
P. shigelloides is distributed worldwide. It is found in aquatic environments, primarily freshwater (including ponds, rivers, and lakes), but it has also been found in estuarine and marine environments. P. shigelloides has been isolated from a wide range of fish, shellfish, birds, and mammals in addition to humans. It is not a member of the human microbiota. The majority of P. shigelloides infections occur in the summer months in tropical and subtropical regions. Infections in the United States are less common, although the true rate of infections in the United States is unknown because it is not a reportable disease.
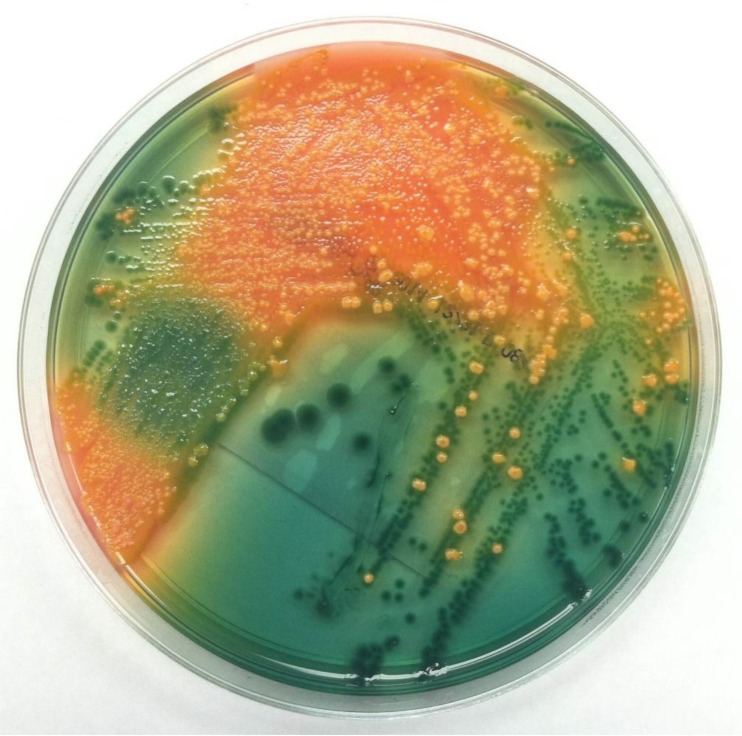
The organism is most commonly recovered in cases of acute gastroenteritis, which is associated with ingestion of contaminated water or shellfish as well as with recent foreign travel (1). The infectious dose is presumed to be greater than 106 CFU, and ingestion does not always cause illness. Gastroenteritis due to P. shigelloides is usually mild and self-limiting, with a duration of 1 to 7 days, although more-severe forms of infection may occur. Routine stool cultures typically do not include selective media for the detection of P. shigelloides. However, the organism can be detected by performing an oxidase test on a swab swept through multiple colonies on blood agar (sweep oxidase). In addition, nonfermenting isolates appear as green colonies on Hektoen enteric agar, with an appearance similar to that of Shigella (Fig. 2). The two organisms can be differentiated by analyses of oxidase, indole, and motility. P. shigelloides is positive for all three, while Shigella is negative. P. shigelloides is easily identified using commercially available identification systems.
FIG 2.
P. shigelloides recovered from a stool culture on Hektoen enteric agar. The green colonies are P. shigelloides, while the orange colonies are normal flora.
P. shigelloides has rarely been isolated in cases of extraintestinal infection, including bacteremia, sepsis, cellulitis, meningitis, ocular infections, septic arthritis, peritonitis, pseudoappendicitis, pancreatic and splenic abscesses, epididymo-orchitis, pyosalpingitis, and pneumonia (1). The most common presentations of extraintestinal infection are sepsis and meningitis, especially in neonates. Risk factors for extraintestinal disease include immunocompromised state, malignancy, blood disorders, hepatobiliary disease, and age (elderly and infants) (1).
Only 2 cases of wound infections, in both of which P. shigelloides was the sole pathogen, have been reported, and both involved freshwater (3, 4). The first patient was a previously healthy 31-year-old male who developed a cutaneous abscess after diving into a freshwater basin near Paris to catch a stray ball. The patient scraped his head, forehead, and chest on rocks at the bottom of the basin, and the wounds subsequently became infected with P. shigelloides (4). The second patient was a previously healthy 13-year-old girl in central Missouri, who developed hyperacute infectious keratitis after being struck in the eye with a rock that came from a creek bed (3). Other bacteria associated with water injuries and subsequent infection include Aeromonas spp., Vibrio spp., Edwardsiella tarda and other Enterobacteriaceae, Shewanella putrefaciens, Pseudomonas aeruginosa, Stenotrophomonas maltophilia, Erysipelothrix rhusiopathiae, Mycobacterium marinum, and others (5). Freshwater wounds are more often associated with Aeromonas hydrophila, while Vibrio spp. are more commonly associated with saltwater wounds.
Isolated in the laboratory, P. shigelloides isolates may be beta-hemolytic or nonhemolytic. Isolates are positive for oxidase, indole, and motility. In addition, most P. shigelloides isolates do not ferment lactose, but rare isolates may ferment lactose. P. shigelloides is unique in that it is positive for lysine and ornithine carboxylase and arginine dihydrolase. The majority of bacteria are positive for only 1 or 2 of these enzymes.
In regard to antimicrobial susceptibility testing breakpoints, Plesiomonas was previously included in the Aeromonas and Plesiomonas breakpoints in the CLSI M45 document. However, in 2015, the Clinical and Laboratory Standards Institute (CLSI) moved Plesiomonas from the Aeromonas and Plesiomonas breakpoints in the M45 document to the Enterobacteriaceae breakpoints in the M100-S25 document (6). P. shigelloides is intrinsically resistant to penicillins due to the presence of a beta-lactamase. Most isolates are susceptible to cephalosporins, carbapenems, fluoroquinolones, and trimethoprim-sulfamethoxazole. Susceptibility to aminoglycosides is variable. Fluoroquinolones and trimethoprim-sulfamethoxazole are first-line oral agents for gastrointestinal illness when antimicrobial therapy is indicated (1). For extraintestinal illness, a variety of antimicrobials, alone and in combination, have been used (1).
SELF-ASSESSMENT QUESTIONS
- Which oxidase-positive, indole-positive organism is more commonly associated with freshwater injuries?
- (a) Stenotrophomonas maltophilia.
- (b) Aeromonas hydrophila.
- (c) Vibrio vulnificus.
- (d) Edwardsiella tarda.
- If a nonhemolytic, non-lactose-fermenting organism isolated from a stool culture agglutinates with Shigella sonnei antisera, which assay can be used to differentiate quickly between Shigella and P. shigelloides?
- (a) Growth on MacConkey agar.
- (b) Growth on Hektoen enteric agar.
- (c) Oxidase.
- (d) Urease.
- P. shigelloides is intrinsically resistant to which class of antibiotics?
- (a) Penicillins.
- (b) Cephalosporins.
- (c) Fluoroquinolones.
- (d) Aminoglycosides.
(For answers to the self-assessment questions and take-home points, see page 1408 in this issue [doi:10.1128/JCM.02652-15].)
REFERENCES
- 1.Stock I. 2004. Plesiomonas shigelloides: an emerging pathogen with unusual properties. Rev Med Microbiol 15:129–139. doi: 10.1097/00013542-200410000-00002. [DOI] [Google Scholar]
- 2.Nolte FS, Poole RM, Murphy GW, Clark C, Panner BJ. 1988. Proctitis and fatal septicemia caused by Plesiomonas shigelloides in a bisexual man. J Clin Microbiol 26:388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klatte JM, Dastjerdi MH, Clark K, Harrison CJ, Grigorian F, Stahl ED. 2012. Hyperacute infectious keratitis with Plesiomonas shigelloides following traumatic lamellar corneal laceration. Pediatr Infect Dis J 31:1200–1201. doi: 10.1097/INF.0b013e318266b61f. [DOI] [PubMed] [Google Scholar]
- 4.Herve V, Bigaillon C, Duhamel P, Petit MP, Soler C. 2007. Cutaneous abscess due to Plesiomonas shigelloides consecutive to a trauma in fresh water. Med Mal Infect 37:840. doi: 10.1016/j.medmal.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Murphey DK, Septimus EJ, Waagner DC. 1992. Catfish-related injury and infection: report of two cases and review of the literature. Clin Infect Dis 14:689–693. doi: 10.1093/clinids/14.3.689. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]