Abstract
Plasmodium cynomolgi is a malaria parasite that typically infects Asian macaque monkeys, and humans on rare occasions. P. cynomolgi serves as a model system for the human malaria parasite Plasmodium vivax, with which it shares such important biological characteristics as formation of a dormant liver stage and a preference to invade reticulocytes. While genomes of three P. cynomolgi strains have been sequenced, genetic diversity of P. cynomolgi has not been widely investigated. To address this we developed the first panel of P. cynomolgi microsatellite markers to genotype eleven P. cynomolgi laboratory strains and 18 field isolates from Sarawak, Malaysian Borneo. We found diverse genotypes among most of the laboratory strains, though two nominally different strains were found to be genetically identical, We also investigated sequence polymorphism in two erythrocyte invasion gene families, the reticulocyte binding protein and Duffy binding protein genes, in these strains. We also observed copy number variation in rbp genes.
Keywords: Plasmodium cynomolgi, Plasmodium vivax, genetic variation, microsatellite repeats, evolution
1. Introduction
The apicomplexan parasite Plasmodium cynomolgi causes malaria in Asian monkeys as well as experimental and rare zoonotic infections in humans (Coatney et al., 1971; Eyles et al., 1960; Garnham, 1966; Ta et al., 2014). It shares important biologic features with its sister taxon, the human malaria parasite Plasmodium vivax, including a dormant liver stage (Krotoski et al., 1982a; Krotoski et al., 1982b), a preference for invading immature red blood cells (Warren et al., 1966), early formation of infectious sexual stages (Dissanaike et al., 1965), modifications of the infected erythrocyte membrane known as Schuffner's stippling (Aikawa et al., 1975), and tertian periodicity (Mulligan, 1935). P. vivax is a serious global health problem that threatens more than 50% of the world's population (Guerra et al., 2010). While P. vivax cannot be cultured in vitro, severely hampering laboratory studies, P. cynomolgi has been successfully adapted to short-term in vitro culture (Nguyen-Dinh et al., 1981). Thus P. cynomolgi represents an ideal model system for investigating P. vivax biology, evolution, and pathology, and for identifying novel drugs against the dormant liver stage.
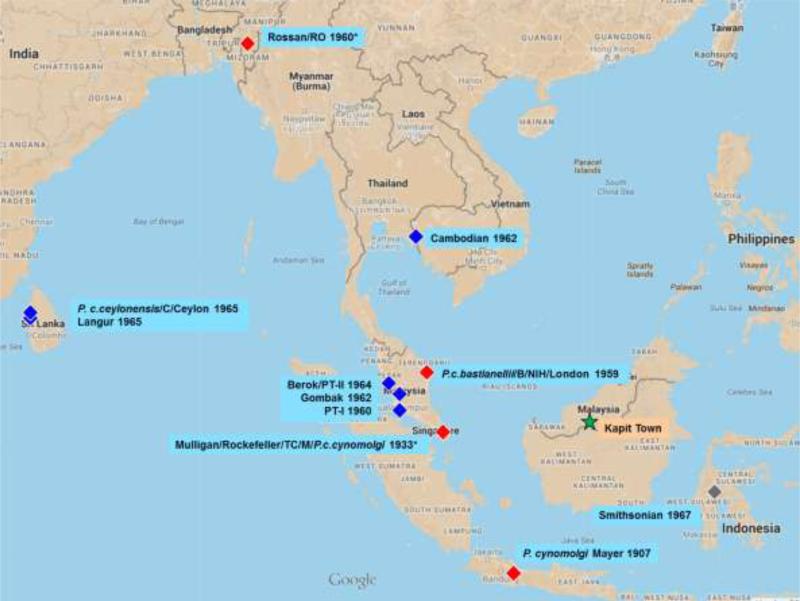
P. cynomolgi was first described by Mayer in 1907, though this ‘type strain’ was not preserved (Mayer, 1907). In 1935 H.W. Mulligan comprehensively re-described the species and maintained his isolate as the first laboratory strain (Mulligan, 1935). A second laboratory strain of P. cynomolgi was described and established by Garnham in 1959, to which he gave subspecies status as P. cynomolgi bastianellii (Garnham, 1959). In the decades following its isolation, stocks of Mulligan's strain were distributed among labs worldwide, and a proliferation of alternate strain names appeared in the literature. Initially it was referred to as the ‘Rockefeller’ strain (Garnham, 1959), or the ‘TC’ (‘typical cynomolgi’) strain (Eyles, 1960). In 1961 it as designated the ‘M’ strain in honor of Mulligan (Coatney et al., 1961; Schmidt et al., 1961), though it has also been referred to simply as ‘Mulligan’ (Cochrane et al., 1985). Adding further confusion, it was also designated as the ‘neotype’ strain of P. cynomolgi and given subspecies status as P. cynomolgi cynomolgi (Eyles et al., 1963). Meanwhile P cynomolgi bastianellii came to be called the B strain (Contacos et al., 1962), because malariologists at the National Institutes of Health did not think the differences between it and the neotype strain were significant enough to warrant subspecies status. Later, a line of the B strain was termed the ‘NIH’ strain and also referred to as the ‘London’ strain; however, these two strains were found to be different from each other, and the NIH strain was found to be identical to the M strain (Cochrane et al., 1985; Enea et al., 1986; Galinski et al., 1987). Identity of ostensible B and M stocks was more recently noted during genome sequencing of several P. cynomolgi strains (Tachibana et al., 2012). Between 1960 and 1967 eight more strains of P. cynomolgi were isolated from monkeys captured in Southeast Asia and Sri Lanka and some of these, too, have borne multiple strain names in the literature and sample repositories (e.g., strain Berok has the alias PT-II in the ATCC catalog, apparently after its original pig-tailed macaque host). Geographic origins and aliases of known strains are shown in Figure 1.
Figure 1. Geographical origins of P. cynomolgi strains.
Strain name, aliases, and year of isolation are shown. Blue, red, and grey markers indicate specific, approximate, and dubious origins respectively, gleaned from the literature (see Supplementary Materials for references). The reported Sulawesi (Celebes) origin for the Smithsonian strain is dubious because the island lies outside of the range of the host monkey species Macaca speciosa. Asterisk (*) indicates that the site was the shipping point of the host monkey, but the location of capture of the monkey is unknown. A star shows the location of Kapit Town in Malaysian Borneo, where P. cynomolgi field isolates genotyped in this paper were collected.
In addition to exhibiting similar biologic features, there is a strong body of evidence that demonstrates a close taxonomic relationship between P. cynomolgi and P. vivax. In 1991, there were hints of homology between these parasites with respect to the 135-kDa Duffy Binding Protein (DBP) (Fang et al. 1991). Between 1992 and 1998 this homology would be explicitly demonstrated across several proteins, including the small subunit (SSU) ribosomal RNA (Water et al. 1993), reticulocyte binding protein (RBP) (Galinski et al. 1992), circumsporozoite protein (Escalante et al. 1995), and cytochrome b from the linear mitochondrial gene (Escalante et al. 1998). These early investigations set the stage for extensive research that would occur over the next two decades, including the recent publication of the genome of the P. cynomolgi B strain from Peninsular Malaysia, which serves as the reference genome for the species, as well as draft genomes of strains Berok and Cambodian from Peninsular Malaysia and western Cambodia, respectively (Tachibana et al., 2012). The genome size of P. cynomolgi is ~26.3 Mb, with an estimated GC content of 40.4%, and ~5,700 genes predicted on 14 chromosomes. Approximately 96% of genes are orthologous between P. cynomolgi and P. vivax, with ~3,800 genes inferred to be under purifying selection and only 83 genes inferred to be under positive selection, indicating that P. cynomolgi will be a useful model for studying P. vivax (Tachibana et al., 2012).
An understanding of genetic diversity in Plasmodium parasites is crucial for vaccine development, malaria control, and elimination. The population genetics of P. vivax has been widely studied in endemic regions of the world (Arnott et al., 2012; Carlton et al., 2013), while the genetic diversity and population structure of P. cynomolgi has received much less attention. Diversity in microsatellite (MS) markers is generated by replication slippage/slipped-strand mismatch repair, which causes expansions or contractions of repeat. Approximately 180 polymorphic intergenic MS were identified in P. cynomolgi B and Berok strains by comparative genomics (Tachibana et al., 2012). Along with their utility for characterizing and comparing the genetic diversity and structure of populations, MS markers can be particularly useful for describing and resolving phylogeographic data, population bottlenecks, gene flow, and evolutionary history (Anderson et al., 1999, 2000; Branch et al., 2011; Oliveira et al., 1998; Sutton et al., 2011).
The observation that P. cynomolgi and P. vivax target immature erythrocytes (reticulocytes) for invasion has focused research on receptor-ligand interactions underlying this preference (Warren et al., 1966). The RBL (Reticulocyte Binding-Like) gene superfamily encodes large ligand proteins (230 - 350 kDa) that are localized to the apical membrane of the invasive stage of the parasite (Galinski et al., 1992; Rayner et al., 2004). Though all RBL genes share a conserved two-exon structure, subfamilies have different target cell preferences in different Plasmodium species. The Reticulocyte Binding Proteins (RBPs), an RBL subfamily found in some Plasmodium species, are thought to specifically recognize the reticulocyte subpopulation of erythrocytes (Galinski et al., 1992). RBPs in P. vivax are promising targets for vaccines that block receptor-ligand interactions; however, high sequence polymorphism and copy number variations of rbp genes could hinder vaccine effectiveness (Rayner et al., 2005), making it important to consider diversity in these genes in P. vivax and P. cynomolgi strains. The rbp genes can be classified into rbp1, rbp2, and rbp3 subgroups based on sequence similarity. In both P. vivax and P. cynomolgi, the gene pairs rbp1a/rbp1b and rbp2c/rbp2d are adjacent to each other on chromosome 7 and chromosome 5, respectively, whereas other rbp genes are located on different chromosomes. All three RBP groups have putatively functional representatives in the P. cynomolgi genome, whereas P. vivax and the zoonotic monkey malaria species P. knowlesi lack predicted functional RBP3 and RBP1 genes, respectively (Carlton et al., 2008; Pain et al., 2008; Tachibana et al., 2012). Therefore, rbp gene family diversification seems to have arisen before speciation, but losses have occurred since, suggesting a possible evolution of multiple species-specific erythrocyte invasion mechanisms. Variation in rbp gene repertoire extends beyond interspecies difference, as our genomic analysis of three P. cynomolgi strains revealed rbp1b to be present in the Berok strain but absent in the B and Cambodian strains (Tachibana et al., 2012), the first reported case of strain-specific copy number variation (CNV) for an rbp gene in Plasmodium. The rbp2e gene was subsequently found to be missing in the Salvador I strain of P. vivax but present in other strains (Hester et al., 2013). The receptors on reticulocytes are still largely unknown for the various expressed RBP proteins.
Erythrocyte Binding-Like (EBL) proteins comprise a second superfamily of erythrocyte-binding invasion ligands (Chitnis and Miller, 1994; Siddiqui et al., 2012; Sim et al., 1994) in Plasmodium. In P. cynomolgi and P. vivax, an EBL subfamily of Duffy-Binding Proteins (DBPs) are released during parasite invasion, where they bind to Duffy Antigen Receptor for Chemokines (DARC) (Adams et al., 1990; Adams et al., 1992; Batchelor et al., 2011) on reticulocyte surfaces. Because DARC-negative individuals typically are resistant to P. vivax, the DBP/DARC interaction was thought for many years to be an absolute requirement for P. vivax malaria (Miller et al., 1976; Miller et al., 1975). However, the recent discovery of P. vixax malaria in DARC-negative individuals indicates that the parasite can exploit heretofore unknown pathways for erythrocyte invasion (Ntumngia et al., 2012). As with RBPs, DBPs show species- and strain-specific CNV. The reference strain of P. vivax (Salvador I) has one dbp gene while P. cynomolgi strains B, Berok, and Cambodian have two very similar DBP paralogs (Carlton et al., 2008; Neafsey et al., 2012; Tachibana et al., 2012). A recent study revealed tandem duplication of dbp in several P. vivax field isolates from Madagascar (Menard et al., 2013). The duplication was prevalent in the regions where the highest frequencies of P. vivax infected Duffy-negative people were observed, suggesting that P. vivax might overcome the barrier to infection presented by Duffy negativity through the duplication of the dbp gene. Moreover, the two copies of dbp in P. vivax are identical except for one non-synonymous difference in the signal peptide, which is even greater sequence similarity than dbp1 vs. dbp2 of P. cynomolgi. The highly conserved nature of the P. vivax DBP duplication in these patients from Madagascar indicated that it was a recent event. Another dbp-like gene distantly related to DBP1/DBP2 also exists in three P. cynomolgi strains and in strains of P. vivax, except for the Salvador I strain (Chan et al., 2015).
Here we describe development and validation of a panel of 14 P. cynomolgi microsatellite (MS) markers using the three P. cynomolgi reference genome strains and eleven nominally different laboratory reference strains collected between 1933 and 1967. We subsequently used the panel to explore the diversity of 18 field samples taken from wild monkeys in Malaysian Borneo to determine their effectiveness at strain differentiation. We show that the 14 MS are capable of characterizing the genetic diversity of laboratory strains and field isolates. We also surveyed the diversity of RBPs and DBPs in all samples, comparing the non-neutral diversity in these functionally important genes to the purported neutral diversity observed in the MS. Also, we discovered CNV of rbp genes among the nine P. cynomolgi strains. Finally, we were able to identify potential strain archiving errors and address some nomenclature discrepancies that have arisen during the long-term maintenance of P. cynomolgi laboratory stocks.
2. Materials and Methods
2.1 P. cynomolgi reference strains
DNA samples from 20 cryostabilate specimens representing eleven laboratory-maintained reference strains of P. cynomolgi were obtained from the Centers for Disease Control (Dr. John Barnwell), the American Type Cell Culture (ATCC), and the Malaria Reagent Reference Resource Center (MR4; Table 1). Samples were supplied as DNA, blood spots, or whole blood. DNA was extracted from blood spots or cryopreserved whole blood using a Qiagen DNeasy Blood and Tissue Kit (Qiagen®, Valencia, CA). All DNA samples were whole genome amplified (WGA) using the Qiagen REPLI-G Mini Kit (Qiagen®, Valencia, CA). WGA DNA was used to amplify microsatellite loci, while non-WGA DNA was used for multigene family amplification and sequencing.
Table 1. Plasmodium cynomolgi laboratory strains.
A unique identifier was assigned to each sample for this study. Alternating grey and white row shading indicates stocks nominally from the same strain. The sample name, sample code, and archive date (date on which the sample was isolated and frozen) were data supplied by the source. Original monkey host information was supplied by source or mined from literature, as were the historical aliases (see Supplementary Materials for references). The revised names for extant US stocks reflect strain identities indicated by our data in the case of M and B strains, and otherwise our preference where multiple aliases exist.
| No. | Unique sample identifier for this study | Sample source | Name on sample | Sample code or Monkey number | Sample archive date | Original monkey host | Aliases in literature and repositories | Revised name |
|---|---|---|---|---|---|---|---|---|
| 1 | Bastianellii_T644 | CDC | CynoB | Monkey #T644 | 21-Mar-69 | Macaca fascicularis | P. c. bastianellii; B strain; NIH; London | M/B |
| 2 | Bastianellii_9903 | CDC | B-Strain | Monkey #9903 | 12-Apr-01 | |||
| 3 | Bastianellii_MRA-350G | MR4 | Bastianellii | MRA-350G (DNA) | unknown | |||
| 4 | Berok_446 | CDC | Berok | Monkey # 84-46 | 9-Jan-01 | Macaca nemestrina | Berok; PT-II | Berok |
| 5 | PT-II_Monkey-8-42 | CDC | PT-II (Berok) | Monkey # 8-42 | 1-Dec-83 | |||
| 6 | Cambodian_9903 | CDC | Cambodian | Monkey #9903 | 31-Jul-01 | Macaca fascicularis | Cambodian | Cambodian |
| 7 | Ceylonensis_ATCC-30144 | ATCC | Ceylonensis | ATCC 30144, Lot 67687 | 17-Mar-71 | Macaca sinica | P.c. ceylonensis; C strain | Ceylon |
| 8 | Ceylonensis_Monkey-8-11 | CDC | Ceylonensis | Monkey 8-11 | 30-Nov-82 | |||
| 9 | Gombak MRA-550 | MR4 | Gombak | MRA-550 | unknown | none* | Gombak | Gombak |
| 10 | Gombak_RH0002 | CDC | Gombak | Monkey #RH0002 | 12-Nov-02 | |||
| 11 | Langur_RH0001 | CDC | Langur | Monkey #RH0001 | 12-Nov-02 | Presbytis entellus thersites | Langur | Langur |
| 12 | Mulligan_ATCC-30155 | ATCC | M Strain | ATCC 30155, Lot 67713 | 5-May-71 | Macaca fascicularis | Mulligan; Rockefeller; TC; M strain; P. c. cynomolgi | M/B |
| 13 | Mulligan_Monkey-18IX97 | CDC | Mulligan | Monkey #89-04 | 18-Sep-97 | |||
| 14 | Mulligan_30037 | ATCC | M-Strain | ATCC 30037, Lot 67427 | Mar-70 | |||
| 15 | Mulligan_T426 | CDC | CynoM | Monkey #T426 | 4-Aug-67 | |||
| 16 | NIH-U-53_Monkey-U-53 | CDC | NIH | Monkey #U-53 | 13-May-87 | unknown | NIH | M/B |
| 17 | PT-I_RH0001 | CDC | PT-I | Monkey #RH0001 | 27-Apr-04 | Macaca nemestrina | PT-I | PT-I |
| 18 | RO_ATCC-30146 | ATCC | RO Strain | ATCC 30146, Lot 67691 | 12-Mar-71 | Macaca mulatta | RO; Rossan | Rossan |
| 19 | RO_RNQ9 | CDC | Rossan | Monkey #RNQ9 | 17-Apr-12 | |||
| 20 | Smithsonian_MRA-351G | MR4 | Smithsonian | MRA-351G (DNA) | unknown | Macaca speciosa | Smithsonian | Smithsonian |
Abbreviations: CDC: Centers for Disease Control; MR4: Malaria Research and Reference Reagent Resource Centre; ATCC: American Type Culture Collection.
isolated from mosquito vector.
Note: ATCC and MR4 strain isolates originated from archived cryopreserved strains maintained by CDC.
2.2 P. cynomolgi field isolates
Sampling of wild long-tailed (M. fascicularis) and pig-tailed (M. nemestrina) macaques, two of the natural host species for P. cynomolgi, was conducted as previously described (Lee et al., 2011). Permission was granted and the study protocol for the capture, blood collection, and forest release of macaques was approved by the Sarawak Forestry Department (Permit Numbers: NPW.907.4.2-32, NPW.907.4.2-97, NPW.907.4.2-98, 57/2006 and 70/2007) and Sarawak Biodiversity Centre (Permit Number: SBC-RP-0081-BS). A total of 37 long-tailed macaques and seven pig-tailed macaques were sampled within a 30 km radius of Kapit town, Sarawak, Malaysian Borneo from 2004 to 2008. The macaques were anaesthetized by intramuscular injection of tiletamine and zolazepam and venous blood was drawn by a veterinarian. Macaques were then tagged with a microchip to prevent re-sampling. All efforts to collect blood samples from macaques were made at the trap sites and the macaques were released immediately into the forest after the blood samples had been obtained. Whole blood samples were frozen in a liquid nitrogen dry shipper and subsequently transferred to the Malaria Research Centre, University Malaysia Sarawak (UNIMAS) for further experiments. DNA was extracted from whole blood samples using the QIAamp DNA Blood Mini Kit (Qiagen®, Germany). Species detection of P. cynomolgi, P. knowlesi, P. fieldi, P. coatneyi, and P. inui was performed by nested PCR assays using species-specific primers as previously described (Lee et al., 2011). All procedures (DNA extraction and detection of species DNA by nested PCR assays) were conducted at UNIMAS.
2.3 Microsatellite genotyping
Microsatellite loci and primers were selected from a set of 175 (83 intergenic, 92 intragenic) published by Tachibana et al., (Tachibana et al., 2012), and NCBI Primer Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) used to confirm their uniqueness. Seventy MS were selected for optimization and tested at DNA concentrations of 0.5 ng/μL, 0.05 ng/μL, 0.005 ng/μL, and 0.0005 ng/μL, with ≤0.5ng/μL chosen as the optimal DNA concentration for our non-nested PCR protocol. Primer pairs were tested for specificity on nine simian malaria parasite species: P. simium (MRA-353), P. simiovale (MRA-488), P. fieldi (MRA-553), P. coatneyi (MRA-445), P. fragile (MRA-352), P. inui Mulligan (MRA-508), P. inui Taiwan I (MRA-551), P. inui shortii (MRA-551), and P. gonderi (MRA-447); three region-specific P. vivax reference strains Indonesia XIX (MRA-378), ONG (MRA-341), Pakchong (MRA-342G); and ten nominal P. cynomolgi strains: Berok (PT-II), Gombak, B (P.c. bastianellii), M (Mulligan), Smithsonian, PT-I, Rossan (RO), Cambodian, Ceylon (P. c. ceylonensis), and Langur (Table 1). All 14 validated MS markers had a 20 nucleotide tag sequence (5 ′- CCACGACGTTGTAAAACGAC-3′) added to the 5′ end of the forward primer. A universal primer was synthesized with the same nucleotide tag sequence and 5’-labeled with the phosphoramidite conjugate 6-FAM (Sigma-Aldrich Co. LLC, St. Louis, MO). This fluorescently-labeled tag sequence anneals to each forward primer during amplification, reducing the cost of synthesizing a probe specific for each primer-pair combination.
PCR amplifications (25 μL volume total) contained 2.5 μM forward primer, 5 μM reverse primer, 2.5 μM of the primer 6-FAM-5′- CCACGACGTTGTAAAACGAC-3', 2X Green GoTaq® Master Mix Reaction Buffer (pH 8.5) (Promega Corporation, Madison, WI), molecular grade water, and DNA (adjusted to 10–20 ng/μl) or whole-genome amplified DNA adjusted similarly. PCR cycling in an Eppendorf Mastercycler (Westbury, NY) was as follows: denaturation at 95°C for 5 min; followed by 40 cycles of denaturation at 95°C for 20 sec, annealing at 58°C for 45 sec, extension at 60°C for 45 sec; and a final extension at 72 °C for 5 min. PCR products were electrophoresed on 1.5% Agarose HS (Denville Scientific Inc., Metuchen, NJ) and visualized under ultraviolet.
Primer optimization and MS genotyping of laboratory strains was performed at New York University (NYU) (New York, NY, USA), while the MS genotyping of the field isolates was performed at the London School of Tropical Medicine and Hygiene (London, UK). To ensure synchronization between sites, identical positive controls served as a means to standardized allele scores by grouping each allele into locus- and size-specific bins. Any inconsistency in allele size between the identical positive controls resulted in a score adjustment which was applied to each of the field isolates tested. To minimize data analysis error, data generated at each site was analyzed by a single individual.
Amplicon sizes were determined using an ABI 3730 genetic analyzer (Applied Biosystems, Foster, CA).The allele length of each MS marker was determined using internal size standards (GeneScan 500 LIZ Size Standard, Applied Biosystems) with GeneMapper v4.0 software (ABI, Foster City, CA). Only alleles detected with a peak height ≥ 200 fluorescent units were considered. Mixed genotype infections were defined as two or more MS loci containing multiple alleles, and minor peaks were defined as peaks equal to one-third the height of the major peak. Negative controls included reactions containing water instead of DNA to monitor for contamination, and the use of P. vivax and P. falciparum DNA to rule out spurious bands that may be generated by non-specific hybridization of human DNA.
2.4 Population genetic and phylogenetic analyses
HE (expected heterozygosity) was calculated using the formula where p is the frequency of the ith allele and n is the number of alleles sampled, as implemented in Arlequin 3.11 (Excoffier et al., 2005). Distance matrices were developed using Nei's DA calculation for genetic distance via POPTREE2 (Takezaki et al., 2010).
2.5 P. cynomolgi RBP and DBP amplification and sequencing
Primers for P. cynomolgi rbp1a, rbp2, rbp3, dbp1, and dbp2 genes were designed manually by inspecting conserved regions of each sequence from P. cynomolgi strains B, Berok, and Cambodian. All sequences were obtained from GenBank (PCYB_033090, PCYB_063270, PCYB_071060, PCYB_147650, JQ422036, JQ422035, JQ422037, JQ422043, AB617789, AB617788, JQ422044, JQ422050) and aligned in Sequencher (version 5.3, Gene Codes Corporation, Ann Arbor, MI USA) or Geneious Software v6.1.6 (Kearse et al., 2012). Primers and PCR conditions are shown in Supplementary Table 3 and Supplementary Table 4, and schematics showing the combinations and locations of these primers are provided in Supplementary Figure 1 and Supplementary Figure 2. GenBank accession numbers correlated to individual isolates and genes can be found in Supplementary Table 5.
PCR amplifications were performed in a total volume of 20 μl in the presence of 10X LA PCR Buffer II (Mg2+free), 2.5mM MgCl2, 400 μM dNTPs, 0.2 μ M primers and 2.5 U/50 μl of TaKaRa LA Taq Polymerase (TaKaRa Bio Inc, Mountain View, CA). The PCR products were analyzed on a 1% agarose gel and purified (QIAquick Gel Extraction Kit, Valencia, CA). Purified products were sequenced using BigDye v3.1 Terminator (Applied Biosystems, Grand Island, NY) and then analyzed on an ABI machine (Applied Biosystems, Grand Island, NY). Sequences were assembled using Sequencher 5 (Gene Codes, Ann Arbor, MI).
2.6 RBP and DBP phylogenetic analyses
Sequence alignments were constructed using Sequencher and then adjusted manually. Regions that could not be unambiguously aligned were omitted from subsequent phylogenetic analyses. Trees were constructed from nucleotide sequences of rbp1a (1,965 bp), rbp3 (4,200 bp), dbp1 (1,866 bp), dbp2 (1,026 bp), and concatenated nucleotide sequence of rbp1a, rbp3, dbp1 and dbp2 (9,057 bp) using Neighbor Joining in the Geneious software package with bootstrap support of 1000 replicates using the HKY substitution model.
3. Results
3.1 Validation of a panel of P. cynomolgi microsatellite markers in laboratory strains
We developed a panel of 14 microsatellite markers from a set of 175 originally mined from the P. cynomolgi strain B and strain Berok genomes (Tachibana et al., 2012). After excluding loci on the basis of primer sequence similarity to P. vivax and missing sequence data using NCBI Primer BLAST, a total of 70 were selected for optimization that: (1) conformed to motif size >1 - ≤4 bp, (2) were expected to have a fragment length between 100 – 400 bp, (3) included both purines and pyrimidines, and (4) were distributed across the 14 P. cynomolgi chromosomes. None of the 70 MS markers were amplified from three P. vivax DNA strains tested. However, amplification of bands from simian malaria species P. simium, P. simiovale, P. fieldi, P. coatneyi, P. fragile, P. inui, and P. gonderi resulted in the exclusion of 36 MS loci. Of the remaining 34 MS markers, 20 were discarded due to non-specific amplification of nine P. cynomolgi reference strains, leaving 14 MS markers. The characteristics of the 14 MS markers including the primers used to amplify them are shown in Supplementary Table 1.
We tested the final panel of 14 MS markers on a total of 20 specimens nominally representing 11 P. cynomolgi reference strains. These strains have been used for as long as 80 years in research laboratories worldwide. Figure 1 illustrates their geographical origins and years of isolation. Six strains were sampled more than once, including three strain B samples, four M samples, two Berok samples, two Gombak samples, two Ceylon samples, and two Rossan samples (Table 1). Some of these strains are archived at the ATCC and/or the MR4, while others were contributions from the Primate Malaria Archive at the Centers for Disease Control and Prevention (CDC), where all P. cynomolgi strains and other primate malaria species are maintained.
Our genotyping showed the specimens nominally representing B, M, and NIH strains to be genetically identical (Table 2), and hereafter we designate these stocks collectively as the ‘M/B strain’. Our subsequent data analysis thus encompassed nine unique laboratory strains. The average number of alleles per locus across the 14 MS loci for the nine laboratory strains was 5.85 ± 0.95, with a range of from four to seven alleles per locus (Table 3). The HE was 0.87 ± 0.09, with a minimum of 0.58 and a maximum of 0.94. Our panel of 14 P. cynomologi MS was thus highly polymorphic and potentially useful as markers for genetic diversity studies.
Table 2. Allele scoring for P. cynomolgi laboratory strains.
The ‘allele score’ is the size, in nucleotides, of the amplicon for each of the 14 MS markers (columns) from each sample (rows).
| 1.307 | 2.36 | 3.574 | 4.41 | 4.462 | 5.956 | 6.455 | 7.1006 | 8.1086 | 10.179 | 10.621 | 11.1096 | 12.286 | 14.429 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bastianellii_9903 | 315 | 215 | 274 | 287 | 264 | 212 | 220 | 292 | 208 | 135 | 289 | 167 | 206 | 341 |
| Bastianellii_MRA-350G | 315 | 215 | 274 | 287 | 264 | 212 | 220 | 292 | 208 | 135 | 289 | 167 | 206 | 341 |
| NIH-U-53_Monkey-U-53 | 315 | 215 | 274 | 287 | 264 | 212 | 220 | 292 | 208 | 135 | 289 | 167 | 206 | 341 |
| Mulligan_Monkey-18IX97 | 315 | 215 | 274 | 287 | 264 | 212 | 220 | 292 | 208 | 135 | 289 | 167 | 206 | 341 |
| Mulligan_ATCC-30155 | 315 | 215 | 274 | 287 | 264 | 212 | 220 | 292 | 208 | 135 | 289 | 167 | 206 | 341 |
| Mulligan_30037 | 315 | 215 | 274 | 287 | 264 | 212 | 220 | 292 | 208 | 135 | 289 | 167 | 206 | 341 |
| Berok_446 | 300 | 192 | 218 | 290 | 252 | 164 | 214 | 308 | 193 | 121 | 241 | 140 | 155 | 331 |
| PT-II_Monkey-8-42 | 300 | 192 | 218 | 290 | 252 | 164 | 214 | 308 | 193 | 121 | 241 | 140 | 155 | 331 |
| Cambodian_9903 | 327 | 190 | 282 | 293 | 255 | 218 | 223 | 244 | 208 | 125 | 281 | 155 | 209 | 333 |
| Ceylonensis_Monkey-8-11 | 327 | 225 | 302 | 287 | 258 | 233 | 223 | 292 | 214 | 125 | 250 | 150 | 206 | 337 |
| Ceylonensis_ATCC-30144 | 327 | 225 | 302 | 287 | 258 | 233 | 223 | 292 | 214 | 125 | 250 | 150 | 206 | 337 |
| Langur_RH0001 | 312 | 225 | 242 | 287 | 249 | 218 | 223 | 344 | 208 | 125 | 250 | 155 | 206 | 337 |
| Smithsonian_MRA-351G | 294 | 190 | 266 | 290 | 264 | 212 | 211 | 284 | 214 | 133 | 285 | 159 | 206 | 339 |
| PT-I_RH0001 | 294 | 190 | 302 | 296 | 264 | 209 | 211 | 296 | 223 | 133 | 289 | 163 | 206 | 335 |
| Gombak_RH0002 | 318 | 190 | 210 | 281 | 270 | 170 | 232 | 236 | 247 | 119 | 253 | 140 | 158 | 321 |
| RO_ATCC-30146 | 312 | 232 | 302 | 293 | 267 | 218 | 217 | 292 | 214 | 121 | 285 | 155 | 206 | 333 |
Table 3. Characteristics of 14 MS loci amplified from laboratory strains and field isolates.
The number of alleles, allele size range, and expected heterozygosity for the 14 MS loci is shown for nine lab strains, 18 field isolates, and then all strains and isolates combined. MS loci 2.36, 4.462, 11.1096, and 12.286 consistently failed to amplify the field isolates and were excluded from subsequent analyses.
| ID | Chr. no. | Motif | No. of alleles | Allele size range | Expected heterozygosity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lab strains | Field isolates | All strains | Lab strains | Field isolates | All strains | Lab strains | Field isolates | All strains | |||
| 1.307 | 1 | TCT | 6 | 6 | 11 | 294-327 | 285-351 | 285-351 | 0.92 | 0.76 | 0.84 |
| 2.36 | 2 | CA | 5 | ND | ND | 190-232 | ND | ND | 0.81 | ND | ND |
| 3.574 | 3 | GTGA | 7 | 10 | 14 | 210-302 | 206-282 | 206-302 | 0.92 | 0.93 | 0.95 |
| 4.41 | 4 | TCT | 5 | 10 | 13 | 281-296 | 273-315 | 273-315 | 0.86 | 0.94 | 0.93 |
| 4.462 | 4 | TCC | 7 | ND | ND | 249-270 | ND | ND | 0.92 | ND | ND |
| 5.956 | 5 | AAG | 6 | 4 | 10 | 164-233 | 167-206 | 164-233 | 0.89 | 0.65 | 0.87 |
| 6.455 | 6 | AGG | 6 | 4 | 7 | 211-232 | 211-229 | 211-232 | 0.89 | 0.47 | 0.74 |
| 7.1006 | 7 | ATAC | 7 | 6 | 13 | 236-344 | 153-260 | 153-344 | 0.92 | 0.82 | 0.90 |
| 8.1086 | 8 | GAG | 5 | 9 | 11 | 193-247 | 184-220 | 184-247 | 0.83 | 0.91 | 0.90 |
| 10.179 | 10 | TA | 5 | 5 | 8 | 119-135 | 117-127 | 117-135 | 0.86 | 0.71 | 0.85 |
| 10.621 | 10 | TGTA | 6 | 10 | 14 | 241-289 | 233-305 | 233-305 | 0.92 | 0.92 | 0.95 |
| 11.1096 | 11 | TGTA | 6 | ND | ND | 140-167 | ND | ND | 0.89 | ND | ND |
| 12.286 | 12 | AAT | 4 | ND | ND | 155-209 | ND | ND | 0.58 | ND | ND |
| 14.429 | 14 | AT | 7 | 8 | 15 | 321-341 | 311-329 | 311-341 | 0.94 | 0.90 | 0.95 |
ND: not determined.
MS allele frequency data was used to calculate a distance matrix for the nine genetically distinct laboratory strains (Supplementary Table 2). With the caveat that the sample size is small, Berok and Gombak appear more distant from other Peninsular Malaysian isolates (M/B, PT-I, and Smithsonian), and the four non-Malaysian isolates (Cambodian, Ceylon, Langur, and Rossan appear more closely related to one another than to the isolates from Peninsular Malaysia.
3.2 Allelic diversity of microsatellite markers in P. cynomolgi field isolates
We next used the 14 MS loci to genotype extant P. cynomolgi isolates collected from long-tailed and pig-tailed macaques in the Malaysian state of Sarawak on the island of Borneo during 2004-2008. A total of 27 isolates were collected from eight sites within a 30 km radius of Kapit Town, Sarawak (Figure 1). MS loci 2.36, 4.462, 11.1096, and 12.286 consistently failed to amplify the field isolates and were excluded from subsequent analyses (Table 3). Largely due to host DNA contamination, low parasitemia and sample volume are characteristic of clinically-isolated Plasmodium species infections. In addition to high host contamination, numerically high mixed-species and mixed-genotype infection rates are expected to further lower the DNA yield for individual species and clones within the infection. The amplification failures observed in this study were likely due to the low DNA quality and quantity attributable to these factors. However, pathological parasite suppression by competing species and/or clones could also play a role.
Six (33.3%) of the 18 field isolates were identified as mixed genotype infections. These isolates were not excluded in subsequent analyses because the major clone could be clearly identified. No shared haplotypes were identified in the 18 field isolates and the average number of alleles per MS locus was 7.20 ± 2.49, with a range of four to ten alleles per locus. The average HE across the 10 MS loci was 0.80 ± 0.15, with a minimum of 0.47 and a maximum of 0.94, very similar to that observed in the laboratory strains (average HE 0.87 ± 0.09, with a minimum of 0.58 and a maximum of 0.94). Haplotype information for the field isolates are shown in Table 4.
Table 4. Allele scoring for P. cynomolgi field isolates.
Alleles scored in each of the field isolates genotyped at 10 microsatellite loci. Italicized isolate names represent field-collected isolates. Highlighted (light grey) allele scores represent the predominant allele within a mixed genotype infections (n = 6 mixed-genotype infections), while single unmixed alleles are not highlighted. Missing genotype data is reflected by gaps.
| Strain/Isolate | 1.307 | 3.574 | 4.41 | 5.956 | 6.455 | 7.1006 | 8.1086 | 10.179 | 10.621 | 14.429 |
|---|---|---|---|---|---|---|---|---|---|---|
| LT040 | 258 | 315 | 173 | 229 | 240 | 211 | 123 | 257 | ||
| LT042 | 315 | 246 | 203 | 229 | 205 | 117 | 265 | 323 | ||
| LT043 | 285 | 282 | 167 | 229 | 214 | 127 | 265 | 325 | ||
| LT058 | 285 | 282 | 293 | 217 | 248 | 205 | 265 | 311 | ||
| LT072 | 294 | 246 | 167 | 229 | 244 | 202 | 123 | 281 | ||
| LT051 | 294 | 262 | 300 | 229 | 240 | 184 | 117 | 241 | 325 | |
| LT073 | 288 | 234 | 306 | 206 | 229 | 252 | 214 | 233 | 325 | |
| LT082 | 294 | 258 | 290 | 203 | 217 | 240 | 202 | 123 | 265 | |
| PTK012 | 351 | 206 | 275 | 229 | 256 | 193 | 117 | 237 | 311 | |
| LT020 | 285 | 266 | 278 | 203 | 211 | 244 | 214 | 123 | 257 | 315 |
| LT049 | 297 | 254 | 300 | 203 | 217 | 240 | 208 | 123 | 257 | 319 |
| LT055 | 294 | 254 | 303 | 203 | 220 | 244 | 202 | 123 | 281 | 319 |
| LT065 | 297 | 246 | 296 | 203 | 229 | 248 | 199 | 119 | 277 | 311 |
| LT070 | 294 | 242 | 300 | 167 | 229 | 260 | 202 | 125 | 273 | 329 |
| LT084 | 294 | 246 | 309 | 203 | 229 | 244 | 211 | 123 | 305 | 315 |
| LT089 | 294 | 270 | 290 | 206 | 229 | 244 | 220 | 123 | 233 | 327 |
| LT091 | 294 | 266 | 296 | 203 | 229 | 240 | 211 | 125 | 297 | 317 |
| PTK014 | 306 | 206 | 273 | 170 | 229 | 153 | 193 | 121 | 237 | 311 |
3.3 Diversity of RBP and DBP genes in P. cynomolgi laboratory strains
We next surveyed the diversity of P. cynomolgi reticulocyte binding proteins (RBPs) and Duffy binding proteins (DBPs) in P. cynomolgi laboratory strains. The status of rbp1, rbp2, rbp3, dbp1, and dbp2 genes was determined by designing primers (Supplementary Table 3, Supplementary Figure 1) for the flanking regions of each gene, such that its presence or absence could be determined from the size of the PCR product. Similar to our previous results that found rbp1b in the Berok strain but not in the M/B and Cambodian strains (Tachibana et al., 2012), we found rbp1b in the Gombak strain but not in the Rossan, Smithsonian, Ceylon, or Langur strains (Table 5). No consistent amplification could be obtained from the PT-I strain. Furthermore, P. cynomolgi strains that lack the rbp2a gene (Berok and Gombak) contained an rbp1b gene, and while those that lacked rbp1b contained rbp2a. No copy number variation was observed for any of the other rbp genes, or for dbp1 and dbp2 genes.
Table 5. Presence of rbp and dbp genes in nine P. cynomolgi laboratory reference strains.
Strain names are the Revised Names from Table 1. Results for rbp1b in bold were previously published by us.
| Berok* | Gombak* | M/B* | Smithsonian | Rossan* | Ceylon* | Langur | Cambodian | PT-I | |
|---|---|---|---|---|---|---|---|---|---|
| rbp1a | + | + | + | + | + | + | + | + | ND |
| rbp1b | + | + | - | - | - | - | - | - | ND |
| rbp2a | - | - | + | + | + | + | + | + | + |
| rbp2b | + | + | + | + | + | + | + | + | ND |
| rbp2c | + | + | + | + | + | + | + | + | ND |
| rbp2d | + | + | + | + | + | + | + | + | + |
| rbp2e | + | + | + | + | + | NP | + | + | ND |
| rbp2f | + | + | + | + | NP | NP | + | + | + |
| rbp3 | + | + | + | + | + | + | + | + | ND |
| dbp1 | + | + | + | + | + | + | + | + | + |
| dbp2 | + | + | + | + | + | + | + | + | + |
represents multiple samples analyzed
+: gene present; -: gene absent; ND: not determined due to insufficient DNA; NP: no product;
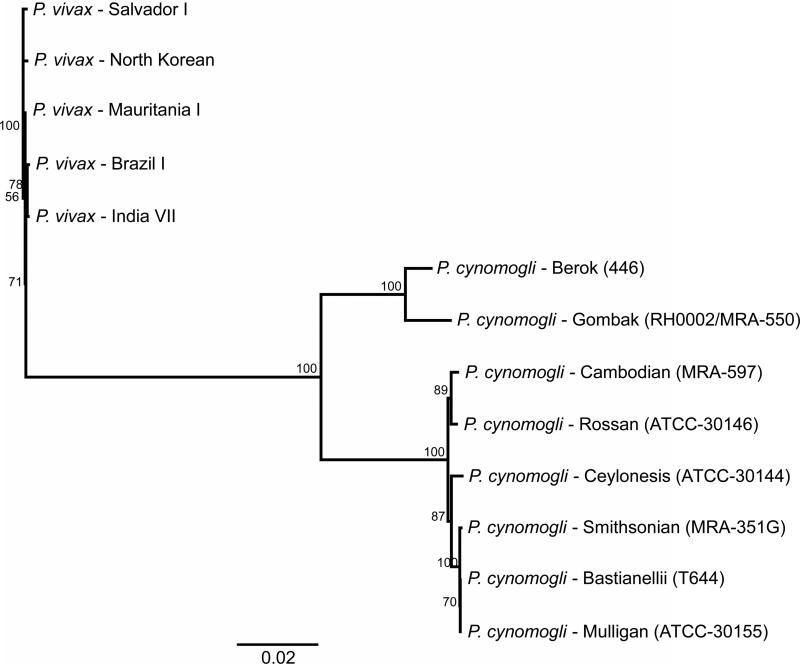
We sampled the genetic diversity of RBP and DBP genes by partially sequencing amplification products of rbp1a, rbp3, dbp1, and dbp2 genes in the P. cynomolgi laboratory strains. For rbp1a and rbp3, five and six different combinations of primers, respectively, were used to amplify the whole gene in fragments. For dbp1 and dbp2, we designed primers targeting the N-terminal cysteine-rich region, where the binding domain is located. We generated a phylogenetic tree using concatenated nucleotide sequences of these genes from eight P.cynomolgi samples and five P. vivax reference strains (Figure 2). Two main clades of P. cynomolgi were apparent, one containing the Berok and Gombak strains, and a second containing six strains in two subclades: the two M/B sequences plus Smithsonian and Ceylon in one branch, and the Cambodian and Rossan strains in a second branch. Sequencing and trees furthermore confirmed 100% identity of B strain and M strain samples. We also generated dendrograms for each gene (Supplementary Figure 3). Berok and Gombak strains consistently branch together in these trees as well.
Figure 2. Dendrogram showing inter-isolate differentiation based on concatenated P. cynomolgi nucleotide sequences from four genes.
Dendrogram based on distance matrices from 8031 bp of concatenated nucleotide sequence of rbp1a, rbp3, and dbp1 from eight P. cynomolgi strains and five P. vivax reference strains built by Neighbor Joining with 1000 bootstraps using the HKY substitution model. The numbers shown along nodes represent bootstrap values. Identical topology was obtained using Maximum Likelihood (not shown).
4. Discussion
The publication of multiple reference genomes for P. cynomolgi and P. vivax presented an opportunity to develop molecular tools for understanding the global diversity within each species. We used data from three P. cynomolgi reference genomes (Tachibana et al., 2012) to develop genetic tools to assess genetic variation of this species. First, we developed a panel of 14 P. cynomolgi-specific microsatellite (MS) markers. Our results indicate that the 14 MS loci optimized in this study are robust genetic markers for measuring diversity. Distributed across 12 of 14 P. cynomolgi chromosomes, the MS panel was shown to be sensitive, specific, and capable of differentiating laboratory strains isolated from various sources. An earlier publication developed a panel of 13 P. vivax microsatellite loci and tested them in 108 P. vivax samples from eight localities in Asia, Africa, South America, and New Guinea, as well as in nine other Plasmodium species (Leclerc et al. 2004). These authors demonstrated significant overlap of microsatellite loci between P. vivax and several other species, including P. cynomolgi, concluding that the flanking regions of their microsatellite loci appear to be genetically similar, and thus evolutionarily conserved. To avoid such overlap, species-specificity of each marker, which can be a concern when deploying this methodology on field isolates that can contain more than one Plasmodium species, was tested and confirmed using nine different simian malaria species and three P. vivax laboratory strains. To ensure the utility of these MS loci as a genotyping tool for field collected isolates, 18 non-human primate samples (collected from Kapit Town, Sarawak, Malaysian Borneo from 2004 to 2008) were tested. High genetic diversity among P. cynomolgi isolates combined with the high mixed species prevalence (specifically with P. knowlesi) reflects the high parasite transmission within the macaque population. However, limitations in the study design prevented the consideration of high genetic diversity attributable to chronic infections and hypnozoite relapse. Whether or not P. cynomolgi in the macaque population is associated with different macaque hosts, as described in P. knowlesi populations (Divis et al., 2015), needs to be determined by more sampling.
Observation of strain-specific CNV of rbp and dbp genes, noted first during genome sequencing of P. cynomolgi (Tachibana et al. 2012) is described more comprehensively here, We note the most variation in the rbp1b and rbp2a. We observed that all P. cynomolgi strains which contain one of this pair lack the other, e.g., the Berok and Gombak strains lack rbp2a but contain rbp1b. During the invasion of reticulocytes by P. cynomolgi, perhaps the presence of rbp1b could compensate for the absence of rbp2a and vice versa. The parasite might first lose rbp1b and then gain rbp2a or the other way around. Such changes could underlie important phenotypic differences. Polymorphism of a gene encoding an erythrocyte binding protein in rodent malaria P. yoelii yoelii was associated with disease virulence (Otsuki et al., 2009; Pattaradilokrat et al., 2009). Particular alleles of two erythrocyte binding protein genes in P. knowlesi were found to be associated with specific disease progression profiles, such as differences in parasitaemia, and markers of disease severity such as hemoglobin levels, platelet levels, renal dysfunction, etc. in a population of knowlesi malaria patients (Ahmed et al., 2014). These two genes are also known to show differential binding between macaque and human erythrocytes (Semenya et al., 2012). Strain-specific CNV has also been found in the antigen gene merozoite surface protein 3 (MSP3) of P. cynomolgi strains B and Berok (Rice et al. 2014).
All P. cynomolgi strains that we tested have two very similar dbp genes (~92% DNA identity) and one other closely related gene. A duplication of dbp in P. vivax, which typically contains only one dbp gene, was prevalent in Madagascar where the highest frequencies of P. vivax-infected, Duffy-negative people were reported, raising the possibility that P. vivax may be able to invade Duffy-negative erythrocytes through the duplication of dbp (Menard et al., 2013). Similarly, P. cynomolgi might enable the parasite to infect Duffy-negative red blood cells of macaques, although the apparent rarity of Duffy-negative macaques in the wild (Palatnik and Rowe, 1984) argues against this.
We observed two subclades of P. cynomolgi, one of which contains Berok and Gombak, in phylogenetic trees of rbp and dbp genes. This pattern in P. cynomolgi has been reported for several antigen genes, including circumsporozoite protein gene (CSP) (Pacheco et al., 2012b), merozoite surface protein 1 (MSP1) (Tanabe et al., 2007), MSP3 (Rice et al., 2014), MSP8, and MSP10 (Pacheco et al., 2012a), suggesting that the fact that the genetic differences shown by Berok and Gombak might be due to the geographical location or host variation. Further sampling of this widespread parasite of macaques is clearly needed to investigate in detail whether there is geographical population genetic structure in P. cynomolgi, and whether particular polymorphic genes are under selection. We suggest that the loci that we present here will be useful for this purpose, and will allow interesting comparisons between patterns seen in this and other parasite species.
Finally, during the MS optimization process, we observed that stocks of some commonly available strains of P. cynomolgi are genetically identical. As early as the mid-1980s, published immunological, electrophoretic, and genetic evidence (Cochrane et al., 1985) suggested that stocks maintained at NIH of P. c. bastianellii (also known as the B or NIH strain) from stock originally sent to the USA from London by Professor P.C.C. Garnham in 1959 (Bennett et al., 1966), and the Mulligan strain (also known as the M, Rockefeller, or TC strain, and at one point given the subspecific name P. c. cynomolgi) (see Supplemental Materials), originally from stock imported from India to the USA during World War II) (Eyles et al., 1963; Schmidt et al., 1949) were similar if not identical. This confusion of the B and M strains had been noted briefly by one of us (JWB) before (Tachibana et al., 2012), and probably resulted from an error committed over the course of the complex monkey passage and stock exchange history of these two strains at NIH and between laboratories since 1959. The oldest samples of B and M strains archived at the CDC, which originated from the old NIH/NIAID Laboratory of Parasite Chemotherapy, date from 1969 and 1967, respectively. As these two specimens are genetically identical in our study, we conclude that this archiving/strain mix-up must have occurred no later than 1967. We here refer to these M and B samples as the ‘M/B strain’ to acknowledge the ambiguity. However, review of the M and B strain literature, and of US specimen archives, and public database sequences annotated as M or B strain (and aliases thereof), and comparison with our sequences, suggests that all B and M stocks currently available from US sources, as well as public database ‘B strain’ sequences derived from US stock (including the reference P. cynomolgi strain B genome), are all probably M strain. Public database analyses, however suggest that uncontaminated B strain stock may still exist in India (SAS, unpublished). Definitive sorting of M and B stocks and sequences will require sequencing of such uncontaminated B strain stock, if it still exists.
5. Conclusions
The 14 P. cynomologi MS described here were validated against nine unique laboratory strains as being highly polymorphic, and thus useful as markers for genetic diversity studies of the species, as indicated by successful genotyping of field isolates. Diversity in this species is also manifested structurally by strain-specific presence/absence of some rbp genes, as well as by the sequences of rbp and dbp genes. Our study further reveals that extant US stocks of strains M and B, the first two P. cynomolgi strains isolated, are genetically identical, likely reflecting a long-standing archiving error.
Supplementary Material
Highlights.
We generated the first panel of Plasmodium cynomolgi microsatellite markers.
High genetic diversity revealed in 18 wild Malaysian monkey isolates.
Sequenced invasion genes confirmed genetic diversity and copy number variation.
We identified strain archiving errors and address some nomenclature discrepancies.
Acknowledgements
ZL is supported by the MacCracken Program in the Graduate School of Arts and Science at New York University (New York, NY). This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) International Centers of Excellence in Malaria Research Grant U19AI089676 to JMC. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JH, Hudson DE, Torii M, Ward GE, Wellems TE, Aikawa M, Miller LH. The Duffy Receptor Family of Plasmodium knowelsi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. A family of erythrocyte binding proteins of malaria parasites. Proc Nat Acad Sci USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AM, Pinheiro MM, Divis PC, Siner A, Zainudin R, Wong IT, Lu CW, Singh-Khaira SK, Millar SB, Lynch S, Willmann M, Singh B, Krishna S, Cox-Singh J. Disease progression in Plasmodium knowlesi malaria is linked to variation in invasion gene family members. PLoS Neglect Trop D. 2014;8:e3086. doi: 10.1371/journal.pntd.0003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa M, Miller LH, Rabbege J. Caveola--vesicle complexes in the plasmalemma of erythrocytes infected by Plasmodium vivax and P cynomolgi. Unique structures related to Schuffner's dots. Am J Pathol. 1975;79:285–300. [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitol. 1999;119:113–25. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Arnott A, Barry AE, Reeder JC. Understanding the population genetics of Plasmodium vivax is essential for malaria control and elimination. Malar J. 2012;11:14. doi: 10.1186/1475-2875-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nature Struct Mol Biol. 2011;18:908–914. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GF, Warren M, Cheong WH. Biology of the Simian Malarias of Southeast Asia. II. The Susceptibility of Some Malaysian Mosquitoes to Infection with Five Strains of Plasmodium cynomolgi. J Parasitol. 1966;52:625. [PubMed] [Google Scholar]
- Branch OH, Sutton PL, Barnes C, Castro JC, Hussin J, Awadalla P, Hijar G. Plasmodium falciparum genetic diversity maintained and amplified over 5 years of a low transmission endemic in the Peruvian Amazon. Mol Biol Evol. 2011;28:1973–1986. doi: 10.1093/molbev/msq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Das A, Escalante AA. Genomics, population genetics and evolutionary history of Plasmodium vivax. Advances in parasitology. 2013;81:203–222. doi: 10.1016/B978-0-12-407826-0.00005-9. [DOI] [PubMed] [Google Scholar]
- Chan ER, Barnwell JW, Zimmerman PA, Serre D. Comparative analysis of field-isolate and monkey-adapted Plasmodium vivax genomes. PLoS Neglect Trop D. 2015;9:e0003566. doi: 10.1371/journal.pntd.0003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coatney GR, Collins WE, Warren M, Contacos PG. The Primate Malarias. U. S. Government Printing Office; Washington, D.C.: 1971. [Google Scholar]
- Coatney GR, Elder HA, Contacos PG, Getz ME, Greenland R, Rossan RN, Schmidt LH. Transmission of the M strain of Plasmodium cynomolgi to man. Amer J Trop Med Hyg. 1961;10:673–678. doi: 10.4269/ajtmh.1961.10.673. [DOI] [PubMed] [Google Scholar]
- Cochrane AH, Gwadz RW, Ojo-Amaize E, Hii J, Nussenzweig V, Nussenzweig RS. Antigenic diversity of the circumsporozoite proteins in the Plasmodium cynomolgi complex. Mol Biochem Parasitol. 1985;14:111–124. doi: 10.1016/0166-6851(85)90110-0. [DOI] [PubMed] [Google Scholar]
- Contacos PG, Elder HA, Coatney GR, Genther C. Man to man transfer of two strains of Plasmodium cynomolgi by mosquito bite. Amer J Trop Med Hyg. 1962;11:186–193. doi: 10.4269/ajtmh.1962.11.186. [DOI] [PubMed] [Google Scholar]
- Dissanaike AS, Nelson P, Garnham PCC. Two new malaria parasites, Plasmodium cynomolgi ceylonensis subsp. nov. and Plasmodium fragile sp. nov., from monkeys in Ceylon. Ceylon J Med Sci. 1965;14:1–9. [Google Scholar]
- Divis PC, Singh B, Anderios F, Hisam S, Matusop A, Kocken CH, Assefa SA, Duffy CW, Conway DJ. Admixture in Humans of Two Divergent Plasmodium knowlesi Populations Associated with Different Macaque Host Species. PLoS Path. 2015;11:e1004888. doi: 10.1371/journal.ppat.1004888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enea V, Galinski M, Schmidt E, Gwadz R, Nussenzweig RS. Evolutionary profile of the circumsporozoite gene of the Plasmodium cynomolgi complex. J Mol Biol. 1986;188:721–726. doi: 10.1016/s0022-2836(86)80017-1. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinf Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Eyles DE. The exoerythrocytic cycle of Plasmodium cynomolgi and P. cynomolgi bastianellii in the rhesus monkey. Amer J Trop Med Hyg. 1960;9:543–555. doi: 10.4269/ajtmh.1960.9.543. [DOI] [PubMed] [Google Scholar]
- Eyles DE, Coatney GR, Getz ME. Vivax-type malaria parasite of macaques transmissible to man. Science. 1960;131:1812–1813. doi: 10.1126/science.131.3416.1812. [DOI] [PubMed] [Google Scholar]
- Eyles DE, Dunn FL, Warren M, Guinn E. Plasmodium Coatneyi from the Philippines. J Parasitol. 1963;49:1038. [PubMed] [Google Scholar]
- Fang XD, Kaslow DC, Adams JH, Miller LH. Cloning of the Plasmodium vivax Duffy receptor. Mol Biochem Parasitol. 1991;44:125–132. doi: 10.1016/0166-6851(91)90228-x. [DOI] [PubMed] [Google Scholar]
- Galinski MR, Arnot DE, Cochrane AH, Barnwell JW, Nussenzweig RS, Enea V. The circumsporozoite gene of the Plasmodium cynomolgi complex. Cell. 1987;48:311–319. doi: 10.1016/0092-8674(87)90434-x. [DOI] [PubMed] [Google Scholar]
- Galinski MR, Medina CC, Ingravallo P, Barnwell JW. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- Garnham PCC. A new sub-species of Plasmodium cynomolgi. Riv di Parassit. 1959;20:273–278. [Google Scholar]
- Garnham PCC. Malaria Parasites and Other Haemosporidia. Blackwell Scientific Publications; Oxford, U.K.: 1966. [Google Scholar]
- Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, Temperley WH, Kabaria CW, Tatem AJ, Manh BH, Elyazar IR, Baird JK, Snow RW, Hay SI. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Neglect Trop D. 2010;4:e774. doi: 10.1371/journal.pntd.0000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester J, Chan ER, Menard D, Mercereau-Puijalon O, Barnwell J, Zimmerman PA, Serre D. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Neglect Trop D. 2013;7:e2569. doi: 10.1371/journal.pntd.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krotoski WA, Bray RS, Garnham PC, Gwadz RW, Killick-Kendrick R, Draper CC, Targett GA, Krotoski DM, Guy MW, Koontz LC, Cogswell FB. Observations on early and late post-sporozoite tissue stages in primate malaria. II. The hypnozoite of Plasmodium cynomolgi bastianellii from 3 to 105 days after infection, and detection of 36- to 40-hour pre-erythrocytic forms. Amer J Trop Med Hyg. 1982a;31:211–225. [PubMed] [Google Scholar]
- Krotoski WA, Garnham PC, Bray RS, Krotoski DM, Killick-Kendrick R, Draper CC, Targett GA, Guy MW. Observations on early and late post-sporozoite tissue stages in primate malaria. I. Discovery of a new latent form of Plasmodium cynomolgi (the hypnozoite), and failure to detect hepatic forms within the first 24 hours after infection. Amer J Trop Med Hyg. 1982b;31:24–35. [PubMed] [Google Scholar]
- Leclerc MC, Durand P, Gauthier C, Patot S, Billotte N, Menegon M, Severini C, Ayala FJ, Renaud F. Meager genetic variability of the human malaria agent Plasmodium vivax. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14455–14460. doi: 10.1073/pnas.0405186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, Cox-Singh J, Singh B. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS pathogens. 2011;7:e1002015. doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. Ueber malaria beim affen. Med Klin Berl. 1907;3:579–580. [Google Scholar]
- Menard D, Chan ER, Benedet C, Ratsimbasoa A, Kim S, Chim P, Do C, Witkowski B, Durand R, Thellier M, Severini C, Legrand E, Musset L, Nour BY, Mercereau-Puijalon O, Serre D, Zimmerman PA. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Neglect Trop D. 2013;7:e2489. doi: 10.1371/journal.pntd.0002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. New Eng J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- Mulligan HW. Descriptions of two species of monkey plasmodium isolated from Silenus irus. Arch f Protist. 1935;84:285–314. [Google Scholar]
- Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, Saif S, Gujja S, Goldberg JM, Young S, Zeng Q, Chapman SB, Dash AP, Anvikar AR, Sutton PL, Birren BW, Escalante AA, Barnwell JW, Carlton JM. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nature Genet. 2012;44:1046–1050. doi: 10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Dinh P, Gardner AL, Campbell CC, Skinner JC, Collins WE. Cultivation in vitro of the vivax-type malaria parasite Plasmodium cynomolgi. Science. 1981;212:1146–1148. doi: 10.1126/science.7233207. [DOI] [PubMed] [Google Scholar]
- Ntumngia FB, King CL, Adams JH. Finding the sweet spots of inhibition: understanding the targets of a functional antibody against Plasmodium vivax Duffy binding protein. Int J Parasitol. 2012;42:1055–1062. doi: 10.1016/j.ijpara.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RP, Broude NE, Macedo AM, Cantor CR, Smith CL, Pena SD. Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc Nat Acad Sci USA. 1998;95:3776–3780. doi: 10.1073/pnas.95.7.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki H, Kaneko O, Thongkukiatkul A, Tachibana M, Iriko H, Takeo S, Tsuboi T, Torii M. Single amino acid substitution in Plasmodium yoelii erythrocyte ligand determines its localization and controls parasite virulence. Proc Nat Acad Sci USA. 2009;106:7167–7172. doi: 10.1073/pnas.0811313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Elango AP, Rahman AA, Fisher D, Collins WE, Barnwell JW, Escalante AA. Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in Plasmodium spp. Infect Genet Evol. 2012a;12:978–986. doi: 10.1016/j.meegid.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Reid MJ, Schillaci MA, Lowenberger CA, Galdikas BM, Jones-Engel L, Escalante AA. The origin of malarial parasites in orangutans. PloS One. 2012b;7:e34990. doi: 10.1371/journal.pone.0034990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain A, Bohme U, Berry AE, Mungall K, Finn RD, Jackson AP, Mourier T, Mistry J, Pasini EM, Aslett MA, Balasubrammaniam S, Borgwardt K, Brooks K, Carret C, Carver TJ, Cherevach I, Chillingworth T, Clark TG, Galinski MR, Hall N, Harper D, Harris D, Hauser H, Ivens A, Janssen CS, Keane T, Larke N, Lapp S, Marti M, Moule S, Meyer IM, Ormond D, Peters N, Sanders M, Sanders S, Sargeant TJ, Simmonds M, Smith F, Squares R, Thurston S, Tivey AR, Walker D, White B, Zuiderwijk E, Churcher C, Quail MA, Cowman AF, Turner CM, Rajandream MA, Kocken CH, Thomas AW, Newbold CI, Barrell BG, Berriman M. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik M, Rowe AW. Duffy and Duffy-Related Human-Antigens in Primates. J Hum Evol. 1984;13:173–179. [Google Scholar]
- Pattaradilokrat S, Culleton RL, Cheesman SJ, Carter R. Gene encoding erythrocyte binding ligand linked to blood stage multiplication rate phenotype in Plasmodium yoelii yoelii. Proc Nat Acad Sci USA. 2009;106:7161–7166. doi: 10.1073/pnas.0811430106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Huber CS, Galinski MR, Barnwell JW. Rapid evolution of an erythrocyte invasion gene family: the Plasmodium reichenowi Reticulocyte Binding Like (RBL) genes. Mol Biochem Parasitol. 2004;133:287–296. doi: 10.1016/j.molbiopara.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Tran TM, Corredor V, Huber CS, Barnwell JW, Galinski MR. Dramatic difference in diversity between Plasmodium falciparum and Plasmodium vivax reticulocyte binding-like genes. Amer J Trop Med Hyg. 2005;72:666–674. [PubMed] [Google Scholar]
- Rice BL, Acosta MM, Pacheco MA, Carlton JM, Barnwell JW, Escalante AA. The origin and diversification of the merozoite surface protein 3 (msp3) multi-gene family in Plasmodium vivax and related parasites. Mol Phylogenet Evol. 2014;78:172–184. doi: 10.1016/j.ympev.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LH, Genther CS, et al. Development of resistance to chlorguanide (paludrine) during treatment of infections with Plasmodium cynomolgi. J of pharmacology and experimental therapeutics. 1949;95:382–398. [PubMed] [Google Scholar]
- Schmidt LH, Greenland R, Genther CS. The transmission of Plasmodium cynomolgi to man. Amer J Trop Med Hyg. 1961;10:679–688. doi: 10.4269/ajtmh.1961.10.679. [DOI] [PubMed] [Google Scholar]
- Semenya AA, Tran TM, Meyer EV, Barnwell JW, Galinski MR. Two functional reticulocyte binding-like (RBL) invasion ligands of zoonotic Plasmodium knowlesi exhibit differential adhesion to monkey and human erythrocytes. Malar J. 2012;11:228. doi: 10.1186/1475-2875-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui AA, Xainli J, Schloegel J, Carias L, Ntumngia F, Shoham M, Casey JL, Foley M, Adams JH, King CL. Fine specificity of Plasmodium vivax Duffy binding protein binding engagement of the Duffy antigen on human erythrocytes. Infection and immunity. 2012;80:2920–2928. doi: 10.1128/IAI.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by plasmodium falciparum.pdf. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- Sutton PL, Torres LP, Branch OH. Sexual recombination is a signature of a persisting malaria epidemic in Peru. Malar J. 2011;10:329. doi: 10.1186/1475-2875-10-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J. 2014;13:68. doi: 10.1186/1475-2875-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana S, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, Arisue N, Palacpac NM, Honma H, Yagi M, Tougan T, Katakai Y, Kaneko O, Mita T, Kita K, Yasutomi Y, Sutton PL, Shakhbatyan R, Horii T, Yasunaga T, Barnwell JW, Escalante AA, Carlton JM, Tanabe K. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nature Genet. 2012;44:1051–1055. doi: 10.1038/ng.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N, Nei M, Tamura K. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol. 2010;27:747–752. doi: 10.1093/molbev/msp312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M, Skinner JC, Guinn E. Biology of the simian malarias of Southeast Asia. I. Host cell preferences of young trophozoites of four species of Plasmodium. J Parasitol. 1966;52:14–16. [PubMed] [Google Scholar]
- Waters AP, Higgins DG, McCutchan TF. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol. 1993;10:914–923. doi: 10.1093/oxfordjournals.molbev.a040038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.