Abstract
Peroxiredoxin 6 (Prdx6) is a unique 1-Cys member of the peroxiredoxin family with both GSH peroxidase and phospholipase A2 (PLA2) activities. It is highly expressed in the lung where it plays an important role in antioxidant defense and lung surfactant metabolism. Glutathionylation of Prdx6 mediated by its hetrodimerization with GSH S-transferase π (πGST) is required for its peroxidatic catalytic cycle. Recombinant human Prdx6 crystallizes as a homodimer and sedimentation equilibrium analysis confirmed that this protein exists as a high affinity dimer in solution. Based on measurement of molecular mass, dimeric Prdx6 that was oxidized to the sulfenic acid formed a sulfenylamide during storage. After examination of the dimer interface in the crystal structure, we postulated that the hydrophobic amino acids L145 and L148 play an important role in homodimerization of Prdx6 as well as in its heterodimerization with πGST. Oxidation of Prdx6 also was required for its heterodimerization. Sedimentation equilibrium analysis and the Duolink proximity ligation assay following mutation of the L145 and L148 residues of Prdx6 to Glu indicated greatly decreased dimerization propensity reflecting the loss of hydrophobic interactions between the protein monomers. Peroxidase activity was markedly reduced by mutation at either of the Leu sites and was essentially abolished by the double mutation, while PLA2 activity was unaffected. Decreased peroxidase activity following mutation of the interfacial leucines presumably is mediated via impaired heterodimerization of Prdx6 with πGST that is required for reduction and re-activation of the oxidized enzyme.
Keywords: Duolink assay, phospholipid hydroperoxide glutathione peroxidase activity, sedimentation equilibrium analysis, hydrophobic interactions, mass spectroscopy, leucine mutation
Introduction
The peroxiredoxins (Prdx) are a widely distributed family of enzymes that use the thiol groups of their cysteines (Cys) to catalyze the reduction of peroxides (1). Thus, unlike the glutathione (GSH) peroxidases, they do not rely on a selenocysteine moiety or other metallic co-factors for their catalytic activity. The Prdx family has been divided into 3 groups based on the mechanism for reducing the oxidized cysteine following interaction with an oxidant. In the typical 2-Cys enzymes (2 conserved Cys moieties per monomer), a disulfide that forms between the “catalytic” Cys of one monomer and the “resolving” Cys of the second monomer is then reduced by thioredoxin (1). The atypical 2-Cys Prdx enzymes are also thioredoxin dependent, but the resolving Cys occupies the same monomer as the catalytic Cys, forming an internal disulfide on its oxidation. On the other hand, the 1-Cys peroxiredoxins have a single conserved Cys on each monomer so that resolution of the oxidized Cys requires interaction with another reductant such as GSH. For peroxiredoxin 6 (Prdx6), the only mammalian 1-Cys member of the peroxiredoxin family, resolution requires formation of a mixed disulfide with GSH (i.e., glutathionylation) as facilitated by GSH S-transferase π (πGST) followed by reduction mediated by GSH (2).
There are several important properties that distinguish Prdx6 from the other mammalian peroxiredoxins. The first, as just mentioned, is the mechanism for activity that depends on a single conserved Cys in the protein monomer, heterodimerization of the oxidized protein with πGST as part of the redox cycle, glutathionylation rather than disulfide formation with another (identical) monomer, and the requirement for GSH rather than thioredoxin to complete the catalytic cycle and to regenerate the active enzyme (2–5). A second important difference from other peroxiredoxins is that oxidized phospholipids can bind to Prdx6 with positioning relative to the active site to allow their reduction, i.e., phospholipid hydroperoxide GSH peroxidase (PHGPx) activity (2, 6, 7). A third difference, also related to its positional binding of phospholipids, is the ability of Prdx6 to hydrolyze the sn-2 fatty acyl bond; that is, Prdx6 expresses phospholipase A2 (PLA2) activity (8, 9). The protein shows a low but significant level of PLA2 activity at acidic pH with non-oxidized phospholipid; there is less PLA2 activity of the protein at cytosolic pH but oxidation of the phospholipid substrate significantly increases its binding to Prdx6 and PLA2 activity at pH 7 (7, 10). PLA2 activity of Prdx6 is increased to an even greater extent by phosphorylation of threonine at residue 177 resulting in approximately equal hydrolase activity at pH 4 and pH 7 (11).
Since phospholipids are a major structural component of all cell membranes, their peroxidation represents a severe threat to cellular integrity. Therefore, the ability of Prdx6 to bind to and reduce phospholipid hydroperoxides is essential to its important role in antioxidant defense (6, 7, 10, 12). Our previous study showed that lungs of Prdx6 null mice are significantly more sensitive to oxidant stress than those from GPx1 null mice, suggesting that Prdx6 may play a more important physiological role than GPx1 in anti-oxidant defense, at least with respect to lungs and lung cells (13). Of note, Prdx6 is expressed in essentially all tissues but at particularly high levels in lung, brain, eye, and testes (1, 2, 14, 15). The difference in anti-oxidant function between GPx1 and Prdx6 is likely due to the fact that although both enzymes can reduce H2O2 and short chain hydroperoxides, Prdx6 but not GPx1 can promote membrane repair by scavenging phospholipid hydroperoxides (13). This scavenging process can occur either by reduction of the oxidized fatty acyl moiety (peroxidase activity) or by hydrolysis to liberate the oxidized fatty acyl moiety (PLA2 activity) followed by reacylation of the lysophospholipid that is generated concurrently. We recently have shown that this latter activity (acyl transferase) also is catalyzed by Prdx6 (16). Our previous studies have shown that both of these pathways are utilized by Prdx6 in the repair of oxidized cell membranes (10, 12).
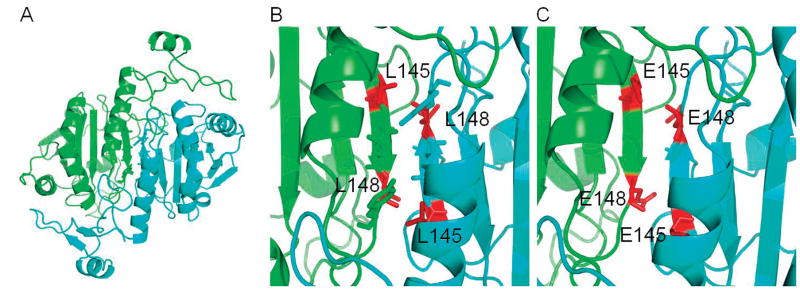
Examination of the crystal structure of oxidized human Prdx6 (with C91 mutated to Ser) indicates that the protein crystallizes as a homodimer with discrete domains in each monomer for the GPx and PLA2 activities (17). The N-terminal domain has an important role in GPx activity while the thioredoxin fold in the C-terminal domain functions in dimerization. The hydrophobic residues (Leu 145, Ile 147, Leu 148 and Tyr 149) between β-strands (βb7 from each monomer) may play an important role in stabilizing the protein dimer as shown in Fig. 1 (17). Our previous analysis has indicated that the interface of the Prdx6 homodimer overlaps the interface for its heterodimerization with πGST (4); thus, these hydrophobic residues in Prdx6 are also likely to contribute to the stability of the Prdx6-πGST heterodimer.
Figure 1. Cartoon representation of the Prdx6 structure.
The images are based on the published crystal structure (17). (A) One monomer is shown in green and the other monomer in blue. Side changes of residues 145–149 at the dimer interface are shown as sticks. (B) A zoomed in image of panel A shows the dimer interface with L145 and L148 highlighted as red sticks. (C) The dimer interface with residues L145 and L148 both mutated to E.
For this study, we focused on the influence of the hydrophobic interface on the monomer- dimer equilibrium for both the Prdx6 homodimer and the Prdx6: πGST heterodimer and determined the consequent effects of perturbing dimerization on Prdx6 enzymatic activities. The hydrophobic interface was disrupted by mutation of the uncharged Leu 145 and 148 residues to negatively charged (hydrophilic) Glu (Fig. 1). Our results indicate that both the dimerization state of Prdx6 and its activity as a peroxidase, but not as a PLA2, are altered by disrupting the hydrophobic dimer interface.
Materials and methods
Reagents
GSH, glutathione reductase, NADPH, 15-lipoxygenase, molecular mass standards, and polybrene (hexadimethrine bromide) were from Sigma-Aldrich (St. Louis, MO). Lipofectamine 2000 was from Invitrogen (Carlsbad, CA). Phospholipids including 1-palmitoyl-2-linoleoyl-sn-glycero-3- phosphocholine (PLPC) were from Avanti Polar Lipids (Birmingham, AL); PLPC hydroperoxide (PLPCOOH) was prepared by treatment of PLPC with 15-lipoxygenase and was purified as described previously (6). Radiolabeled dipalmitoyl phosphatidylcholine (3H-DPPC) was from American Radiochemicals (St Louis, MO). Unless indicated otherwise, oligonucleotide primers were purchased from Eurofins Genomics (Huntsville, AL).
Protein preparation in E. coli
A plasmid expressing human Prdx6 that had been codon optimized for expression in E. coli was purchased from DNA 2.0 (Menlo Park, CA). The codon-optimized sequence of human Prdx6 was inserted into the plasmid pJExpress414 between the NdeI site and XhoI sites. This plasmid codes for ampicillin resistance. Expression of the human Prdx6 gene is driven by an upstream T7 promoter and there is also a T7 transcription terminator located after the 3′ end of the coding region. The optimized Prdx6 coding sequence is:
ATGCCAGGCGGTTTACTTTTAGGTGACGTAGCCCCGAATTTTGAAGCGAATACGACAGTGGGCCGCATCCGCTTTCATGACTTCTTGGGCGATAGCTGGGGCATCCTGTTCTCTCACCCGCGTGACTTCACCCCGGTTTGCACCACGGAACTGGGCCGTGCTGCGAAACTGGCACCGGAGTTTGCAAAACGCAACGTCAAACTGATTGCTCTGAGCATCGATAGCGTCGAGGATCACCTGGCGTGGAGCAAGGACATTAACGCATACAACTGTGAAGAGCCGACTGAGAAGTTGCCGTTCCCGATTATTGACGATCGTAATCGTGAACTGGCAATCCTGCTGGGTATGTTGGATCCGGCGGAGAAAGACGAGAAGGGTATGCCGGTCACCGCGCGTGTCGTTTTTGTGTTTGGTCCTGACAAGAAACTGAAGCTGAGCATCCTGTACCCGGCCACGACGGGTCGTAACTTCGACGAGATTCTGCGTGTGGTTATTTCGCTGCAGCTGACCGCGGAGAAACGCGTTGCCACCCCAGTGGACTGGAAAGATGGCGATTCCGTGATGGTTCTGCCGACCATCCCTGAAGAAGAGGCGAAGAAGCTGTTCCCGAAAGGTGTTTTCACCAAAGAACTGCCGAGCGGTAAGAAGTATCTGCGTTATACCCCGCAACCGTAA.
The leucine codons at positions 145 and 148 of this construct were mutated to glutamic acid by changing them from CTG to GAA using the QuikChange Lightning mutagenesis kit (Agilent Technologies, Santa Clara, CA). The primers used for mutagenesis were designed using the tool on the Agilent website and consisted of complementary pairs for each mutation. The sequence of the member of the primer pair corresponding to the coding strand for each mutant construct was:
5′-CCTGACAAGAAACTGAAGGAAAGCATCCTGTACCCGGCC-3′ for L145E,
5′-ACTGAAGCTGAGCATCGAATACCCGGCCACGACGG-3′ for L148E, and
5′-GGTCCTGACAAGAAACTGAAGGAAAGCATCGAATACCCGGCCACGACGGGTC-3′ for the double mutant, L145E/L148E. The sequence of the member of the primer pair corresponding to the noncoding strand of each mutant construct was:
5′-GGCCGGGTACAGGATGCTTTCCTTCAGTTTCTTGTCAGG-3′ for L145E,
5′-CCGTCGTGGCCGGGTATTCGATGCTCAGCTTCAGT-3′ for L148E, and
5′-GACCCGTCGTGGCCGGGTATTCGATGCTTTCCTTCAGTTTCTTGTCAGGACC-3′ for the double mutant, L145/L148. For all constructs described in this section, the mutated codons are indicated in bold letters.
We also generated human Prdx6 in which the C91 residue was mutated to Ser, corresponding to the protein that was used previously for crystallization (17). This mutation utilized a plasmid that we have described previously for generation of his-tagged human Prdx6 (8). The plasmid was mutated for generation of C91S Prdx6 without the his-tag by insertion of a nonsense codon into the nucleotide sequence before the his-tag. The primers for C91S mutation (purchased from Sigma-Aldrich) were:
5-CAATGCTTACAATAGCGAAGACCCACAG-3′ (forward
and 5′-CTGTGGGCTCTTCGCTATTGTAAGCATTG-3′ (reverse).
For the insertion of a STOP codon between the C-terminal amino acid and the his-tag, the primers were:
5′-CACCCCAGCCTTAAGAGCACCACCAC-3′ (forward) and
5′-GTGGTGGTGCTCTTAAGGCTGGGGTG-3′(reverse).
Human wild type (WT) and mutant Prdx6 proteins (L145E, L148E, and C91S) were expressed as described previously (8, 9). Prdx proteins were purified using ion-exchange chromatography (2, 8, 9) resulting in a homogeneous product as determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Human glutathione S-transferase P1-1 was prepared from a plasmid containing a πGST cDNA clone, a kind gift of Dr. Roberta Colman (University of Delaware), and purified on an S-hexylglutathione-agarose column, essentially as described (18).
Analytical ultracentrifugation
The oligomeric state of human Prdx6 was determined using sedimentation equilibrium in an Optima XLI analytical ultracentrifuge. Immediately before the sedimentation experiment, aggregates were removed from protein samples by gel filtration using two Superdex75 columns (GE Healthcare, Milwaukee, WI) connected in series, equilibrated with 20 mM Tris, 130 mM sodium chloride, 0.1 mM EDTA, 5% glycerol (pH 7.4) buffer solution and maintained at 4°C. To study oxidized protein, WT Prdx6 was incubated with 60 μM H2O2 in 20 mM Tris, 130 mM sodium chloride, 0.1 mM EDTA, pH 7.4 for 1 h at room temperature. After incubation, the sample was immediately gel-filtered into a solution containing 20 mM Tris, 130 mM sodium chloride, 0.1 mM EDTA, and 5% glycerol (pH 7.4) using two Superdex 75 columns connected in series.
For each experiment, three different initial concentrations of protein and two rotor speeds were analyzed. Wild type and mutant Prdx6 were analyzed at both 4°C and 30°C; oxidized wild type Prdx6 was analyzed only at 30°C. Samples (110 μl) were loaded into Epon double sector centerpieces. A water blank scan was acquired immediately prior to sample analysis and was used to correct for window distortion in the fringe displacement data (19). Fringe displacement data were collected every 4 h until equilibrium was obtained as determined from comparison of successive scans using the WinMatch v0.99 program. The blank scan correction was edited using the WinReed v0.999 program. Global data analysis was performed using the WinNonlin v0.99 program. The programs WinMatch, WinReed and WinNonlin are available from the www.rasmb.org website. The apparent molecular mass of proteins was determined and compared to the mass calculated from the known amino acid composition.
Circular Dichroism
Circular dichroism (CD) of wild type and mutant proteins was measured as a check on their secondary structure. Spectra of proteins (2.5 μM) in potassium phosphate buffer (40 mM, pH 7.4) were obtained by the Protein and Proteomic Core Facility of Children’s Hospital of Philadelphia. Recording of spectra and analysis were done with the Jasco J810 Circular Dichroism Spectrapolarimeter (Jasco Analytical Instruments, Easton, MD) that is controlled by Jasco’s Spectra Manager ™ software. The program automatically subtracts the background and has a built- in formula to calculate molecular ellipticity. The contributions of α-helix and β-strand to the structure were compared for the different proteins by use of the K2D3 server (http://cbdm-01.zdv.uni-mainz.de/~Andrade/h2d3) (20).
Mass Spectroscopy (ESI MS)
Purified recombinant proteins were separated using two Superdex 75 (GE Healthcare) columns connected in series and equilibrated in 0.1 M NaC1 with 10 mM Tris (TBS), pH 7.4, that was maintained at 4°C. The peak fraction for each sample was diluted to 0.55 mg/ml in TBS, incubated for 1h at 37°C, and stored at 0°C overnight. For analysis, TCEP (5 mM final concentration) was added to half of each sample; samples were incubated for an additional 30 min at 37°C and then stored on ice until injected. Samples were diluted to 0.1 mg/ml immediately prior to injection of 1 μl onto the LC-MS system. A Dionex C4 Trap column and a self-packed Poros C8 analytical column (20 cm) were interfaced directly with an Orbitrap XL mass spectrometer operating at 100K resolution. Deconvolution of the monoisotopic masses was performed using the Thermo Xtract software (ver. 3.063).
Cell culture
Mice were used as a source of pulmonary microvascular endothelial cells (PMVEC). The protocols for this procedure have been approved by the University of Pennsylvania Animal Care and Use Committee. Wild type C57Bl/6J mice were obtained from Jackson Laboratories (Bar Harbor, ME). The generation of Prdx6 null mice on the C57Bl/6J background has been described previously (13, 21). PMVEC were isolated from the lungs of mice by digestion with collagenase followed by adherence to magnetic beads coated with mAb to platelet endothelial cell adhesion molecule (10, 12, 22, 23). The endothelial phenotype of the PMVEC preparation was confirmed by evaluating cellular uptake of 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate-acetylated low-density lipoprotein (DiI-Ac-LDL) and by immunostaining for PECAM-1 and vascular endothelial cadherin (VE-cadherin). Cells previously isolated from the mice are maintained in our laboratory and no mice were specifically sacrificed to provide cells for this study. Cells at passage 4 or 5 were used for these experiments. Cells were cultured in low glucose DMEM medium (GIBCO Laboratories, Grand Island, NY) supplemented with 10% fetal bovine serum plus penicillin/streptomycin (each 1%). The 293T cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in high glucose DMEM containing 10% fetal bovine serum and 1% each of glutamine and sodium pyruvate. Cells were incubated under 5% CO2 in air at 37 °C.
Lentiviral Prdx6 constructs
HMD lentiviral transfer plasmid, packaging plasmid pCMV-dR8.2, and envelope plasmid pCMV-VSVG were a generous gift from Dr. Mitchell Weiss (The Children’s Hospital of Philadelphia). WT human Prdx6 was recloned from the hPrdx6 PET21b expression plasmid (8) into a HMD lentivirus transfer plasmid expressing EGFP between the Xho1 and EcoR1 restriction sites. The mutant Prdx6 proteins were generated by overlap PCR. The forward primers for making the HMD lentiviral transfer plasmids for mutants L145E, L148E, and the double mutant were:
5′TGATAAGAAGCTGAAGGAGTCTATCCTCTACCCAGCT 3′,
5′AGCTGAAGCTGTCTATCGAGTACCCAGCTACCACTGGCA 3′, and
5′AAGAAGCTGAAGGAGTCTATCGAGTACCCAGCTACCA 3′, respectively. The reverse primers for making these three mutant HMD lentiviral transfer plasmids were:
5′AGCTGGGTAGAGGATAGACTCCTTCAGCTTCTTATCA 3′,
5′TGCCAGTGGTAGCTGGGTACTCGATAGACAGCTTCAGCT 3′, and
5′TGGTAGCTGGTACTCGATAGACTCCTTCAGCTTCTT 3′, respectively. Nucleotides corresponding to the mutations in the above constructs are in bold. The mutation-specific primers were used with the following general primers in overlap PCR:
5′ ACACCTCGAGGCCACCATGCCCGGAGGTCTGCTTCTCGG 3′ (forward; used with specific reverse primers above), and
5′ ACACGAATTCTTAAGGCTGGGGTGTGTAGCGGAGG 3′ (reverse; used with specific forward primers above). Letters in bold italics for the general primers represent restriction enzyme cleavage sites (forward, XhoI; reverse EcoRI) that were used for cloning.
Lentiviral infection
HMD transfer plasmids containing WT or L145E or L148E mutant Prdx6 plasmids were packaged with second generation packaging plasmid pCMV-dR8.2 and envelope plasmid pCMV-VSVG in 293T cells using Lipofectamine 2000. The medium containing virus was collected at 48 h and 72 h after infection. Prdx6 null PMVEC were infected with the virus in medium with added polybrene (~8μg/ml). EGFP expression was maximal at 72 – 96 hours after infection. Empty HMD lentiviral transfer plasmid and HMD lentiviral transfer plasmid containing WT Prdx6 were used as negative and positive controls, respectively.
Measurement of enzymatic activities
Peroxidase and PLA2 activities were measured in PMVEC that had been lysed as described previously (12). Peroxidase activities of recombinant Prdx6 and cell lysates of lentiviral infected Prdx6 null PMVEC were determined by the oxidation of NADPH in the presence of GSH and glutathione reductase (6). The reaction buffer was 50 mM Tris–HC1 (pH 8.0), 0.1 mM EDTA, 0.3 mM NADPH, 0.36 mM GSH, and 0.23 units/ml glutathione reductase. Recombinant Prdx6 was pre-incubated at room temperature for 30 min with equimolar πGST. Fluorescence was continuously recorded at 460 nm (340 nm excitation). After a steady baseline was achieved, the reaction was started by the addition of substrate (7.6 μM H2O2 or 100 μM PLPCOOH plus 0.1% Triton X-100) and the linear change in fluorescence indicating utilization of NADPH for reduction of oxidized GSH was recorded for 5–10 min. The change in fluorescence was corrected for the relatively small baseline of non-enzymatic oxidation of NADPH.
PLA2 activity was measured by the liberation of a free fatty acid (palmitate) from DPPC. Activity of recombinant protein and of PMVEC homogenates was measured at pH 4 in Ca2+-free buffer as described previously (12, 24). Mixed unilamellar liposomes labeled with [3H]-DPPC (9, 10-3H-palmitate in the sn-2 position of DPPC) were used as substrate during 1 h incubation. Disintegrations per minute (dpm) were measured in the nonesterified fatty acid (palmitate) spot obtained by thin layer chromatography and was used to calculate activity. Protein content for both the peroxidase and PLA2 assays was measured by the Bradford assay using Coomassie blue (BioRad, Richmond, CA) with bovine γ-globulin as standard.
Proximity Ligation Assay (Duolink)
This assay was used to evaluate the effect of Leu and Cys mutations of Prdx6 on its homodimerization and on the heterodimerization of Prdx with πGST in intact cells. The experimental protocol utilized Prdx6 null endothelial cells that were infected with lentiviral vector constructs to express WT and mutant Prdx6. Heterodimers of Prdx6 with πGST in intact cells were detected with the in situ Proximity Ligation Assay using the Duolink® In Situ Probemaker kit (Olink Bioscience, Uppsala, Sweden) according to the manufacturer’s protocol as described previously (5). The kit includes 4′, 6-diamidino-2-phenylindole (DAPI) for nuclear staining. Cells were incubated with 200 μM t-BOOH for 2 h prior to assay since demonstration of the interaction of Prdx6 with πGST requires the pretreatment of cells with an oxidant (5). To determine recovery from oxidative stress, cells were washed at the end of 2 h exposure to t-BOOH with oxidant-free medium and incubation was continued for an additional 10 min, 30 min, 60 min, or 6 h. The assay for the Prdx6: πGST heterodimer utilized a monoclonal (mouse) antibody to Prdx6 (EMD Millipore, Billerica, MA) and a polyclonal (rabbit) antibody to πGST (anti-human GST P1-1, Oxford Biomed, Rochester Hills, MI), each conjugated to one of the two arms of the oligonucleotide supplied with the assay kit. We also utilized the DuoLink kit to detect intracellular homodimers of Prdx6, by conjugating both arms of the oligonucleotide supplied with the kit directly to the anti- Prdx6 monoclonal antibody. In this latter case, cells were not pre-treated with oxidant.
For both the homodimer and heterodimer assays, PMVEC cultured on glass coverslips were rinsed with PBS and fixed with 4% paraformaldehyde for 10 min on ice, followed by 10 min incubation with 1% Triton X-100 solution in PBS and further 40 min blocking in a 3% solution of bovine serum albumin in PBS containing 0.2% Triton X-100. Cells were immunolabeled with the oligonucleotide-conjugated primary antibody (1:100 dilution in 0.2% Triton X-100 solution in PBS) for 1 h at room temperature. Cells were observed by confocal microscopy at 600× magnification. Dimerization is indicated by the red fluorescence signal, indicating that the two proteins within cells are separated by <40 nm (25).
Immunoblot analysis
Recombinant Prdx6 or C91S Prdx6 protein was heated at 95 °C for 10 min before SDS/PAGE (10 mM Tris-HCl, 100 mM NaCl, pH 7.4 with or without addition of 5 mM TCEP) using the Nu-Page system (Invitrogen). The samples were transferred to 0.45 um pore size nitrocellose membranes (Invitrogen) for western blot. The blots were probed with the antibody to over-oxidized Prdx6 (Thermo Fisher Scientific, Pittsburgh, Pa, cat.#LF-PA0005) as the primary antibody; the secondary antibody was goat anti-rabbit Ig labeled with IrDye 800 (Rockland, Gilbertsville, PA) for imaging on the green (800 nm) channel. After imaging, the blots were “stripped” using Restore™ Western Blot Buffer (Pierce, Rockford, IL) and reprobed with the mouse monoclonal antibody against human Prdx6 (EMD Millipore) as the primary and goat anti-mouse IgG as the secondary antibody. The blots were analyzed using the two-color Odyssey (LI-COR, Lincoln, NE) technique as described previously (23, 26).
Statistical analysis
Data are expressed as mean ± SE. Statistical significance was assessed with Sigma Stat software (Jandel Scientific, San Jose, CA). Group differences were analyzed by one-way ANOVA or Student’s t-test as appropriate. Differences between mean values were considered statistically significant at p < 0.05.
Results
Mutations at L145 and L148 sites disrupt the dimerization of Prdx6
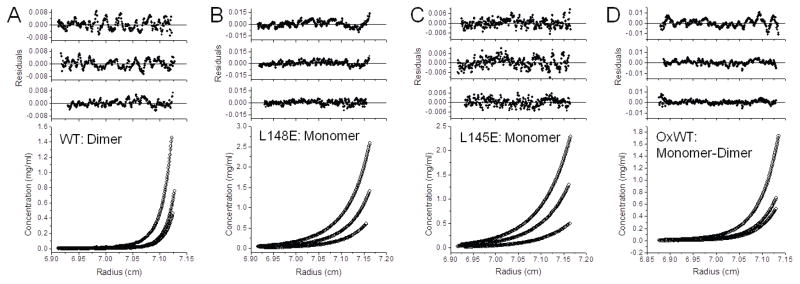
Sedimentation equilibrium by analytical ultracentrifugation was used to evaluate the monomeric/dimeric state of WT Prdx6 and mutant proteins. The observed molecular mass for WT Prdx6 shows that this protein exists as a high affinity homodimer under the conditions studied with no detectable monomer (Fig. 2A; Table 1). Since no monomers were detected, a precise Kd could not be calculated but was estimated as <20 nM. Analyses of the L145E or L148E mutant proteins for the concentration of protein versus radius (Fig. 2B,C) were fitted with various models of self-association using nonlinear regression (27). The results for both mutants are best described by a model consisting predominantly of monomers (Table 1). Results for WT and mutant Prdx6 were similar at 4°C (Table 1) and 30°C (not shown); the results for WT Prdx6 at 30°C have been reported previously (28). These results for sedimentation equilibrium analysis clearly show disruption of Prdx6 dimerization in solution by mutation at either L145 or L148.
Figure 2. Sedimentation equilibrium analysis of Prdx6.
Proteins in 20 mM Tris, 130 mM NaCl, 0.1 mM EDTA, 5% glycerol, pH 7.4 were centrifuged at 26,200 rpm until equilibrium was achieved; EDTA was not present in the buffer for the experiment described in (A). For all conditions, the lower panel shows the raw data (circles) and the line shows the global ideal fit. The upper panels show the residuals of the fitted curve to the data points for each concentration, highest to lowest, top to bottom, respectively. (A) Wild type Prdx6 was analyzed at three different initial loading concentrations (0.4, 0.2 and 0.1 mg/ml) at 4°C. The best fit for the sample data was a dimer model and no significant monomer was detected under these conditions. These results are consistent with a Kd in the low nanomolar range or less. Prdx6 L148E (B) and Prdx6 L145E (C) were analyzed at three different initial loading concentrations (0.8, 0.4 and 0.2 mg/ml) at 4°C. The data for both were best fit by a monomer model. (D) WT Prdx6 was oxidized by H2O2 and then analyzed at three different initial loading concentrations (0.8, 0.4, and 0.2 mg/ml) at 30°C. The data from this sample was best fit by a monomer-dimer model although a single equilibrium constant for each concentration could not be fitted (0.8 mg/ml, Kd ~ 0.8 μM; 0.4 mg/ml, Kd ~ 0.07 μM; and 0.2 mg/ml, no monomer was detected). The inability to fit a single Kd at all concentrations indicates a non-ideal equilibrium.
Table 1.
Molecular mass of Prdx6 proteins determined by sedimentation equilibrium analysis (experimental mass) and mass calculated from the amino acid composition
| Experimental Mass | Calculated Mass* | ||
|---|---|---|---|
| Monomer | Dimer | ||
| Daltons | Daltons | ||
| Wild Type (WT) | 51,060 | 24,904 | 49,808 |
| Oxidized WT† | 25,016/49,232# | 24,920 | 49,840 |
| L145E | 24, 813 | 24,920 | 49,840 |
| L148E | 24,254 | 24,920 | 49,840 |
Mass is calculated for the sulfenic acid form for oxidized WT and for the reduced form for the other proteins using the average atomic mass for each element.
Prdx6 was treated with 60 mM H2O2 for 1 h at room temp.
Both species present.
Sedimentation equilibrium analysis of WT Prdx6 that was oxidized by treatment with H2O2 showed a molecular mass that is compatible with a monomer/dimer equilibrium (Fig. 2D, Table 1), although the distribution between the two species varied moderately with experimental conditions suggesting a heterogeneous population. The H2O2-treated sample is referred to as “over-oxidized protein”. Over-oxidized protein showed lower dimer affinity (Kd ~ 1 μM) as compared to the untreated protein; a single dissociation constant could not be applied to all concentrations analyzed indicating a non-ideal equilibrium.
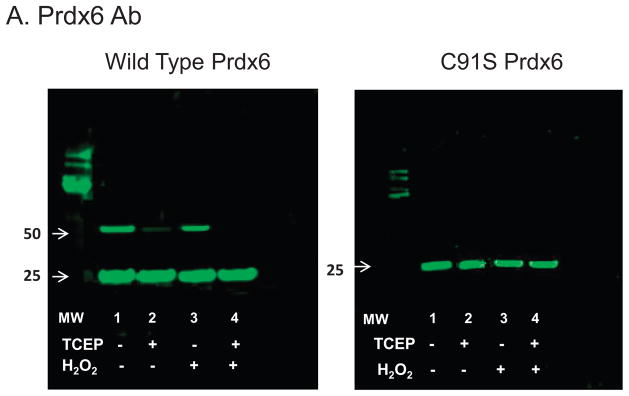
Western blot analysis of Prdx6 using the anti-Prdx6 Ab following SDS-PAGE showed predominantly monomer at ~ 25 kDa under both non-reducing and reducing conditions (Fig 3A left, lanes 1& 2). This indicates that the dimer shown on analytical ultracentrifugation represents predominantly non-covalent rather than covalent interaction; the presence of SDS in the buffer is expected to abolish dimerization associated with hydrophobic interactions. The western blots also showed a minor dimer band at ~50 kDa that was reduced by treatment with TCEP (Fig 3A left, lanes 1 & 2). Over several studies, the minor dimer band appeared to be related to the duration of protein storage and frequently was not seen in freshly isolated wild type protein (not shown). There was no significant change in the Prdx6 dimer band or its reduction by TCEP after treatment of the protein with oxidant (1 mM H2O2 for 10 min) (Fig 3A left, lanes 3&4). The dimer band was not seen with the C91S mutant protein, either before or after oxidant treatment (Fig 3A right). Thus, the minor dimer band appears to represent homodimerization through disulfide formation involving the C91 residues in the two monomers or, less likely, the C91 residue of one monomer and the C47 residue of the other.
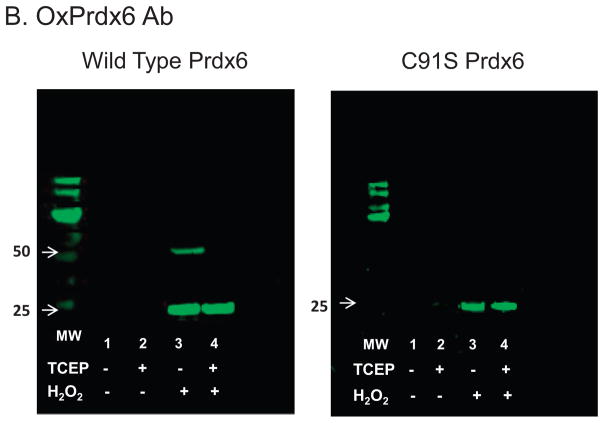
Figure 3. Western blot analysis to evaluate possible covalent interactions and oxidative state of Prdx6.
Western blots of human wild type Prdx6 and C47S Prdx6 (0.25 mg/ml in 100 mM NaCl, 10 mM Tris, pH 7.4) used the antibody to over-oxidized Prdx6 (B); the gel was stripped and re-probed with the antibody to Prdx6 (A). Note that the gels are presented in the reverse order relative to how the experiment was conducted. Proteins with or without prior treatment with 1 mM H2O2 were analyzed with or without added 5 mM TCEP as indicated. The molecular mass standards are shown in the lanes labeled MW; the position of the 25 and 50 kDa (some blots only) markers is indicated by the arrows.
Over-oxidized protein as determined by western blot was not seen with either wild type or C91S Prdx6 prior to oxidant treatment (Fig 3B left & right, lanes 1&2). After H2O2 treatment of WT protein, the antibody to over-oxidized Prdx6 (-SOOH/-SOOOH) demonstrated a major band at ~25 kDa, indicating that overoxidized protein was predominantly monomeric (Fig. 3B left, lane 3). Again, there was a minor band indicating an overoxidized dimer at ~50 kDa that was reduced by the presence of TCEP (Fig. 3B left, lanes 3& 4). On the other hand, dimers were not seen either before or after oxidant treatment of the C91S protein (Fig. 3B, right lanes 3&4). These results indicate that the protein was over-oxidized at position C47 by oxidant treatment allowing it to react with the antibody but that the disulfide bond at C91 was not affected by treatment with the oxidant. This mechanism for dimerization at C91would not occur in Prdx6 proteins that contain a single cysteine, e.g., rat or bovine Prdx6 (3, 29), and was not further investigated as it is unlikely to be physiologic.
Characterization of redox state and secondary structure of Prdx6 proteins
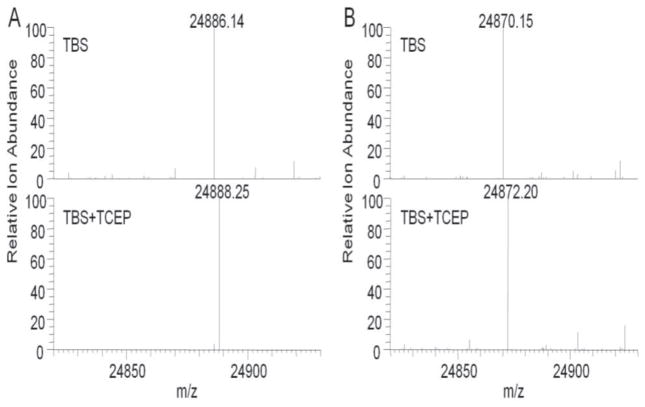
Redox state of the WT proteins used for sedimentation equilibrium analysis was investigated further by determination of molecular mass of the WT and C91S proteins by ESI MS (Fig. 4). The observed monoisotopic mass of WT Prdx6 in the absence of a reducing agent was 2.01 Da less than the expected monoisotopic mass of non-oxidized Prdx6 (Table 2). A similar result (−2.02 Da compared to predicted) was observed for the non-oxidized C91S mutant protein (Fig. 4). However, the expected mass for reduced Prdx6 was observed for both proteins (WT and C91S Prdx6) in the presence of a reducing agent, TCEP (Fig. 4, Table 2). A possible explanation for the observed result in the absence of TCEP is the formation of an internal disulfide between C47 and C91of the same WT polypeptide chain. However, the similar result for the C91S mutant excludes this possibility since an internal disulfide could not occur with the mutant protein. An alternative possibility to account for the observed mass of Prdx6 is the loss of 2 H+ resulting in the formation of a thioether, but that appears to be relatively rare in non-engineered proteins and a mechanism for its possible formation is not clear (30). The most likely possibility for the observed molecular mass of non-reduced Prdx6 is the auto-oxidation of Prdx6 to the sulfenic acid (-Cys S-OH) and its subsequent dehydration with formation of a sulfenylamide bond between the sulfur atom of C47 and the nitrogen of an adjacent amino acid. Formation of a sulfenylamide has been proposed previously for other enzymes including protein tyrosine phosphatase (PTP1B) and Prdx3 (31–33). The oxidation and molecular rearrangement of Prdx6 to form the sulfenylamide occurred presumably during preparation and storage of the recombinant protein and was reversed by the addition of the reducing agent, TCEP. Sulfenylamidation stabilized the oxidized Cys (sulfenic) in Prdx6 and prevented its overoxidation under control conditions (Fig. 3B). Based on the results with analytical ultracentrifugation, this sulfenylamide form of Prdx6 is dimeric. The possible occurrence of sulfenylamide formation in Prdx6 under physiological conditions has not yet been investigated.
Figure 4. Protein mass analysis to evaluate oxidation state of reactive Cys.
ESI-MS was used to determine monoisotopic masses of the intact protein for wild type Prdx6 and C91S Pdx6 in TBS (10 mM Tris, 100mM NaC1, pH7.4) and TBS+TCEP (5 mM). (A) Prdx6 WT protein showing a molecular mass of 24,886.14 Da in the non-reduced protein and 2.01 Da greater in the protein reduced with TCEP; the expected monoisotopic mass is 24,888.15 Da. (B) C91S Prdx6 protein showing a molecular mass of 24,870.15 Da in the non-reduced protein and 2.02 Da greater in the presence of TCEP; the expected monoisotopic mass is 24,872.17 Da.
Table 2.
Molecular mass of wild type and C91S Prdx6 determined by electron spray ionization mass spectroscopy (ESI MS)
| hPrdx6, Da | C91S Prdx6, Da | |||||
|---|---|---|---|---|---|---|
| measured | calculated | Δ mass* | measured | calculated | Δ mass* | |
| non-Reduced | 24,886.14 | 24,888 | −2.01 | 24,870.15 | 24,872.17 | −1.98 |
| Reduced | 24,888.25 | 24,888 | 0.10 | 24,872.20 | 24,872.17 | 0.03 |
Samples were measured in the absence (non-reduced) or presence (reduced) of 1 mM TCEP. Calculated value is the monoisotopic mass based on the most abundant isotope for each element.
Measured mass minus calculated mass.
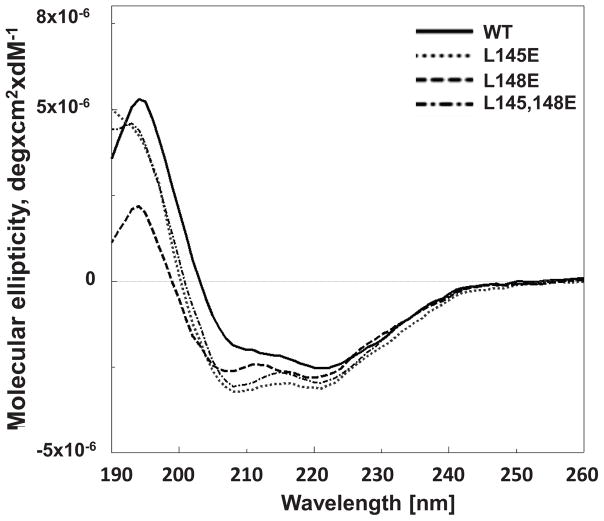
To determine whether changes in conformation of the mutant proteins could be responsible for their failure to homodimerize, their secondary structure was evaluated. CD analysis of the wild type protein resulted in spectra that were similar to our previous report (9). The CD spectra of the single mutant proteins (L145E, L148E) or the double mutant (L145/148E) protein were not significantly different from that of the wild type (Fig. 5) and analysis by the K2D3 server program (20) indicated no appreciable differences among the proteins.
Figure 5. Analysis of peroxiredoxin 6 (Prdx6) secondary structure by circular dichroism (CD).
The CD spectra are for human codon optimized recombinant wild type (WT) and L145E, L148E, and L145/148E mutant Prdx6 proteins.
Mutation of L145E or L148E inhibits Prdx6 dimerization in cells
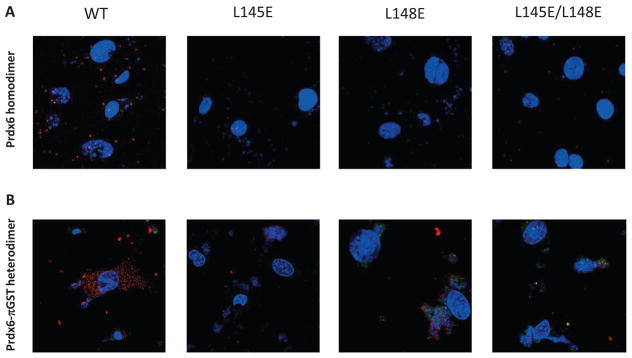
The observations made with proteins in vitro that mutation of L145 or L148 disrupts Prdx6 protein dimerization was evaluated further with intact PMVEC. Prdx6 null cells were infected with lentivirus expressing WT or mutant Prdx6 (L145E, L148E, or L145/148E) and evaluated by the Duolink in situ Proximity Ligation Assay. A positive fluorescence signal was detected in control (Prdx6 null cells that were infected with WT Prdx6) indicating close proximity (<40 nm) of the two labeled monomers, compatible with the demonstrated homodimerization of Prdx6 (Fig. 6A). Cells infected with mutant L145E or L148E Prdx6 did not show a fluorescence signal with the Duolink test. These results indicate that mutation of either the L145 or the L148 amino acid disrupts homodimerization of Prdx6 in intact cells.
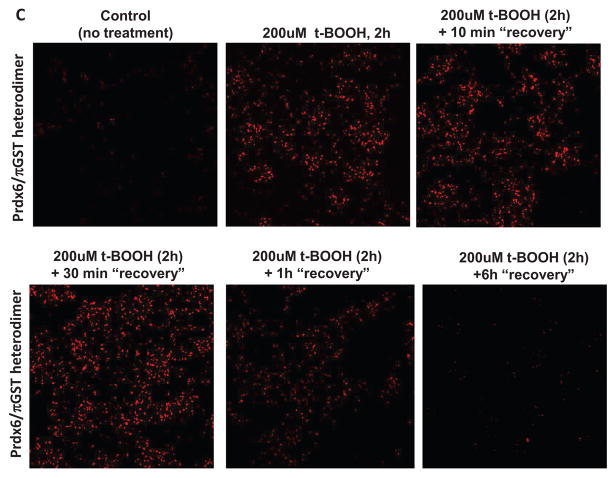
Figure 6. Effect of mutation on the dimerization of Prdx6 determined by the Proximity Ligation Assay (Duolink) test.
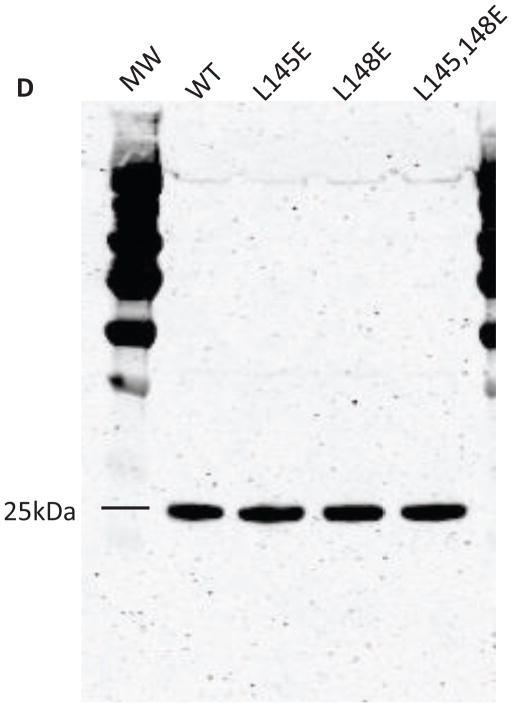
Prdx6 null mouse pulmonary microvascular endothelial cells expressing WT or mutant Prdx6 were fixed and probed by the DuoLink reaction according to the manufacturer’s protocol. Cells in (A) and (B) were counter-stained with 4′, 6–diamidino-2-phenylindole (DAPI) to show nuclei. Images in (A), (B), and (C) are representative of 3 or more fields that were examined for each of n=3 independent incubations. (A) Assay for homodimerization. Both arms of the DuoLink probe were conjugated to the primary antibody against Prdx6. Red fluorescence indicates proximity of two Prdx6 monomers. (B) Assay for heterodimerization. Cells were incubated with 200 μM t-BOOH for 2 h, and then processed using primary antibodies against Prdx6 and πGST for the 2 arms of the DuoLink probe. Red fluorescence indicates proximity of the Prdx6 and πGST monomers. (C) Reversibility of Prdx6: πGST heterodimerization. Experimental conditions as in (B). Panels show control (no t-BOOH), treatment with 200 μM t-BOOH for 2 h, and “recovery” for 10 min, 30 min, 60 min, or 6 h after removal of t-BOOH from the incubation medium. (D) Immunoblot analysis. Western blot of wild type (WT) and mutant (L145E, L148E, L145/148E) Prdx6 indicating that the mutant proteins react with the anti-Prdx6 antibody. MW, molecular mass markers.
Based on analysis of the crystal structures of Prdx6 and πGST (17, 34), we postulated that the molecular site on Prdx6 for heterodimerization of these two proteins would be similar to the Prdx6 site for homodimerization and that association of oxidized Prdx6 with πGST occurs essentially by “swapping” of subunits. This was assessed in intact cells by a Duolink experiment using probes for Prdx6 and πGST. Cells were pre-treated with oxidants since Prdx6 oxidation is necessary in order to see heterodimerization of Prdx6 with πGST (5). Mutation of either L145 or L148 abrogated the interaction of oxidized Prdx6 with πGST indicating the importance of the hydrophobic surface for heterodimerization (Fig. 6B).
The decrease of dimerization following removal of the oxidant was evaluated at intervals by the DuoLink assay in order to assess the reversibility of the process. The association of wild type Prdx6 and πGST essentially was unchanged at 10 and 30 min after removal of the oxidant from the incubation medium but there was a marked decrease in the DuoLink signal at 1 h and full reversal at 6 h (Fig. 6C). The reversibility indicates that intracellular Prdx6, unlike Prdx6 in vitro, is not irreversibly overoxidized by treatment with t-BOOH, presumably related to the presence of intracellular πGST/GSH.
Because lack of reactivity of the Prdx6 antibody to mutated Prdx6 could have influenced the dimeric reactions seen by Duolink assay, western blot analysis was used to evaluate the ability of the Prdx6 Ab to recognize the protein after mutation at the L145 and L148 sites. The mouse monoclonal antibody that was used to probe for Prdx6 reacted similarly to wild type and mutated protein (Fig. 6D) indicating that altered affinity of the antibody for L145E or L148E mutant Prdx6 was not responsible for the observed effects.
Effect of mutations on enzymatic activities of Prdx6
Assay of WT recombinant protein demonstrated the presence of both the peroxidase (at pH 7) and the PLA2 (at pH 4) activities of Prdx6 (Table 3). Peroxidase activity was undetectable in the absence of πGST (not shown), consistent with previous results (2). Mutation of either the L145 or the L148 amino acids of Prdx6 markedly decreased the GPx activity of the recombinant protein with H2O2 substrate while mutation of both sites completely abolished this activity; in contrast, these same mutations had no effect on PLA2 activity (Table 3). PLA2 activity requires binding of the enzyme to the surface of the liposomal substrate (7, 9) while catalysis requires the close association of amino acid residues constituting the catalytic triad, H26, S32, and D140 (11). The normal enzymatic activity provides evidence that the tertiary structure of Prdx6, at least as related to its lipid binding site and its Prdx6 catalytic domain, was not disturbed by the amino acid mutations.
Table 3.
Effect of mutation at L145 or/and L148 on the peroxidase and PLA2 activities of recombinant Prdx6 protein
| Enzymatic activity nmol/min/mg protein | ||
|---|---|---|
| Peroxidase | PLA2 | |
| WT | 3900 ± 300 | 99.7 ± 3.5 |
| L145E | 1100 ± 100* (28%) | 102 ± 4.7 (102%) |
| L148E | 1300 ± 200* (33%) | 98.5 ± 1.2 (99%) |
| L145/148E | 300 ± 100*† (8%) | 99.9 ± 0.9 (100%) |
Peroxidase activity was measured at pH 7 using H2O2 as substrate; PLA2 activity was measured at pH 4 in the absence of calcium with [3H]-dipalmitoylphosphatidylcholine as substrate. πGST (equimolar to Prdx6) was added for peroxidase assay; GSH (5μM) was added for both assays. Values are mean ± SE for n=4. Numbers in parentheses indicate % of corresponding wild type (WT) value.
P<0.05 vs WT;
P<0.05 vs either single mutant protein.
The activities of Prdx6 also were measured in Prdx6 null PMVEC that had been infected with lentiviral vectors expressing WT or mutant (L145E, L148E, or L145/L148E) Prdx6. Examination of cells by microscopy showed relatively similar levels of GFP expression (not shown) indicating similar infection levels as reported previously for other Prdx6 mutant constructs (10). We have previously reported the values (mean ± SE, n=3) for peroxidase and PLA2 activities of WT (non-infected) MPMVEC homogenates as indicated in the legend to Table 4. The peroxidase activity of disrupted lentiviral infected cells was 5% less than the value for WT cells while the PLA2 activity of WT MPMVEC was 15% less (Table 4). These differences of infected cells from WT should not affect the interpretation of the results.
Table 4.
Effect of mutation at L145 and/or L148 generated through infection of Prdx6 null MPMVEC with lentiviral vectors on cellular peroxidase and PLA2 activities
| Enzymatic activity nmol/h/mg protein | |||
|---|---|---|---|
| Peroxidase | PLA2 | ||
| H2O2 | PLPCOOH | ||
| Vector | 7990 ± 220 | 60 + 12 | 0.02 ± 0.01 |
| WT | 14750 ± 60 | 774 ± 42 | 7.85 ± 0.13 |
| L145E | 9530 ± 440 (23%) | 197 + 84 (19%) | 7.17 ± 0.15 (91%) |
| L148E | 9380 ± 260 (21%) | 117 + 18 (8%) | 7.28 ± 0.23 (93%) |
| L145/148E | 7810 ± 147 (0%) | 73 + 6 (2%) | 6.88 ± 0.23 (88%) |
Activities were measured in cell lysates and are expressed per h of incubation. Peroxidase activity was measured with H2O2 or 1- palmitoyl -2- linoleoyl- sn- glycerol -3- phosphocholine hydroperoxide (PLPCOOH) as substrate. PLA2 activity was measured as in Table 3. Values are mean ± SE for n = 3. The number in parenthesis indicates the % of the WT value after subtracting the baseline (vector). Previously reported values for wild type cells under control conditions are 816±95 nmol/h/mg protein for peroxidase activity (PLPCOOH substrate) and 9.28±0.52 nmol/h/mg protein for PLA2 activity (12).
Peroxidase activity of disrupted lentiviral-infected Prdx6 null cells was evaluated with both H2O2 and PLPCOOH as substrate. Cells infected with the lentiviral vector alone showed very low peroxidase activity with PLPCOOH substrate (Table 4) indicating the relative absence of other peroxidases in PMVEC with the ability to reduce this substrate. On the other hand, activity with H2O2 substrate was considerably higher, compatible with the presence of other H2O2-reducing enzymes such as catalase, GSH peroxidases, and other peroxiredoxins. Infection with lentiviral vector to express WT Prdx6 considerably increased the peroxidase activity with either substrate. Infection of PMVEC with either L145E or L148E Prdx6 resulted in markedly lower peroxidase activity as compared to the WT construct and activity was further decreased by infection with the double mutant (Table 4). With the single mutation, peroxidase activity compared to WT was lower by 77–92%, after correcting for the baseline control (vector alone) value; activity with the double mutant with either substrate was reduced to the level seen with the vector alone. Thus, the results with recombinant protein (Table 3) and cells (Table 4) demonstrate that Prdx6 dimerization is essential for peroxidase activity. Since Prdx6 has limited peroxidase activity in the absence of πGST (2–4), the present results provide strong evidence that heterodimerization of the two proteins is required for the Prdx6 peroxidatic catalytic cycle. In contrast to the effects on peroxidase activity, neither the single nor the double mutation had an appreciable effect on PLA2 activity as compared to WT indicating that this activity does not require either homo- or heterodimerization (Tables 3, 4).
Discussion
The present study utilizing sedimentation equilibrium analysis indicates that WT Prdx6 in solution exists as a high affinity homodimer. Based on the results with Leu mutations, hydrophobic interactions in the WT protein appear to be the major mechanism for homodimerization. As an alternative, disulfide formation between monomers is possible but unlikely on theoretical grounds since the protein contains a single conserved Cys residue and the two conserved Cys residues in the dimer (one per monomer) are separated by a considerable distance (17, 28). Although, the vicinal Cys91 possibly could be involved in interchain disulfide formation of human Prdx6, that is unlikely to be a major mechanism based on the western blots. Further, crystallization of human Prdx6 demonstrated that dimerization of the protein occurs in the absence of C91 and without disulfide formation (17). There was evidence of covalent dimerization in some experiments, but the fraction of total Prdx6 that was covalently dimerized was minor and involved a Cys residue (C91) that is present in human Prdx6 but is not conserved in the Prdx6 proteins of other species that have been sequenced (14, 29, 35).
The peroxidase activity of Prdx6 requires heterodimerization with πGST to regenerate the reduced protein as part of the catalytic cycle as demonstrated previously (2–4) and confirmed in the present study. Dissociation of both the oxidized Prdx6 homodimers as well as the πGST homodimers is required for formation of the heterodimer. Although dissociation of the πGST monomer is controversial (36, 37), we have shown that πGST in solution is in monomer-dimer equilibrium (4); heterodimerization of a πGST monomer with Prdx6 would shift the equilibrium leading to further dissociation of the πGST homodimers. In our previous study, the πGST: Prdx6 heterodimer retained GST activity although its activity was reduced with addition of GSH (3). The preponderance of evidence indicates that Prdx6 and πGST interact to form heterodimers resulting in glutathionylation of Prdx6 and subsequent reduction of the protein with restoration of peroxidase activity.
We propose here that oxidation of Prdx6 results in moderate destabilization of the homodimer resulting in an increased rate of Prdx6 monomer formation. Evidence obtained in this study for oxidant-induced dissociation of Prdx6 homodimers is the increase of Prdx6 monomers by sedimentation equilibrium analysis after treatment of Prdx6 in solution with an oxidant. Under these conditions, there was an increase in the calculated Kd following oxidation indicating decreased affinity of the monomers for association as compared to the untreated protein. Evidence for the role of oxidation in the formation of Prdx6 monomers under physiological conditions is provided by our previous Duolink analysis of mouse PMVEC that expressed C47S Prdx6 (5). These cells, unlike those expressing WT Prdx6, failed to show Prdx6: πGST heterodimerization following oxidative stress, compatible with the requirement for oxidation of the C47 amino acid in Prdx6 in order to promote dissociation of the homodimer. Monomers of C47S Prdx6 and πGST generated in solution by treatment with hexanediol were able to heterodimerize, indicating that the C47S mutation per se did not impair the binding of Prdx6 with πGST once monomers had been formed (3). Although it is the sulfenic form that is the initial product formed with oxidant treatment, the in vitro studies of oxidized protein described in this report, as well as in most prior reports, presumably were carried out with overoxidized Prdx6. So, an important question is whether dissociation of Prdx6 homodimers occurs with oxidation to the sulfenic state or only with overoxidation to sulfinic/sulfonic forms.
The present results do not provide direct evidence regarding the oxidation state of Prdx6 associated with monomer formation, but these results along with previous studies provide strong evidence that dissociation of the Prdx6 homodimer does occur with oxidation of the C47 residue in the protein to the sulfenic acid. The reasoning is that the peroxidase activity of inactive Prdx6 is restored in vitro by the addition of πGST:GSH, as shown in our previous studies (2); this re-activation requires a monomer of oxidized Prdx6 to heterodimerize with πGST as a prelude to GSH-mediated reduction of the oxidized Cys47. That reacting monomer must be the sulfenic acid form since the reduction of overoxidized C47 (the sulfenic/sulfonic forms) is not possible, according to our present understanding (38). Compatible with the in vitro results, the reversal of heterodimerization of Prdx6 in PMVEC beginning between 30–60 min following removal of tBOOH from the incubation medium was shown by DuoLink experiments in the present study. Further, we have shown that lipid peroxidation of PMVEC membranes is completely repaired within 1 – 2 h after removal of tBOOH; this repair was dependent in large part on the peroxidase activity of Prdx6 (10, 39). Thus, the reversal of heterodimerization and restoration of peroxidase activity in cells is evidence that the Cys47 of cellular Prdx6 cycled from sulfenic to reduced states during exposure to and recovery from oxidative stress, similar to that observed in vitro.
As mentioned above, the sulfenic form of oxidized Cys (-SOH) is relatively unstable and is a relatively short-lived compound under most circumstances (40). There is now evidence that the sulfenic oxidation state of Prdx6 can be stabilized by several mechanisms. The primary stabilizing influence for Prdx6-SOH under physiological conditions is the πGST-mediated glutathionylation of the protein to form a Prdx6-SSG mixed disulfide (3, 4, 9), analogous to the formation of a protein disulfide that is a step in the reduction of oxidized 2-Cys Prdx proteins (1). As a second mechanism, the Prdx6 crystallization study suggested that Mg2+ could stabilize the sulfenic acid intermediate in vitro by charge interaction and might physically block access of O2 or H2O2 to the -SOH moiety (17). As a third mechanism, the present ESI-MS analysis suggests that dehydration of the sulfenic acid intermediate resulting in formation of a sulfenylamide may stabilize the oxidized protein. Stabilization of the protein can maintain it in the homodimeric state as shown by the crystal structure (17) and the present ESI-MS analyses. In the absence of a stabilizing influence, the -SOH group of C47 is further oxidized to stable but irreversibly oxidized forms (-SO2H/-SO3H).
We propose that dissociation of Prdx6 following its oxidation results from a conformational change in the protein. While oxidized Prdx6 crystallized as a homodimer (17), this most likely reflects the predominance of dimers in the high concentrations of oxidized protein used for crystallization and possibly the effect of stabilizing factors as discussed above. We have recently demonstrated that the structure of Prdx6 in solution, as determined by zero-length chemical cross-linking and homology modeling, shows several regions with substantially different conformation from that of oxidized Prdx6 as determined by crystal structure (28). The protein used for the study of Prdx6 in solution corresponded to the sulfenylamide-stabilized Prdx6 discussed above while the crystallized protein also appears to have been a sulfenic acid but stabilized by another mechanism. The differences between the protein in solution and in the crystalline state would be compatible with structural plasticity for the oxidized (sulfenic) protein.
The results of the present study lead to several important conclusions regarding the characteristics of Prdx6 interactions with πGST. First, Prdx6 in the native reduced state is a high affinity dimer; dimer affinity is weakened after oxidation so that a solution of oxidized Prdx6 will contain a mixed population of monomers and homodimers. Prdx2 and Prdx3 also have been shown to form a mixture of monomers and dimers on exposure to H2O2 (33). Second, oxidized monomers of Prdx6 heterodimerize with GSH-loaded πGST (2–5); this essentially results in a subunit exchange that is facilitated by the decreased homodimer affinity of oxidized Prdx6 and by the presence of GSH (3). Third, both homodimers of Prdx6 and heterodimers of Prdx6 with πGST result from hydrophobic interactions at the dimer interface as demonstrated by mutation of L145/148 in Prdx6. Our previous studies have indicated that Prdx6 and πGST also can heterodimerize via disulfide bond formation although this is a relatively slow reaction of unknown but probably limited physiological relevance (3). Finally, heterodimerization of Prdx6 and πGST through hydrophobic interactions is required for the peroxidase but not the PLA2 activity of Prdx6; this dichotomy reflects the absolute requirement for reduction of oxidized C47 in Prdx6 prior to further peroxidase activity, while a reduced vs oxidized C47 has relatively minor effects on PLA2 activity.
These observations lead to the following proposal for the catalytic mechanism of hydroperoxide reduction by Prdx6 (reactions 1–5); this scheme represents a slight modification of the reaction sequence that we have published previously (2–4). In the resting (non-oxidized state), both Prdx6 and πGST are predominantly homodimeric and do not interact (5). However, with oxidation of Prdx6 (Cys 47→sulfenic acid, reaction 1), its decreased homodimer affinity favors its dissociation and subsequent heterodimerization of oxidized Prdx6 monomer with a GSH bound πGST monomer (reaction 2) (3, 4). Caveats related to the predominantly circumstantial evidence for the role of the Prdx6 sulfenic acid in this reaction are discussed above. Subsequent steps include glutathionylation of Prdx6 that is catalyzed by πGST (reaction 3) followed by reduction of the oxidized enzyme with GSH (reaction 4) resulting in dissociation of the heterodimer and reformation of the Prdx6 and πGST homodimers (reaction 5). The proposed reactions are:
(Prdx6-SH)2 + 2 H2O2 → 2 Prdx6 – SOH + 2 H2O, (oxidation)
Prdx6 – SOH +π GST(GSH)→ Prdx6 – SOH: πGST(GSH), (heterodimerization)
Prdx6 – SOH: πGST(GSH) → Prdx6 – S-SG: πGST + H2O, (glutathionylation)
Prdx6 – S-SG:πGST + GSH → Prdx6-SH + πGST + GSSG, (reduction)
2 Prdx6 – SH → (Prdx6 -SH)2, (homodimerization).
Although the reduction shown in reaction 4 may be mediated by πGST, that has not been demonstrated conclusively. The πGST liberated by reaction 4 rapidly binds free GSH and the liberated GSSG is reduced to GSH by reaction with NADPH mediated by the enzyme glutathione reductase.
In summary, the present results obtained from studies with recombinant proteins in vitro and with intact cells indicate that amino acids L145 and L148 by contributing to the hydrophobic interface are crucial to dimerization of Prdx6 with another Prdx6 (homodimer) or with πGST (heterodimer). Disruption of the Prdx6 hydrophobic surface containing these amino acids has no significant effect on its PLA2 activity (as expected since this activity is relatively independent of the Cys oxidation state) but dramatically decreases its peroxidase activity. This latter effect could result from impaired heterodimerization and glutathionylation of Prdx6 that interferes with reduction of the oxidized cysteine, resulting in the inability to restore the active state of the enzyme.
Highlights.
Peroxiredoxin 6 (Prdx6) exists as a homodimer in aqueous solution.
Mutation of L145 or L148 at the hydrophobic interface prevents homodimerization.
These mutations also prevent heterodimerization of Prdx6 with GSH S-transferase π (πGST).
Heterodimerization is required to restore peroxidase activity of the oxidized protein.
The phospholipase A2 is fully active in the Prdx6 monomer.
Acknowledgments
We thank Dr. Roberta Colman (U Del) for gift of the GSTP1-1 expression plasmid, Dr. Mitchell Weiss for the lentiviral plasmids, Drs. Steven Seeholzer and Hua Ding of the Protein and Proteomics Core of the Children’s Hospital of Philadelphia for the circular dichroism studies, the Proteomics Core of the Wistar Institute for mass spectroscopy analyses, and Dawn Williams for typing the manuscript. This research was supported by research grants R01-HL102016 (to ABF) and P30-CA 10815 (core grant to the Wistar Institute) from the National Institutes of Health. Presented in part at the Experimental Biology meetings in 2014 (San Diego, Ca) and 2015 (Boston, Ma).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- 2.Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc Natl Acad Sci U S A. 2004;101:3780–5. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ralat LA, Manevich Y, Fisher AB, Colman RF. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry. 2006;45:360–72. doi: 10.1021/bi0520737. [DOI] [PubMed] [Google Scholar]
- 4.Ralat LA, Misquitta SA, Manevich Y, Fisher AB, Colman RF. Characterization of the complex of glutathione S-transferase pi and 1-cysteine peroxiredoxin. Arch Biochem Biophys. 2008;474:109–18. doi: 10.1016/j.abb.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S, Lien YC, Shuvaeva T, DeBolt K, Feinstein SI, Fisher AB. Functional interaction of glutathione S-transferase pi and peroxiredoxin 6 in intact cells. Int J Biochem Cell Biol. 2013;45:401–7. doi: 10.1016/j.biocel.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher AB, Dodia C, Manevich Y, Chen JW, Feinstein SI. Phospholipid hydroperoxides are substrates for non-selenium glutathione peroxidase. J Biol Chem. 1999;274:21326–34. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- 7.Manevich Y, Shuvaeva T, Dodia C, Kazi A, Feinstein SI, Fisher AB. Binding of peroxiredoxin 6 to substrate determines differential phospholipid hydroperoxide peroxidase and phospholipase A(2) activities. Arch Biochem Biophys. 2009;485:139–49. doi: 10.1016/j.abb.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J Biol Chem. 2000;275:28421–7. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 9.Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. Structure and phospholipase function of peroxiredoxin 6: Identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res. 2007;48:2306–18. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Benipal B, Zhou S, Dodia C, Chatterjee S, Tao JQ, Sorokina EM, Raabe T, Feinstein SI, Fisher AB. Critical role of peroxiredoxin 6 in the repair of peroxidized cell membranes following oxidative stress. Free Radic Biol Med. 2015;87:356–65. doi: 10.1016/j.freeradbiomed.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Feinstein SI, Manevich Y, Chowdhury I, Pak JH, Kazi A, Dodia C, Speicher DW, Fisher AB. Mitogen-activated protein kinase-mediated phosphorylation of peroxiredoxin 6 regulates its phospholipase A(2) activity. Biochem J. 2009;419:669–79. doi: 10.1042/BJ20082061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lien YC, Feinstein SI, Dodia C, Fisher AB. The roles of peroxidase and phospholipase A2 activities of peroxiredoxin 6 in protecting pulmonary microvascular endothelial cells against peroxidative stress. Antioxid Redox Signal. 2012;16:440–51. doi: 10.1089/ars.2011.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu G, Feinstein SI, Wang Y, Dodia C, Fisher D, Yu K, Ho YS, Fisher AB. Comparison of glutathione peroxidase 1 and peroxiredoxin 6 in protection against oxidative stress in the mouse lung. Free Radic Biol Med. 2010;49:1172–81. doi: 10.1016/j.freeradbiomed.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TS, Dodia C, Chen X, Hennigan BB, Jain M, Feinstein SI, Fisher AB. Cloning and expression of rat lung acidic Ca(2+)-independent PLA2 and its organ distribution. Am J Physiol. 1998;274:L750–61. doi: 10.1152/ajplung.1998.274.5.L750. [DOI] [PubMed] [Google Scholar]
- 15.Singh AK, Shichi H. A novel glutathione peroxidase in bovine eye. Sequence analysis, mRNA level, and translation. J Biol Chem. 1998;273:26171–8. doi: 10.1074/jbc.273.40.26171. [DOI] [PubMed] [Google Scholar]
- 16.Fisher AB, Dodia C, Sorokina EM, Li H, Zhou S, Raabe T, Feinstein SI. A novel lysoPhosphatidylcholine acyl transferase activity is expressed by peroxiredoxin 6. J Lipid Res. 2016 doi: 10.1194/jlr.M064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol. 1998;5:400–6. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 18.Ralat LA, Colman RF. Glutathione S-transferase Pi has at least three distinguishable xenobiotic substrate sites close to its glutathione-binding site. J Biol Chem. 2004;279:50204–13. doi: 10.1074/jbc.M407445200. [DOI] [PubMed] [Google Scholar]
- 19.Yphantis DA. Equilibrium ultracentrifugaton of dilute solutions. Biochemistry. 1964;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]
- 20.Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2012;80:374–81. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 21.Mo Y, Feinstein SI, Manevich Y, Zhang Q, Lu L, Ho YS, Fisher AB. 1-Cys peroxiredoxin knock-out mice express mRNA but not protein for a highly related intronless gene. FEBS Lett. 2003;555:192–8. doi: 10.1016/s0014-5793(03)01199-2. [DOI] [PubMed] [Google Scholar]
- 22.Milovanova T, Chatterjee S, Hawkins BJ, Hong N, Sorokina EM, Debolt K, Moore JS, Madesh M, Fisher AB. Caveolae are an essential component of the pathway for endothelial cell signaling associated with abrupt reduction of shear stress. Biochim Biophys Acta. 2008;1783:1866–75. doi: 10.1016/j.bbamcr.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milovanova T, Chatterjee S, Manevich Y, Kotelnikova I, Debolt K, Madesh M, Moore JS, Fisher AB. Lung endothelial cell proliferation with decreased shear stress is mediated by reactive oxygen species. Am J Physiol Cell Physiol. 2006;290:C66–76. doi: 10.1152/ajpcell.00094.2005. [DOI] [PubMed] [Google Scholar]
- 24.Wu YZ, Manevich Y, Baldwin JL, Dodia C, Yu K, Feinstein SI, Fisher AB. Interaction of surfactant protein A with peroxiredoxin 6 regulates phospholipase A2 activity. J Biol Chem. 2006;281:7515–25. doi: 10.1074/jbc.M504525200. [DOI] [PubMed] [Google Scholar]
- 25.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nature Biotechnology. 2002;20:473–7. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Feinstein SI, Fisher AB. Peroxiredoxin 6 as an antioxidant enzyme: protection of lung alveolar epithelial type II cells from H2O2-induced oxidative stress. Journal of cellular biochemistry. 2008;104:1274–85. doi: 10.1002/jcb.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–88. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivera-Santiago RF, Harper SL, Zhou S, Sriswasdi S, Feinstein SI, Fisher AB, Speicher DW. Solution structure of the reduced form of human peroxiredoxin-6 elucidated using zero-length chemical cross-linking and homology modelling. Biochem J. 2015;468:87–98. doi: 10.1042/BJ20141463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolomeo AM, Carraro A, Bakiu R, Toppo S, Place SP, Ferro D, Santovito G. Peroxiredoxin 6 from the Antarctic emerald rockcod: molecular characterizationof its response to warming. J Comp Physiol B. 2015 doi: 10.1007/s00360-015-0935-3. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30.Lindgren J, Eriksson Karlstrom A. Intramolecular thioether crosslinking of therapeutic proteins to increase proteolytic stability. Chembiochem: a European journal of chemical biology. 2014;15:2132–8. doi: 10.1002/cbic.201400002. [DOI] [PubMed] [Google Scholar]
- 31.Sivaramakrishnan S, Cummings AH, Gates KS. Protection of a single-cysteine redox switch from oxidative destruction: On the functional role of sulfenyl amide formation in the redox-regulated enzyme PTP1B. Bioorg Med Chem Lett. 2010;20:444–7. doi: 10.1016/j.bmcl.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–73. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 33.Haynes AC, Qian J, Reisz JA, Furdui CM, Lowther WT. Molecular basis for the resistance of human mitochondrial 2-Cys peroxiredoxin 3 to hyperoxidation. J Biol Chem. 2013;288:29714–23. doi: 10.1074/jbc.M113.473470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinemer P, Dirr HW, Ladenstein R, Schaffer J, Gallay O, Huber R. The three-dimensional structure of class pi glutathione S-transferase in complex with glutathione sulfonate at 2.3 A resolution. Embo J. 1991;10:1997–2005. doi: 10.1002/j.1460-2075.1991.tb07729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akiba S, Dodia C, Chen X, Fisher AB. Characterization of acidic Ca(2+)-independent phospholipase A2 of bovine lung. Comp Biochem Physiol B Biochem Mol Biol. 1998;120:393–404. doi: 10.1016/s0305-0491(98)10046-9. [DOI] [PubMed] [Google Scholar]
- 36.Gildenhuys S, Wallace LA, Burke JP, Balchin D, Sayed Y, Dirr HW. Class Pi glutathione transferase unfolds via a dimeric and not monomeric intermediate: functional implications for an unstable monomer. Biochemistry. 2010;49:5074–81. doi: 10.1021/bi100552d. [DOI] [PubMed] [Google Scholar]
- 37.Aceto A, Caccuri AM, Sacchetta P, Bucciarelli T, Dragani B, Rosato N, Federici G, Di Ilio C. Dissociation and unfolding of Pi-class glutathione transferase. Evidence for a monomeric inactive intermediate. Biochem J. 1992;285(Pt 1):241–5. doi: 10.1042/bj2850241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo HA, Jeong W, Chang TS, Park KJ, Park SJ, Yang JS, Rhee SG. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. Journal of Biological Chemistry. 2005;280:3125–8. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 39.Krishnaiah SY, Dodia C, Sorokina EM, Feinstein SI, Fisher AB. Binding sites for interaction of peroxiredoxin 6 with surfactant protein A. Biochim Biophys Acta - Proteins and Proteomics. 2015 doi: 10.1016/j.bbapap.2015.12.009. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta V, Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta. 2013;1840:847–75. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]