Abstract
FIKK kinases are a novel family of kinases unique to the Apicomplexa. While most apicomplexans encode a single FIKK kinase, Plasmodium falciparum expresses 21 and piroplasms do not encode a FIKK kinase. FIKK kinases share a conserved C-terminal catalytic domain, but the N-terminal region is highly variable and contains no known functional domains. To date, FIKK kinases have been primarily studied in P. falciparum and Plasmodium berghei. Those that have been studied are exported from the parasite and associate with diverse locations in the infected erythrocyte cytosol or membrane. Deletion of individual P. falciparum FIKK kinases indicates that they may play a role in modification of the infected erythrocyte. The current study characterizes the single FIKK gene in Toxoplasma gondii to evaluate the importance of the FIKK kinase in an apicomplexan that has a single FIKK kinase. The TgFIKK gene encoded a protein of approximately 280 kDa. Endogenous tagging of the FIKK protein with Yellow Fluorescent Protein (YFP) showed that the FIKK protein exclusively localized to the posterior end of tachyzoites. A YFP-tagged FIKK and a Ty-tagged FIKK both co-localized with T. gondii membrane occupation and recognition nexus protein (TgMORN1) to the basal complex and were localized apical to inner membrane complex protein-5 (IMC5) and Centrin2. Deletion of TgFIKK, surprisingly, had no detectable effect on the parasite’s lytic cycle in vitro in human fibroblast cells or in acute virulence in vivo. Thus, our results clearly show that while the FIKK kinase is expressed in tachyzoites, it is not essential for the lytic cycle of T. gondii.
Keywords: FIKK kinase, Toxoplasma, Intracellular pathogen, Parasite, Serine threonine kinases, Basal complex
Graphical Abstract
1. Introduction
Comparative genomic analysis of apicomplexan parasites previously revealed a family of putative kinases collectively designated as FIKK kinases due to the presence of a conserved phenylalanine (F) – isoleucine (I) – lysine (K) – lysine (K) amino acid motif in their kinase subdomain II (Ward et al., 2004). FIKK kinases have C-terminal catalytic kinase domains that are distinct from other serine threonine kinases and a highly variable N-terminal domain. The FIKK kinases, first identified and given their nomenclature by Ward et al. (2004) are highly divergent from any well characterized kinase groups present in higher eukaryotes and do not cluster with any other known kinases (Ward et al., 2004). FIKK kinases are unique to apicomplexans but their numbers differ dramatically between species of the Apicomplexa. The Plasmodium falciparum genome encodes 21 related putative FIKK kinases, the majority of which are predicted to have catalytic activity and to be secreted into the infected erythrocyte (Nunes et al., 2007). In contrast, most other Plasmodium spp. as well as other apicomplexans including Cryptosporidium, Toxoplasma and Neospora are predicted to encode only a single putative FIKK kinase while the piroplasms lack a FIKK kinase (Ward et al., 2004; Talevich et al., 2011). Notably, the piroplasms replicate in the cytoplasm of host cells, unlike other apicomplexans that replicate within a parasitophorous vacuole (PV). Thus, it has been hypothesized that FIKK kinases may play a role in the interaction between the parasitophorous vacuole membrane (PVM) and parasite or PVM and host cell (Talevich et al., 2011).
To date, FIKK kinases have been primarily studied in P. falciparum (Pf) and Plasmodium berghei. Initial studies confirmed kinase activity and indicated a role for FIKK kinases in remodeling of infected erythrocytes (Nunes et al., 2007, 2010; Brandt and Bailey, 2013; Kats et al., 2014). PfFIKK4.1, PfFIKK9.3, PfFIKK9.6 and PfFIKK12 co-localize with Maurer’s clefts proteins, a specialized membrane network created by the parasite to export proteins to the surface of the infected erythrocyte (Nunes et al., 2007). In contrast, PfFIKK9.2 is retained within the parasite, consistent with its lack of a signal peptide (Nunes et al., 2007). Deletion of PfFIKK7.1 and PfFIKK12, which both localize to the inner face of the erythrocyte membrane, did not alter parasite viability or adhesion of infected erythrocytes to diverse substrates, but did result in alterations in the rigidity of infected erythrocytes (Nunes et al., 2010). PfFIKK4.2 has been shown to function as an active kinase and localizes to a distinct compartment in the erythrocyte designated k-dots. Deletion of PfFIKK4.2 affected erythrocyte rigidity but also significantly impaired knob structures on infected erythrocytes, decreasing adhesion (Kats et al., 2014). A genome-wide kinome reverse genetics study in P. berghei that encodes a single FIKK kinase, failed to delete the FIKK kinase, suggesting it may be essential (Tewari et al., 2010).
The fact that FIKK kinases are unique to the Apicomplexa has led to their exploration as potential drug or vaccine targets (Miranda-Saavedra et al., 2012). Thus, it is important to determine whether they play a critical role in the biology and pathogenesis of different apicomplexan pathogens. In the current study, we chose to delete the putative FIKK kinase gene in Toxoplasma gondii, an apicomplexan with a single FIKK kinase and the causative agent of human toxoplasmosis. We show that the TgFIKK protein exclusively localizes to the posterior end of T. gondii and co-localizes with membrane occupation and recognition nexus protein (MORN1) inside the basal complex. Surprisingly, deletion of the gene did not significantly alter the parasite’s lytic cycle in vitro in human foreskin fibroblast cells (HFF) nor impact virulence during acute infection in mice.
2. Materials and methods
2.1. Parasites and cell culture
The RHΔHPT strain of T. gondii with a deletion of its hypoxanthine-xanthine-guanosine phosphoribosyl transferase (HPT) was used for all of the studies, except where mentioned otherwise. The Prugniaud strain of T. gondii with a deletion of the HPT gene was a kind gift from Dr. Laura Knoll (University of Wisconsin School of Medicine, USA). Endogenous tagging of proteins and targeted gene deletion were done in the RHΔKu80ΔHPT strain (a kind gift from Dr. Vern Carruthers, University of Michigan, Ann Arbor, MI, USA). The deletion of the Ku80 gene involved in DNA repair via the non-homologous end joining pathway results in greater levels of homologous recombination, allowing for incorporation of reporter proteins into the 3′ end of the endogenous genes (Huynh and Carruthers, 2009; Fox et al., 2011). Parasite strains were maintained in monolayers of human foreskin fibroblast (HFF) cells (American Type Culture Collection, Manassas, VA, USA) in DMEM high glucose medium supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml of penicillin and 100 mg/ml of streptomycin (D10 medium). Cells were cultured in a humidified 5% CO2 incubator at 37°C.
2.2. Toxoplasma gondii FIKK mRNA and protein analysis
Total RNA was isolated from RHΔHPT tachyzoites harvested from HFF cells. RNA was isolated using the RNeasy Plus Mini Kit (QIAGEN, Valencia, CA, USA) as per the manufacturer’s instructions. cDNA was synthesized using SuperScript III First-Strand Synthesis Kit (Life Technologies, Grand Island, NY, USA). The primers used for amplifying the FIKK cDNA were designed based on gene prediction and expression data available at the time at ToxoDB.org (www.toxodb.org, version 4.3) for the T. gondii FIKK gene (TGME49_289050) and are listed in Supplementary Table S1. The 5′ and 3′ ends of the cDNA were determined using a First Choice RLM-RACE Kit (Life Technologies) and Accuprime Pfx DNA polymerase (Life Technologies) according to manufacturers’ instructions. The primer sets for 5′ rapid amplification of cDNA ends (RACE) experiments were: outer – 5′ - GTC GTG TGT CAG CTG TCT TT – 3′ and inner – 5′ - GGA CAG TGT GTG GAT GTA TGG – 3′. The primers used for the 3′ RACE were: 5′-CGG ACT AAA TCG GTT GGC CAG AAT -3′ and 5′-ACA TCC CGA TAA CCC TGA AAG CGA -3′. All PCR products were cloned into the pCR2.1-TOPO cloning vector (Life Technologies) and sequenced at Genewiz (South Plainfield, NJ, USA) using the M13 primer sites present in the pCR II-TOPO plasmid. DNA sequence alignments and analysis was done using Lasergene 10 core suite (DNASTAR, Inc. Madison, WI, USA). Identified sequences for isoform-1 and isoform-2 are available through GenBank™ under the accession numbers KP901266 and KP901267, respectively.
Conceptual translation of the identified cDNA was done using the ‘Translate’ tool at the Expert Protein Analysis System (ExPASy) proteomics server (http://www.expasy.org) to identify the encoded protein(s). Mass spectrometry (MS) peptide data available for the gene was used to confirm the identified protein (Kissinger et al., 2003; Gajria et al., 2008). Protein motif searches were performed using the CD-Search which is the NCBI interface for searching the Conserved Domain Database (CDD). Protein alignments were done using Clustal Omega (McWilliam et al., 2013) with manual alignment and visualized using BOXSHADE 3.3.1 (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::boxshade). Sequences for Plasmodium FIKK kinases were obtained from PlasmoDB.org version 11.1 (Aurrecoechea et al., 2009).
The probe and primers used for quantification of total TgFIKK transcript (isoform-I and 2 by real time quantitative reverse transcriptase PCR (qRT-PCR) were Probe -/56-FAM/AAA CAA TTC/ZEN/AGC GGC ATC TAC GCC/3IABkFQ/; Forward - CAA TCA ACC GCG CAA CTT T; Reverse - GAC ACC GAG CAT ATA GAC ATC AG. A probe specific for only the putative 3′ catalytic domain in isoform-1 was used for qRT-PCR to detect only isoform-1: Probe -/56-FAM/TCT TCG ATA/ZEN/GTG CAG ATC CGG AGG T/3IABkFQ/; Forward - CAA GTG TTC GGA CAA GGA GAT; Reverse - GAA TGG AGG GAG ACG AGA ATT T. Parasite actin was used as a reference control.
2.3. Endogenous tagging of the TgFIKK gene
The endogenous FIKK gene (TGGT1_289050) in RHΔKu80ΔHPT was tagged with the Yellow Fluorescent Protein (YFP) gene on the 3′ end to enable expression and trafficking studies using IFA microscopy. A 1.5 kb region of the 3′ end of the TgFIKK gene, until right before the stop codon, was amplified using primers 5′-TAC TTC CAA TCC AAT TTA GCG GCG TCC AAA CGA AGC CTT CAA TA-3′ and 5′-TCC TCC ACT TCC AAT TTT AGC GGA AGT CAG CGC AGC CTC TTG TCC-3′, and inserted into Pac1 digested vector pyfp-lic-hpt (a kind gift from Dr. Vern Carruthers) using an In-Fusion PCR cloning system (Clontech, Mountain View, CA, USA) in frame with the YFP gene. The resulting construct was linearized with BsiWI and 25 μg of this construct was used for electroporation. Parasites were cultured in HFF cells with D10 media supplemented with 50 mg/ml of mycophenolic acid and 50 mg/ml of xanthine to select for the HPT gene in the vector pyfp-lic-hpt in transformed parasites. The population was cloned using limiting dilution in 96 well tissue culture plates containing HFF cells to obtain single clones. PCR was performed using primers 5′-ACT GCC TTT AGC GGT AAA CTT GCG-3′ and 5′-ACC ATG AAT TCC CGT CCT CCA CTT-3′ to identify clones where the YFP gene inserted before the stop codon of the native TgFIKK gene by a homologous single cross-over event with the targeting construct. All restriction enzymes used in the study were obtained from New England Biolabs (Ipswich, MA, USA).
For N-terminal Ty-tagging of endogenous FIKK on the RHΔKu80ΔHPT background, 1.9 kb of the TgFIKK 5′ end was amplified, starting at the second exon and cloned via BglII and NotI restriction sites, in the tetO7Sag4-DHFR plasmid (Morlon-Guyot et al., 2014) using primers 5′-CAG AGA TCT CCT GCG AGT TGG TCT CCG CAT TTG C-3′ and 5′-CAG GCG GCC GCA TGG CGG TGA GCG ATC CAC ACC-3′. The plasmid was linearized with MfeI and 50 μg were transfected in RHΔKu80ΔHPT parasites using electroporation. Transgene parasites were selected for with 1 μM pyrimethamine, cloned by limiting dilution, and expression of the Ty-fusion protein verified by IFA using anti-Ty antibody (Ab) (kind gift of Chris de Graffenried, Brown University, Providence, RI, USA).
2.4. Western blot analysis
Denatured lysates from 2×107 parasites containing a protease and phosphatase inhibitor (Thermo Scientific Inc., MA, USA) were loaded per well in a 4–20% Tris-glycine SDS/PAGE gel for protein electrophoresis. The gel was transferred to Immun-Blot polyvinylidene difluoride (PVDF) membrane (Life Technologies) using Tris-glycine buffer without methanol. Mouse anti–GFP Ab (1:1000 dilution) (Roche Diagnostics mixture of clones 7.1 and 13.1 [Roche Diagnostics GmbH, Germany]) was used to stain for YFP labeled TgFIKK protein. Surface associated antigen-1 (SAG1) expression was analyzed as a loading control by using mouse monoclonal antibody (mAb) DG52 hybridoma tissue culture supernatant (1:10 dilution) that reacts to T. gondii surface protein SAG1. Anti-mouse antibody conjugated to alkaline phosphatase as provided in the Western Breeze kit (Life Technologies) was used as a secondary antibody and the blots developed using the same kit as per the manufacturer’s instructions.
2.5. Immunofluorescence analysis
For immunolocalization studies, HFF cells were cultured in 8-well chamber slides (Life Technologies) and infected at a multiplicity of infection of 0.5 with TgFIKK-YFP parasites. Parasites without the YFP tag were used as a negative control. Expression of the endogenously YFP-tagged TgFIKK was analyzed by immunofluorescence at distinct stages of the parasite’s lytic cycle, including expression in freshly lysed extracellular parasites and at 1, 24 and 36 h p.i. At specific time points after infection, monolayers were fixed in 4% formaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) in 1x Dulbecco’s PBS (Life Technologies) for 20 min at room temperature, blocked and permeabilized by incubation in PBS containing 10% FCS and 0.2% TritonX100 (Andwin Scientific, IL, USA) for 30 min. Parasites were visualized using polyclonal antisera to T. gondii (Fitzgerald Industries International, Concord, MA, USA). Alexa-Fluor-594-conjugated goat anti-rabbit IgG (Life Technologies) was used as the secondary antibody to detect the polyclonal antibody to T. gondii. A mouse mAb to GFP was used to detect the endogenously tagged FIKK-YFP protein to increase sensitivity in some studies (Roche Life Sciences). Mouse Anti–TgISP1 (inner membrane complex (IMC) subcompartment protein-1 (ISP1)) Ab was used to mark the apical end of the parasite and was detected using Alexa-Fluor 647 conjugated goat anti-mouse Ab (Life Technologies) as the secondary antibody. The anti-ISP-1 protein was a kind gift from Dr. Peter Bradley (University of California, Los Angeles, USA). Samples were mounted using Vecta-Shield mounting media with DAPI (Vector Laboratories, Burlingame, CA, USA). IFA studies were visualized using a Zeiss inverted Axiovert 200 motorized microscope with a 100X objective (PlanApo 1.4 na oil PH3 objective); Zeiss filter sets 31, 34, 38 and 50 and Axiovision 4.3 software. Series of Z-stack florescent images were deconvolved using the Zeiss 3D deconvolution constrained iterative algorithm provided in the Zeiss 3D deconvolution module (Carl Zeiss International, Oberkochen, Germany). Pictures were taken with the Zeiss Axiocam MRM cool CCD camera. The only adjustments made to pictures post photography were alterations in contrast that were applied to the entire image or devonvolution of z-stack series. To increase resolution, localization of the FIKK YFP protein was also visualized using the Zeiss LSM 880 with Airyscan.
To co-localize Ty-FIKK with marker proteins of the basal complex, 25 μg of pmorn1-mCherry-MORN1, ptub-mCherry-IMC5 or ptub-YFP-Centrin2 plasmid DNA were transiently transfected in parasites expressing the Ty-FIKK fusion protein (Anderson-White et al., 2011). Parasites were fixed with methanol for 15 min at room temperature 18 h after transfection, then blocked with 1% BSA in PBS and incubated with mouse anti-Ty and rabbit anti-mCherry Ab (kind gift of Dr. Iain Cheeseman, Whitehead Institute, Cambridge, MA, USA) in the case of mCherry-MORN1 and mCherry-IMC5. IFAs were incubated with secondary goat antibodies linked to Alexa488 or Alexa594 (Molecular Probes, Oregon, USA). DNA was stained with DAPI. Imaging was performed on a Zeiss Axiovert 200M wide-field fluorescence microscope equipped with standard DAPI, FITC, YFP and TRTC filter sets, a α-Plan-Fluar 100x/1.45 NA oil objective and images were analyzed and processed using Volocity (Perkin-Elmer, Norwalk, CT, USA) software.
2.6. Generation of the ΔTgFIKK clones
The deletion of the entire coding region of the FIKK gene was accomplished by double homologous recombination using a construct derived from the pMini-GFP.hpt knockout vector which contains the selectable HPT marker gene as well as GFP to distinguish homologous and heterologous recombinants (a kind gift from Dr. Gustavo Arrizabalaga, University of Indiana School of Medicine, USA) (Karasov et al., 2005). The 5′ flank and 3′ flank of the construct was amplified from RH T. gondii parasites using primers 5′ ACC GCG GTG GCG GCC GCA GAG GAT TGT GAC CAG TGC CTT GA -3′; 5′-ATC CAC TAG TTC TAG ATG CCA ACC AAC ACA TTT CTC ACC G -3′ and 5′-CGT GCT GAT CAA GCT TTC CGT CAG TTG CCG ATT ATT GGG T -3′; 5′-CGG GCC CCC CCT CGA GTC GAC TGT TTG GCT AGC ACC GAT A -3′, respectively. These genomic flanks were then cloned into pMini-GFP-hpt upstream and downstream of HPT, resulting in the vector pFIKK-KO-HPT. To create the pFIKK-KO-HPT vector, the Flank 1 PCR product was cloned into a NotI and XbaI digested vector pMini-GFP.hpt using Ligation Independent Cloning (LIC). The resulting plasmid was digested with HindIII and XhoI, and Flank 2 was cloned in using LIC. The LIC reactions were performed with Clontech In-Fusion® PCR cloning system (Clontech) according to the manufacturer’s instructions. The resulting targeting vector (pFIKK-KO-HPT) was purified using a QIAGEN miniprep plus kit according to the manufacturer’s instructions. Fifty micrograms of NotI linearized pFIKK-KO-HPT targeting vector was transfected into the RHΔHPTΔKU80 strain. Seven independent electroporations were performed. Stable clones carrying the transfected plasmid were positively selected for the HPT gene using mycophenolic acid in D10 media supplemented with xanthine, as described in Section 2.3, for the creation of the FIKK-YFP tagged clone. To identify clones with the FIKK gene deleted, parasites were single cell cloned, DNA isolated and PCR analysis performed first to confirm the presence of HPT and the absence of the FIKK kinase. Gene-specific crossover, as opposed to random integration of the plasmid, was confirmed with PCR using primers within the selectable marker cassette HPT and the 5′ and 3′ genomic loci upstream and downstream of the FIKK genomic region, but outside the 5′ and 3′ flanking regions in the gene targeting cassette. The PCR products were sequenced to confirm the replacement of the entire FIKK gene with the HPT gene. The 5′ junction was PCR amplified and sequenced using primers 5′-TTGCCAAGATGACTCCTCACAGTG-3′ and 5′-TTGGTTGCTGCTACGACTTCAACG-3′ specific for HPT gene in the vector and for the T. gondii genome sequence adjacent to the 5″ region of the 5′ flank in the plasmid. The 3′ junction was PCR amplified and sequenced using primers 5′-GTG GCG ATT CTC ATC GAC TT- 3′ and 5′-AGA CTG TGT GCC AGG ATC TGA A -3′ specific for HPT gene in the vector and for the T. gondii genome sequence adjacent to the 3′ region of the 3′ flank present in the targeting construct used to generate the gene deletion. Pictures for PCR have been spliced together to eliminate empty lanes between samples. Deletion of the FIKK transcript was also confirmed by qRT-PCR using two primer sets that amplified different portions of the 3′ catalytic region of the FIKK gene. qRT Primer set 1 used primers 5′-TATCCTCTACGGCAGCACAGCCTTT-3′ and 5′-GCGGTGTAATTCCTAGTGCTCTGA-3′. qRT Primer set 2 used primers 5′-GTGCAGCTGAAGGCGTAGATGCCG-3′ and 5′-TTGTCGTGGATCAGAGCAAGTCCA-3′. Parasite actin was used as a control using primers 5′-GTCTATCGTCGGAAAGCCCAAGAA-3′ and 5′-TGACGATACCGTGCTCAATAGGGT-3′.
2.7. Intracellular growth
Freshly egressed parasites were inoculated onto new HFF cell monolayers in 8-well chamber slides (Life Technologies). At 12, 24 and 36 h p.i., cultures were fixed with 4% formaldehyde and stained with polyclonal antisera to T. gondii (Fitzgerald Industries International). Alexa-Fluor-488 conjugated goat anti rabbit (Life Technologies) was used as the secondary antibody. The numbers of parasites per vacuole were counted in 100 vacuoles for each parasite clone/time point. Two to three counts of 100 vacuoles each were performed per experiment.
2.8. Mouse infections
C57BL/6 female mice, 6–9 weeks of age (Jackson Laboratory, Bar Harbor, Maine, USA), were infected i.p. with 500 tachyzoites of wild type (WT) or TgΔFIKK clones derived from independent electroporations. Parasite suspensions used for infections were applied to HFF monolayers in 24-well tissue culture plates to determine the dose of viable parasites in the inoculate used for infection and to ensure approximately equal numbers of viable parasites were injected. The acute infection experiment was performed once using five mice per strain. All animals were housed and bred under specific pathogen-free conditions at New York Medical College (NYMC), USA and all experiments were conducted in accordance with a NYMC Institutional Animal Care and Use Committee approved protocol 1-2-0113H.
3. Results
3.1. Characterization of the T. gondii FIKK kinase
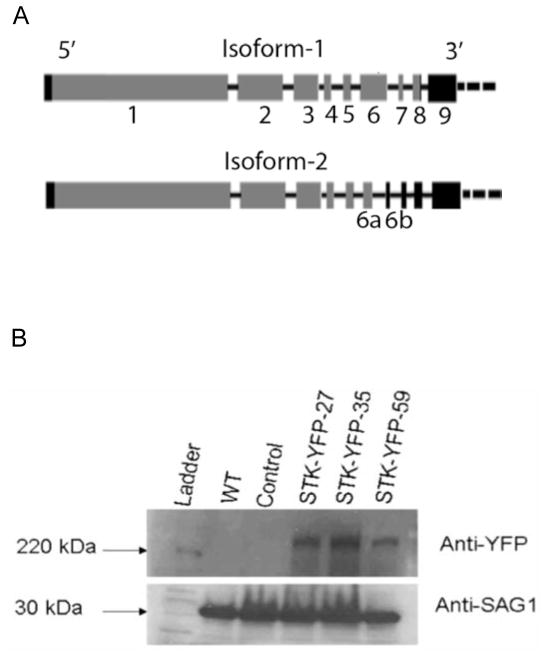
Bioinformatics analysis of the T. gondii genome confirmed the presence of a single putative FIKK kinase gene in T. gondii (TGGT1_289050) located on chromosome IX. Since the gene was hypothetical, the cDNA for the TgFIKK protein was sequenced for the Type I genotype RH strain and is shown in Fig. 1A. The predicted coding region for the FIKK protein based on cDNA sequencing consisted of nine transcribed regions interrupted by introns. The cDNA resulted in the same amino acid sequence as the prediction for the GT1 strain in ToxoDB.org version 10. The 5′ untranslated region (UTR) in the RH strain for the FIKK gene was 103 bp upstream of the predicted exon 2 resulting in a slightly shorter sequence than predicted in ToxoDB.org version 10. Two isoforms of the FIKK cDNA produced by alternative splicing in the sixth exon within the coding region of the gene (Fig. 1A) were identified in both the Prugniaud and RH strain (isoform-1 and 2).
Fig. 1.
Gene structure of the Toxoplasma gondii TgFIKK kinase. (A) Schematic of the cDNA sequence for isoform-1 and isoform-2 of the FIKK gene. The gene encodes two transcripts generated by alternative splicing and are designated isoform-1 and isoform-2. Isoform-2 is truncated due to alternative splicing of exon 6 into 6a and 6b, resulting in a stop codon before the complete kinase domain. Boxes represent transcribed regions (black: untranslated regions (UTRs) and grey: exons), black lines connecting them depict introns. (B) Western blot analysis of the Yellow Fluorescent Protein (YFP)-tagged isoform-1 TgFIKK protein. TgFIKK kinase isoform-1 encodes an approximately 280 kDa protein. Western blot analysis of three distinct Toxoplasma clones (clones STK-YFP-27, 35 and 59) in which the endogenous FIKK protein is tagged with YFP shows expression of a high molecular weight protein of approximately 300 kDa which includes the weight of the YFP tag. No corresponding bands in this region are seen in the wild type (WT) and a control T. gondii clone expressing a different YFP tagged gene to confirm the band is specific for the FIKK-tagged YFP protein rather than YFP in general. The YFP-tagged FIKK protein was detected by a monoclonal antibody (mAb) to GFP that reacts with YFP (mixture of clones 7.1 and 13.1 Roche Diagnostics GmbH, Germany). Expression of T. gondii surface protein SAG1 (30kDa) was detected with mAb DG52 to confirm equal loading of protein samples.
Conceptual translation taking the first AUG triplet occurring in the mRNA sequence identified a gene structure containing eight exons followed by a 3′UTR which was interrupted by a single intron (Fig. 1A), giving rise to a 288 kDa protein encoded by 2762 amino acids as TgFIKK kinase isoform-1. Expression of this isoform was confirmed by endogenous tagging of the native gene with YFP followed by western blot analysis (Fig. 1B). An intron within the sixth exon of the first isoform, resulting in two smaller transcribed regions 6a and 6b, characterized TgFIKK isoform-2 (Fig. 1A). This isoform was present in both the Prugniaud and the RH strain and contained seven exons followed by a long 3′UTR interrupted with multiple introns due to the presence of an in-frame stop codon immediately after the start of the seventh transcribed region (element 6b in Fig. 1A), resulting in a shorter, approximately 263 kDa, protein encoded by 2536 amino acids. The identified protein sequence for isoform-1 is supported by MS identified peptides available for this gene at ToxoDB.org (Hu et al., 2006).
3.2. Analysis of the catalytic domain
FIKK kinases have a highly variable N-terminal extension followed by a catalytic domain that is relatively conserved. An alignment of the predicted catalytic domain for the RH and Prugniaud T. gondii FIKK kinase with the catalytic domains for P. falciparum FIKK kinases with demonstrated kinase activity is shown in Supplementary Fig. S1. The TgFIKK kinase isoform-1 shares most of the conserved residues and motifs that are conserved among the FIKK family members and contains all the residues shown to be critical for catalytic activity for kinases in P. falciparum. The TgFIKK kinase contains motif “FVKK” instead of the hallmark “FIKK” in the second sub-domain, however there are other members within the FIKK kinase family with demonstrated kinase activity with a similar deviation. For example, FIKK 4.2 has demonstrated kinase activity and has “FVKK” instead of “FIKK” (Kats et al., 2014) (Supplementary Fig. S1). TgFIKK kinase isoform-2, in contrast to isoform-1, is interrupted within the catalytic region by a stop codon and encodes an incomplete catalytic domain (Supplementary Fig. S1).
3.3. TgFIKK isoform-1 is the dominant form expressed in tachyzyoites
In order to evaluate the relative expression of isoform-1 and isoform-2 in tachyzoites, qRT-PCR, with primers to regions common to both TgFIKK kinase isoforms-1 and 2 versus the downstream region restricted to TgFIKK kinase isoform-1, was performed at time points throughout the parasite’s lytic cycle. Isoform-1 with the intact kinase domain was the predominant form in both RH and Prugniaud tachyzoites throughout the parasite’s lytic cycle in vitro with no clear contribution of isoform-2 (Supplementary Fig. S2A). The expression level of the isoform-1 transcript was also evaluated during the parasite’s lytic cycle at 12, 24 and 36 h just prior to host cell lysis. Transcription levels of isoform-1 are presented in Supplementary Fig. S2B. The FIKK transcript was constitutively expressed throughout the parasite’s lytic cycle but was slightly lower at 24 h compared with 12 and 36 h, similar to the available data on ToxoDB.org (Behnke et al., 2010).
3.4. TgFIKK is constitutively expressed and localizes to the basal end of tachyzoites
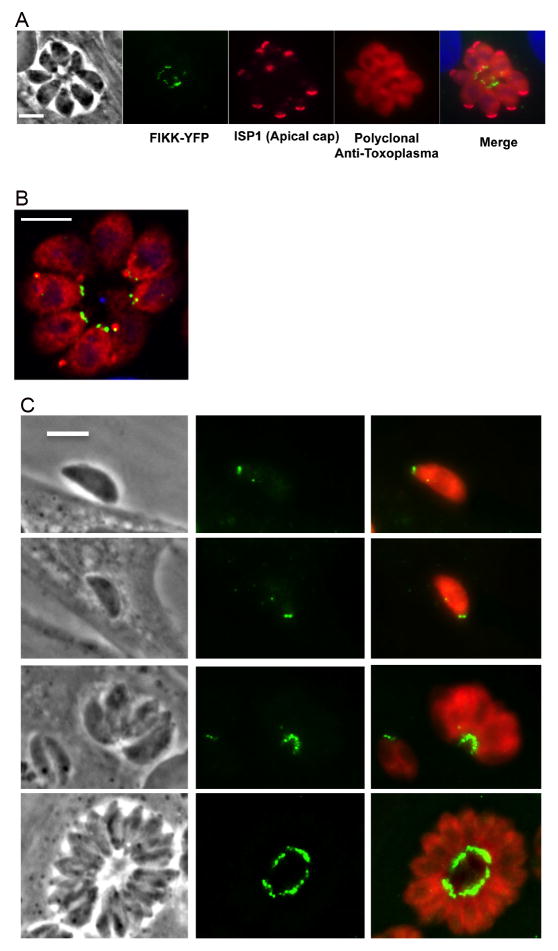
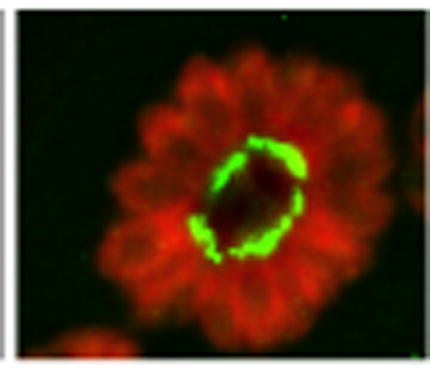
We endogenously tagged the TgFIKK kinase gene isoform-1 with YFP to evaluate FIKK protein expression and its localization and trafficking. Expression of the fusion protein was verified by qRT-PCR and western blot analysis (Fig. 1B). The approximate size of the YFP-tagged FIKK protein (FIKK-YFP) was 300 kDa including the molecular weight of the YFP tag (27 kDa) which is consistent with the estimated size of 288 kDa estimated from its amino acid sequence. IFA analysis indicated that the protein was exclusively localized to the posterior end of intracellular parasites at 24 h p.i. of HFF cells (Fig. 2A). The posterior localization of the FIKK protein was confirmed by co-staining with an antibody to the apical cap protein IMC-ISP1 to mark the apical end of the parasite (Beck et al., 2010). Airyscan super resolution (SR) microscopy, shown in Fig. 2B, confirmed that the FIKK-YFP localized to the basal end of the parasite, reminiscent of staining to the basal complex, and that it could be resolved as two adjacent dots consistent with a circle or ring when cut in half (Gubbels et al., 2006; Hu, 2008; Heaslip et al., 2010; Anderson-White et al., 2011). To determine whether the localization of the FIKK- protein remained consistent throughout the lytic cycle of the parasite, the expression of the FIKK-YFP was examined in parasites following egress, during invasion, and at 12, 24 and 36 h p.i. The FIKK-YFP was exclusively localized to the basal end of the parasite in both extracellular and intracellular parasites throughout the parasite lytic cycle (Fig. 2). The fact that FIKK-YFP remained associated with the basal end of the parasite in extracellular parasites confirmed its localization within the parasite rather than the network between the tachyzoite posterior end and the residual body membrane within the parasite rosette (Muniz-Hernandez et al., 2011).
Fig. 2.
Immune-localization of Toxoplasma gondii TgFIKK kinase endogenously tagged with Yellow Fluorescent Protein (YFP). YFP-tagged protein (YFP-FIKK) was exclusively localized to the posterior end of the parasite throughout the parasite’s lytic cycle. (A) IIFA showing the YFP-tagged FIKK protein (green) in the posterior end of the parasite infecting human foreskin fibroblast (HFF) cells at 24 h p.i. The apical cap portion of the parasite was stained with a monoclonal antibody (mAb) to apical protein inner membrane complex (IMC) subcompartment protein-1 (ISP1) and is shown in pink. Polyclonal rabbit anti Toxoplasma antibody was used to stain for parasites (red). The host nucleus is stained with DAPI (blue). Scale bar = 5 μm. (B) Zeiss LSM 800 with Airyscan image of the YFP-tagged FIKK protein 24 h p.i. The FIKK protein was localized to two adjacent punctate dots in the basal end of the parasite (green). Parasites (red) are stained with a polyclonal antiserum to the parasite. Scale bar = 5 μm. (C) Localization of the FIKK protein during the parasite’s lytic cycle. The FIKK protein was constitutively and exclusively expressed in the basal end of tachyzoites throughout the parasite’s lytic cycle. IFA of the YFP tagged FIKK protein (green) in a parasite during invasion and at 2 and 36 h p.i. Localization of the FIKK kinase at 24 h p.i. is shown in Figs. 4A, C. Scale bar = 5 μm.
3.5. FIKK protein co-localizes with MORN1 in the basal complex
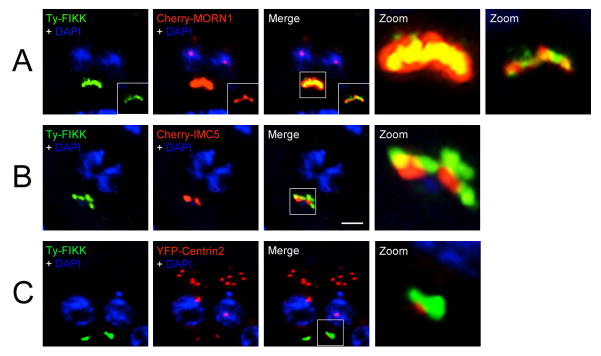
In the absence of a specific antibody to the FIKK kinase, we evaluated the localization of a N-terminal Ty-tagged FIKK protein under the control of the tetO7Sag4 minimal promoter to confirm that the localization of the FIKK protein was independent of the YFP tag. Ty-tagged FIKK (Ty-FIKK) protein showed a similar posterior localization as the YFP-tagged FIKK protein. To confirm localization of the FIKK protein to the basal complex, and to determine to which part of the basal complex Ty-FIKK localizes, we used sub-compartment-specific markers MORN1, IMC5 and Centrin2 (Anderson-White et al., 2011). Ty-FIKK co-localized with the basal complex marker protein MORN1 to the basal end of the parasite (Fig. 3A), apically to IMC5 and posterior to cup marker Centrin2 (Figs. 3B, C). We therefore conclude that Ty-FIKK localizes in the most apical part of the basal complex, together with the previously described proteins MORN1, IMC9 and IMC13 (Hu et al., 2006; Anderson-White et al., 2011)
Fig. 3.
Co-localization analysis of the Toxoplasma gondii N-terminal Ty-tagged FIKK protein with markers of the basal complex. FIKK protein localizes in the basal complex. (A) N-terminal Ty-tagged FIKK co-localizes with membrane occupation and recognition nexus protein (MORN1) inside the basal complex. (B) The FIKK protein sits apical to IMC5. (C) The FIKK protein sits apical to the posterior cup marker Centrin2. Ty-tagged FIKK is pseudo-colored green. MORN1, IMC5 and Centrin2 are pseudo-colored red. Magnification of boxed areas is shown in the zoom. Scale bar = 2 μm.
3.6. Generation of ΔFIKK parasites
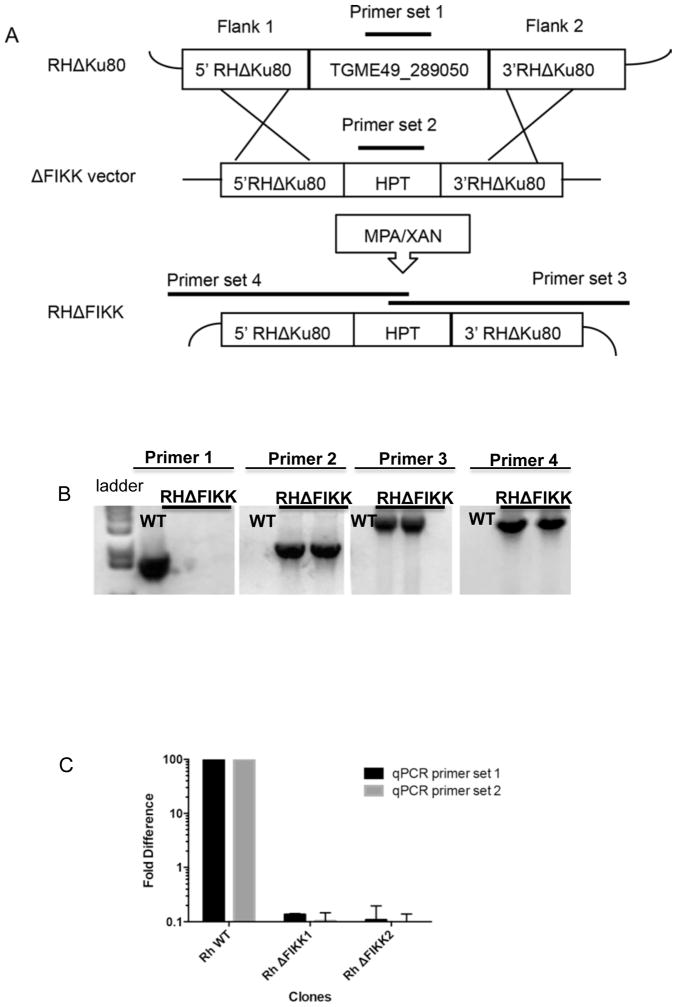
In order to determine whether the FIKK protein was important for any specific step in the parasite’s lytic cycle in vitro or in vivo, we deleted the FIKK kinase gene in the RHΔKu80ΔHPT strain. The FIKK kinase gene was deleted by homologous recombination in which the entire coding region of the FIKK gene was replaced with the HPT gene. The schematic in Fig. 4A shows the approach used for targeted gene deletion and the primer sets used to confirm gene deletion in T. gondii by PCR. Shown in Fig. 4B are PCR results for two independent T. gondii ΔFIKK clones compared with the parental strain (wild type, WT). Primer set 1 for the FIKK gene amplified the FIKK gene only from WT parasites. Primer set 2 amplified the HPT gene only from ΔFIKK clones. Primer set 3 amplified the junction between the genomic region downstream of the targeted FIKK gene deletion and the HPT gene only in ΔFIKK clones, confirming that the homologous recombination event occurred at the targeted locus. The 5′ junction between HPT and the genomic region upstream of the targeted FIKK gene deletion was also confirmed and is shown using primer set 4. The junctional PCR products were sequenced to confirm replacement of the FIKK gene with HPT. qRT-PCR, using two primer sets for different parts of the putative catalytic domain in the 3′ end, confirmed the FIKK transcript was present in WT parasites but absent from the ΔFIKK clones (Fig. 4C). Primers used for PCR, sequencing and qRT-PCR are listed in Section 2.6.
Fig. 4.
Schematic showing approach used to generate Toxoplasma gondii ΔTgFIKK clones and to confirm gene deletion. (A) Schematic showing the generation of the ΔTgFIKK clones by homologous recombination. (B) Confirmation of deletion of TgFIKK gene in the ΔTgFIKK clones by PCR. (C) Real-time quantitative reverse transcription PCR (qRT-PCR) analysis of the FIKK transcript in wild type (WT) versus ΔTgFIKK clones. The FIKK transcript was undetectable in the ΔTgFIKK clones using two different sets of primers. HPT, hypoxanthine-xanthine-guanosine phosphoribosyl transferase; MPA/Xan, mycophenolic acid and xanthine.
3.7. TgFIKK is not essential for the parasite’s lytic cycle
We first evaluated the contribution of the TgFIKK kinase to the parasite’s lytic cycle in vitro in HFF cells. Wild type RH and ΔFIKK clones were compared for their ability to form plaques in vitro in HFF cells after 5 days of culture. The sizes of the plaques in the HFF monolayer are dependent on the ability of the parasite to complete multiple sequential rounds of the parasite lytic cycle including migration, invasion, replication, egress and re-invasion. There was no difference between the RH and ΔFIKK clones in the number and size of plaques they formed in an HFF monolayer (data not shown). We directly evaluated whether the FIKK gene was important for parasite replication in HFF by determining the number of parasites per vacuole over time by florescence microscopy. As shown in Fig. 5, deletion of the FIKK gene did not significantly alter parasite replication in HFF cells. Thus while a role for the FIKK kinase in the parasite’s lytic cycle in vitro in HFF cells including parasite migration, invasion, replication and egress is not ruled out, it is clearly not required.
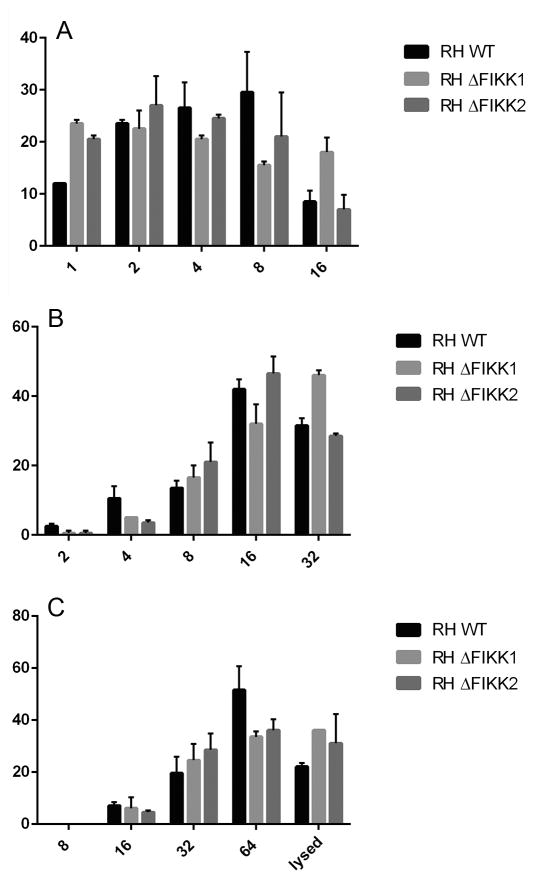
Fig. 5.
The number of wild type (WT) versus ΔTgFIKK Toxoplasma gondii parasites per vacuole at 12, 24 and 36 h p.i. The FIKK kinase is not important for parasite replication in human foreskin fibroblast (HFF) cells. The average number of parasites per vacuole was counted for 100 vacuoles per parasite clone at (A)12 h, (B) 24 h, and (C) 36 h p.i. Each count was performed three times per experiment. The x-axis shows the number of parasites per vacuole and the y-axis shows the percent of vacuoles that contain a given number of parasites per vacuole. Results from a single representative experiment are shown and the experiment was repeated at least twice.
3.8. TgFIKK is not essential for virulence in the RH strain
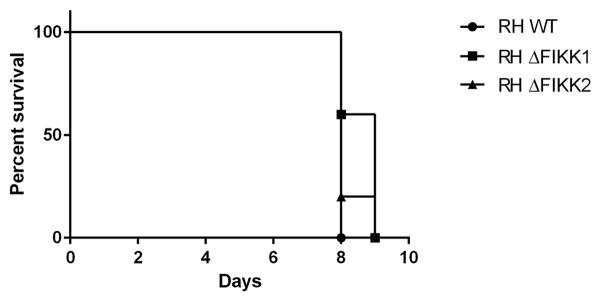
To determine whether the FIKK gene was critical for in vivo infection, we inoculated C57BL/6 mice i.p. with 500 WT or ΔFIKK parasites and evaluated mouse survival. As shown in Fig. 6, infection with WT and ΔFIKK parasites resulted in similar mortality with mice dying between days 8 and 9 p.i. While it is possible that lower doses of parasites might show a difference in virulence between WT parasites and ΔFIKK parasites, it is clear that the FIKK gene is not required for acute infection in the mouse model using the RH strain.
Fig. 6.
Survival curve of mice infected i.p. with 500 wild type (WT) versus ΔTgFIKK Toxoplasma gondii tachyzoites. TgFIKK kinase is dispensable for in vivo acute infection. X-axis represents days p.i. and the Y-axis shows percent survival of mice infected with WT or ΔTgFIKK clones. n = 5 mice per group. The experiment was performed once only.
4. Discussion
To date, the function and localization of FIKK kinases have been primarily studied in P. falciparum, in which the kinase family is expanded relative to other apicomplexans (Ward et al., 2004). The FIKK kinases studied in P. falciparum are predominantly secreted proteins, many of which have demonstrated kinase activity and play a role in remodeling of the infected erythrocyte (Nunes et al., 2007, 2010; Brandt and Bailey, 2013; Kats et al., 2014). Although only non-essential FIKKs have been reported in P. falciparum, their respective knock-out (KO) lines display interesting phenotypes that may be critical for virulence and disease. Furthermore, some of the 21 Pf FIKK may turn out to be essential. An attempt to delete the single FIKK kinase in P. berghei was unsuccessful, suggesting it may be essential (Tewari et al., 2010).
In the current study, we chose to evaluate the single FIKK kinase present in T. gondii to evaluate the role of the FIKK protein in other apicomplexans. We showed that the T. gondii FIKK kinase localized exclusively to the apical section of the basal complex, co-localizing in the complex with MORN1, throughout the parasite’s lytic cycle. Targeted deletion of the FIKK gene in the RH strain of the parasite did not alter the ability of the parasite to transverse its lytic cycle in HFF cells in vitro or impair parasite replication, nor did it affect the overall virulence of T. gondii during acute infection. While our findings indicate that the FIKK protein is not essential for the parasite’s lytic cycle, they do not rule out a critical role for the FIKK protein in other developmental stages including tachyzoite to bradyzoite development. It is also possible that the FIKK protein may contribute to virulence in less virulent strains of T. gondii.
Although FIKK kinases have been proposed as potential therapeutic targets due to their unique restriction to the Apicomplexa, their function and importance in the Apicomplexa remain largely unknown. The evolutionary loss of the FIKK kinases in the piroplasms has led to speculation that they may play a role in apicomplexan biology related to establishment of the PV or in communication between the parasite, PV and host cell, as piroplasms replicate in the cytosol rather than in a PV (Talevich et al., 2011). Studies to date of the FIKK kinases in P. falciparum are consistent with this possibility. The majority of P. falciparum FIKK kinases have export signatures and many of those that have been studied traffic to distinct sites associated with the infected erythrocyte membrane (Schneider and Mercereau-Puijalon, 2005; Nunes et al., 2007, 2010).
In the current study we show that the T. gondii FIKK protein localized exclusively in tachyzoites to the basal complex. MORN1 is a critical organizer of the basal complex of T. gondii (Gubbels et al., 2006; Hu et al., 2006; Hu, 2008; Heaslip et al., 2010; Lorestani et al., 2010). Other members of the basal complex include TgCentrin2 (Hu et al., 2006), and several intermediate filament like proteins (IMC5, IMC8, IMC9, IMC13) (Anderson-White et al., 2011). Co-localization analysis showed the FIKK protein localized with MORN1 inside the basal complex where it sat apical to IMC5 and the posterior cup marker Centrin2. This is consistent with identification of the FIKK protein in studies using MORN1 as bait in BioID proximity-dependent biotinylation studies (K. Engelberg and M.J. Gubbels, unpublished data). To date, the majority of proteins that localize to the basal complex and posterior end of the tachyzoites localize to other locations within the parasite as well. In contrast to MORN1 and these other proteins, TgFIKK localized exclusively to the basal complex and was not evident in nascent daughter cells during endodyogeny, suggesting it is only expressed in mature parasites. Also, in contrast to MORN1, deletion of the TgFIKK did not affect parasite division (Lorestani et al., 2010). The FIKK-YFP analyzed by western blot showed that the size of the tagged protein was consistent with our sequence data, thus it is unlikely that localization was a consequence of protein degradation that can occur with tagged proteins. Furthermore, localization of the FIKK protein was unchanged using a Ty-tagged FIKK protein.
Based on our bioinformatics analysis, TgFIKK kinase has the conserved catalytic residues that have been implicated in kinase activity in Plasmodium FIKK kinases. However, we have been unable to purify the intact FIKK protein or catalytic domain to demonstrate kinase activity (data not shown). Outside the catalytic domain, the FIKK proteins have no homologous regions to other proteins or functional domains. Even the FIKK8 homologs common to members of the Apicomplexa with a single FIKK kinase are not highly conserved. In the current study we showed that in both RH and Prugniaud strains there were two isoforms of the FIKK kinase; the transcript for isoform-1 was longer with the intact catalytic domain and isoform-2 lacked the kinase domain due to alternative splicing that resulted in a stop codon before the catalytic domain. Our results showed that isoform-1 with the catalytic domain was the dominant transcript throughout the parasites’ lytic cycle while transcription of isoform-2 appeared negligible.
It is important to emphasize that our studies only indicate that the FIKK gene is not essential for the lytic cycle of T. gondii in vitro in HFF cells. Protein kinases have clearly been implicated in tachyzoite to bradyzoite conversion and cyst production. Further studies are needed to evaluate the contribution of the FIKK protein to other developmental stages of the parasite as well as in stage differentiation and cyst formation. To more readily address the role of the FIKK protein in conversion to bradyzoites and cyst formation, we have deleted the gene in the more cystogenic Type II genotype of T. gondii in order to evaluate its role in cyst production and chronic infection.
Supplementary Material
Supplementary Fig. S1. Sequence of catalytic domain of Toxoplasma gondii FIKK compared with Plasmodium falciparum FIKK kinases with confirmed kinase activity. (A) Analysis and alignment of the TgFIKK kinase catalytic domain compared with P. falciparum FIKK kinases. The TgFIKK protein shares all the necessary residues shown to be important for kinase activity for P. falciparum FIKK kinases. Alignment shows the similarity of the TgFIKK kinase with P. falciparum kinases with proven kinase activity. Identical and conserved residues among the various sequences are shown on a black background and similar residues are shaded grey. Presence of asterisk symbol at the top of a residue indicates a conserved residue shared by all members of the FIKK family and stretches of such residues or in close vicinity of each other indicate conserved motifs. Residues with double asterisk are those which have been shown to be important for catalytic activity (Ward et al., 2004). Roman numerals indicate the 11 sub-domains that constitute the catalytic domain. (F/L) amino acid has been found at the position indicated by asterisk with a period at top. (B) Alignment of the RH and Prugnaud isoform-1 and isoform-2 transcripts putative catalytic domains. Isoform-1 transcript contains the complete putative catalytic domain while alternative splicing of isoform-2 results in a stop codon and truncation of the catalytic domain. Variant residues have been highlighted. Residues that have been shown to be critical for catalytic activity in FIKK kinases are marked with an asterisk.
Supplementary Fig. S2. Analysis of the FIKK transcript (isoform-1 and isoform-2) during the Toxoplasma gondii parasite’s lytic cycle for both the RH and Prugnaud (Pru) strain of parasite. (A) A comparison of the expression of total transcript for both isoforms versus expression of just the isoform-1 (long form) by real-time quantitative reverse transcriptase PCR (qRT-PCR) reveals isoform-1 is the dominant transcript at all time points in both the RH and Prugnaud (Pru) strain of parasite. Primers were made to the kinase domain specific for isoform-1 (long form) or the 5′ region upstream of the kinase domain conserved in both isoform-1 and isoform-2 (total [long and short forms]) Primers are listed in section 2.2. (B) Transcript levels of TgFIKK isoform-1, relative to parasite actin levels, throughout the parasites lytic cycle in vitro in human fibroblast cells. TgFIKK isoform-1 is expressed throughout the parasite’s lytic cycle but is highest at 12 and 36 h p.i., corresponding to after invasion and just prior to egress. Transcript levels were determined by real-time quantitative reverse transcriptase PCR (qRT-PCR) and expressed relative to parasite actin levels at each time point. Primers are listed in section 2.2.
Highlights.
The Toxoplasma FIKK kinase exclusively localizes to the basal complex
The FIKK kinase is expressed throughout the parasite’s lytic cycle
The FIKK kinase is not required for the parasite’s lytic cycle in vitro
The FIKK kinase is not required for acute infection in a mouse model
Acknowledgments
This work was supported by the National Institutes of Health, USA, grants AI072028, AI107431 and AI108006 to D.G.M., AI081924 and AI110690 to M.J.G., a Deutsche Forschungs-gemeinschaft, Germany, fellowship grant to K.E., and Qatar National Research Fund, Qatar (JSREP: 05-007-3-002, a Junior scientist Fellowship to S.S.).
Footnotes
Note: New nucleotide sequence data reported in this paper are available through GenBank™ under the accession numbers KP901266 and KP901267.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson-White BR, Ivey FD, Cheng K, Szatanek T, Lorestani A, Beckers CJ, Ferguson DJ, Sahoo N, Gubbels MJ. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cell Microbiol. 2011;13:18–31. doi: 10.1111/j.1462-5822.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr, Treatman C, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JR, Rodriguez-Fernandez IA, Cruz de Leon J, Huynh MH, Carruthers VB, Morrissette NS, Bradley PJ. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, Sibley LD, White MW. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One. 2010;5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt GS, Bailey S. Dematin, a human erythrocyte cytoskeletal protein, is a substrate for a recombinant FIKK kinase from Plasmodium falciparum. Mol Biochem Parasitol. 2013;191:20–23. doi: 10.1016/j.molbiopara.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Fox BA, Falla A, Rommereim LM, Tomita T, Gigley JP, Mercier C, Cesbron-Delauw MF, Weiss LM, Bzik DJ. Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot Cell. 2011;10:1193–1206. doi: 10.1128/EC.00297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, Pinney DF, Roos DS, Stoeckert CJ, Jr, Wang H, Brunk BP. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- Heaslip AT, Dzierszinski F, Stein B, Hu K. TgMORN1 is a key organizer for the basal complex of Toxoplasma gondii. PLoS Pathog. 2010;6:e1000754. doi: 10.1371/journal.ppat.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K. Organizational changes of the daughter basal complex during the parasite replication of Toxoplasma gondii. PLoS Pathog. 2008;4:e10. doi: 10.1371/journal.ppat.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Johnson J, Florens L, Fraunholz M, Suravajjala S, DiLullo C, Yates J, Roos DS, Murray JM. Cytoskeletal components of an invasion machine--the apical complex of Toxoplasma gondii. PLoS Pathog. 2006;2:e13. doi: 10.1371/journal.ppat.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov AO, Boothroyd JC, Arrizabalaga G. Identification and disruption of a rhoptry-localized homologue of sodium hydrogen exchangers in Toxoplasma gondii. Int J Parasitol. 2005;35:285–291. doi: 10.1016/j.ijpara.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Kats LM, Fernandez KM, Glenister FK, Herrmann S, Buckingham DW, Siddiqui G, Sharma L, Bamert R, Lucet I, Guillotte M, Mercereau-Puijalon O, Cooke BM. An exported kinase (FIKK4.2) that mediates virulence-associated changes in Plasmodium falciparum-infected red blood cells. Int J Parasitol. 2014;44:319–328. doi: 10.1016/j.ijpara.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Kissinger JC, Gajria B, Li L, Paulsen IT, Roos DS. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31:234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorestani A, Sheiner L, Yang K, Robertson SD, Sahoo N, Brooks CF, Ferguson DJ, Striepen B, Gubbels MJ. A Toxoplasma MORN1 null mutant undergoes repeated divisions but is defective in basal assembly, apicoplast division and cytokinesis. PLoS One. 2010;5:e12302. doi: 10.1371/journal.pone.0012302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Saavedra D, Gabaldon T, Barton GJ, Langsley G, Doerig C. The kinomes of apicomplexan parasites. Microbes Infect. 2012;14:796–810. doi: 10.1016/j.micinf.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Morlon-Guyot J, Berry L, Chen CT, Gubbels MJ, Lebrun M, Daher W. The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell Microbiol. 2014;16:95–114. doi: 10.1111/cmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz-Hernandez S, Carmen MG, Mondragon M, Mercier C, Cesbron MF, Mondragon-Gonzalez SL, Gonzalez S, Mondragon R. Contribution of the residual body in the spatial organization of Toxoplasma gondii tachyzoites within the parasitophorous vacuole. J Biomed Biotechnol. 2011;2011:473983. doi: 10.1155/2011/473983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MC, Goldring JP, Doerig C, Scherf A. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol Microbiol. 2007;63:391–403. doi: 10.1111/j.1365-2958.2006.05521.x. [DOI] [PubMed] [Google Scholar]
- Nunes MC, Okada M, Scheidig-Benatar C, Cooke BM, Scherf A. Plasmodium falciparum FIKK kinase members target distinct components of the erythrocyte membrane. PLoS One. 2010;5:e11747. doi: 10.1371/journal.pone.0011747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider AG, Mercereau-Puijalon O. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics. 2005;6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevich E, Mirza A, Kannan N. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol Biol. 2011;11:321. doi: 10.1186/1471-2148-11-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R, Straschil U, Bateman A, Bohme U, Cherevach I, Gong P, Pain A, Billker O. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Sequence of catalytic domain of Toxoplasma gondii FIKK compared with Plasmodium falciparum FIKK kinases with confirmed kinase activity. (A) Analysis and alignment of the TgFIKK kinase catalytic domain compared with P. falciparum FIKK kinases. The TgFIKK protein shares all the necessary residues shown to be important for kinase activity for P. falciparum FIKK kinases. Alignment shows the similarity of the TgFIKK kinase with P. falciparum kinases with proven kinase activity. Identical and conserved residues among the various sequences are shown on a black background and similar residues are shaded grey. Presence of asterisk symbol at the top of a residue indicates a conserved residue shared by all members of the FIKK family and stretches of such residues or in close vicinity of each other indicate conserved motifs. Residues with double asterisk are those which have been shown to be important for catalytic activity (Ward et al., 2004). Roman numerals indicate the 11 sub-domains that constitute the catalytic domain. (F/L) amino acid has been found at the position indicated by asterisk with a period at top. (B) Alignment of the RH and Prugnaud isoform-1 and isoform-2 transcripts putative catalytic domains. Isoform-1 transcript contains the complete putative catalytic domain while alternative splicing of isoform-2 results in a stop codon and truncation of the catalytic domain. Variant residues have been highlighted. Residues that have been shown to be critical for catalytic activity in FIKK kinases are marked with an asterisk.
Supplementary Fig. S2. Analysis of the FIKK transcript (isoform-1 and isoform-2) during the Toxoplasma gondii parasite’s lytic cycle for both the RH and Prugnaud (Pru) strain of parasite. (A) A comparison of the expression of total transcript for both isoforms versus expression of just the isoform-1 (long form) by real-time quantitative reverse transcriptase PCR (qRT-PCR) reveals isoform-1 is the dominant transcript at all time points in both the RH and Prugnaud (Pru) strain of parasite. Primers were made to the kinase domain specific for isoform-1 (long form) or the 5′ region upstream of the kinase domain conserved in both isoform-1 and isoform-2 (total [long and short forms]) Primers are listed in section 2.2. (B) Transcript levels of TgFIKK isoform-1, relative to parasite actin levels, throughout the parasites lytic cycle in vitro in human fibroblast cells. TgFIKK isoform-1 is expressed throughout the parasite’s lytic cycle but is highest at 12 and 36 h p.i., corresponding to after invasion and just prior to egress. Transcript levels were determined by real-time quantitative reverse transcriptase PCR (qRT-PCR) and expressed relative to parasite actin levels at each time point. Primers are listed in section 2.2.