Abstract
Atractylodes rhizomes have been used as the herbal medicine “Changchul” or “Baekchul,” according to their clinical purpose, in Korea, China, and Japan. Among the Atractylodes species, the medicinal use of Atractylodes japonica has been controversial, as it is categorized as both Changchul and Baekchul in those countries, and, moreover, parts of the rhizome have been differently used, depending on age of the plant, in Korea. Chromatographic fingerprinting by using HPLC combined with chemometric analyses and internal transcribed spacer (ITS) sequencing analysis were conducted to classify and identify 34 crude drugs derived from Atractylodes rhizomes. The identification of the samples, authenticated by their morphological features as A. japonica Koidz. (Changchul and Baekchul), A. chinensis Koidz., and A. macrocephala Koidz., was confirmed as A. japonica, A. chinensis, and A. macrocephala by ITS sequencing. The results from chemometric analyses showed that the chemical components of the crude drugs from A. japonica were significantly different from those from A. macrocephala but were similar to those from A. chinensis. The analyses also suggested that the categorization by age of A. japonica as Changchul or Baekchul is not recommended. The results indicate that A. japonica should be categorized as “Changchul” and should not be further categorized by age.
1. Introduction
The genus Atractylodes (Asteraceae) are perennial herbs distributed in Korea, China, and Japan. Their dried rhizomes have been classified into two kinds of herbal medicines according to their clinical purpose, “Baekchul” (Baizhu in Chinese, Byakujutsu in Japanese) and “Changchul” (Cangzhu in Chinese, Soujutsu in Japanese) [1]. In medicinal applications in Korea, Japan, and China, the rhizomes of A. lancea DC. and A. chinensis Koidz. have been classified as Changchul, while that of A. macrocephala Koidz. has been classified as Baekchul in the pharmacopeias of Korea, China, and Japan [2–4].
However, there has been disagreement between countries in classifying the rhizome of A. japonica Koidz.: Korean and Japanese pharmacopeias, as well as some studies from Korea and Japan, have classified the rhizome of A. japonica as Baekchul, whereas Chinese studies have classified it as “Gwan-Changchul” (Guan-Cangzhu in Chinese), a type of Changchul, which is not even listed in the Chinese pharmacopeia [5, 6]. Moreover, A. japonica is recorded as a synonym of A. lancea in the Flora of China [7]. Confusion also occurs in local herbal markets in Korea, where the rhizomes of A. japonica have been used as Changchul when the fibrous substance has formed after it has grown over two years [8]. The dried rhizomes of Atractylodes species can be identified by their morphological features; however, it is difficult to discriminate them by macroscopic observation, a subjective method, due to their morphological similarity. Consequently, the misuse of Atractylodes rhizomes may occur when identification is based only on their morphological features. Therefore, strict classification is required for the exact use of Atractylodes rhizomes for medicinal purposes.
Genetic identification achieved by hybridization, polymerase chain reaction (PCR), and sequencing techniques is considered more objective, precise, and reliable method for identifying and authenticating herbal species [9, 10]. Internal transcribed spacer (ITS) regions, which are rapidly evolving regions of nuclear ribosomal DNA, have been widely used for the identification of plant materials, and phylogenetic analysis using ITS regions has been conducted to investigate the genetic variability of complicated herbal species [11–13]. It has been reported that the four species of Atractylodes (A. japonica, A. macrocephala, A. lancea, and A. chinensis) can be distinguished by their different genotypes on ITS sequences [14]. In contrast, one study conducted by PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the ITS region on nucleotide ribosome DNA (nrDNA) reported that A. japonica is not different from A. macrocephala [15]. Other studies have demonstrated that the geographical distributions, morphological features, and genetic differentiation between A. lancea and A. chinensis were not consistent, and A. japonica was most closely related to A. lancea, according to the results from ITS and trnL-F sequences [16, 17]. The results of these studies demonstrate that the classification of Atractylodes rhizomes, particularly that of A. japonica, remains controversial.
Chemical fingerprinting by using HPLC is an effective and reliable method for the investigation of chemical components in herbal medicines, owing to its high separation efficiency and high detection sensitivity [18]. HPLC fingerprinting also enables a systematic and comprehensive approach to the identification and quantification of the components in herbal medicines [19]. HPLC fingerprinting, combined with chemometric statistical analysis, has been widely used for the quality control of herbal medicines and related herbal products [20, 21]. Moreover, such techniques have also been used to discriminate interspecies differences in herbal medicines, by incorporating principal components analysis (PCA) [22–24]. In previous studies, A. lancea was chemically differentiated from A. chinensis by HPLC fingerprinting combined with PCA or orthogonal partial least squares-discriminant analysis [25, 26]. However, the chemical discrimination of Atractylodes species, using HPLC fingerprinting and chemometric analysis, has not been conducted.
In the present study, chromatographic fingerprinting and chemometric statistical analyses, including PCA, hierarchical clustering analysis (HCA) analysis, and Pearson's correlation coefficient analysis were conducted to classify the crude drugs derived from Atractylodes rhizomes. The ITS sequences from nrDNA were also examined to identify the Atractylodes species.
2. Materials and Methods
2.1. Plant Materials and Reagents
Thirty-four samples of crude drugs from Atractylodes rhizomes were collected or purchased from the wild, agricultural fields, or local markets in Korea and China. The samples were authenticated by their morphological features through identification criteria by authors. Fifteen rhizomes were identified as A. chinensis (coded as “AC”); seven samples of Baekchul and five samples of Changchul were “young” and “aged” dried rhizomes of A. japonica (coded as “AJB” and “AJC,” resp.); and seven rhizomes were identified as A. macrocephala (coded as “AM”) (Table 1). The voucher specimens have been deposited in the herbarium of the College of Korean Medicine, Woosuk University (Jeonju, Jeonbuk, Korea).
Table 1.
Morphological identification of the crude drugs of Atractylodes rhizomes.
| Code | Species | Origin | Code | Species | Origin |
|---|---|---|---|---|---|
| AC-1 | A. chinensis Koidz. | China | AJB-3 | A. japonica Koidz. | China |
| AC-2 | A. chinensis Koidz. | China | AJB-4 | A. japonica Koidz. | Unclear |
| AC-3 | A. chinensis Koidz. | China | AJB-5 | A. japonica Koidz. | Unclear |
| AC-4 | A. chinensis Koidz. | China | AJB-6 | A. japonica Koidz. | Unclear |
| AC-5 | A. chinensis Koidz. | China | AJB-7 | A. japonica Koidz. | Korea |
| AC-6 | A. chinensis Koidz. | Korea | AJC-1 | A. japonica Koidz. | Korea |
| AC-7 | A. chinensis Koidz. | Korea | AJC-2 | A. japonica Koidz. | Unclear |
| AC-8 | A. chinensis Koidz. | China | AJC-3 | A. japonica Koidz. | Unclear |
| AC-9 | A. chinensis Koidz. | China | AJC-4 | A. japonica Koidz. | Korea |
| AC-10 | A. chinensis Koidz. | China | AJC-5 | A. japonica Koidz. | Unclear |
| AC-11 | A. chinensis Koidz. | China | AM-1 | A. macrocephala Koidz. | China |
| AC-12 | A. chinensis Koidz. | China | AM-2 | A. macrocephala Koidz. | China |
| AC-13 | A. chinensis Koidz. | China | AM-3 | A. macrocephala Koidz. | China |
| AC-14 | A. chinensis Koidz. | China | AM-4 | A. macrocephala Koidz. | China |
| AC-15 | A. chinensis Koidz. | China | AM-5 | A. macrocephala Koidz. | China |
| AJB-1 | A. japonica Koidz. | Korea | AM-6 | A. macrocephala Koidz. | China |
| AJB-2 | A. japonica Koidz. | Korea | AM-7 | A. macrocephala Koidz. | China |
Nine specimens (coded as Arabic number) of Atractylodes plants were collected as species reference samples for identification of Atractylodes rhizomes in Table 1 and those samples were deposited in the Korea Institute of Oriental Medicine as voucher specimens. The classification result of nine dried-voucher specimens is shown in Table 2.
Table 2.
Identification of dried-voucher specimens coded as Arabic numbers.
| Number | Identification |
|---|---|
| 1 | A. japonica Koidzumi |
| 2 | A. lancea de Candolle |
| 3 | A. lancea de Candolle |
| 4 | A. macrocephala Koidzumi |
| 5 | A. japonica Koidzumi |
| 6 | A. japonica Koidzumi |
| 8 | A. lancea de Candolle |
| 9 | A. japonica Koidzumi |
| 17 | A. japonica Koidzumi |
2.2. Preparation of Genomic DNA
The genomic DNA was extracted from the crude drugs of Atractylodes rhizomes according to the manuals of NucleoSpin® Plant II kit (Macherey-Nagel, Dueren, Germany). For some samples, 10% cetyl trimethyl ammonium bromide (CTAB) and 0.7 M NaCl were used to remove the phenolic compounds and polysaccharides.
2.3. PCR Amplification
For ITS amplification, PCR was performed using T-personal cycler (Biometra, Germany). In brief, 600 nM of primer set of ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [27], 1x BluePreMIX-HF (Macrogen, Korea), and 50 ng of genomic DNA were used for PCR amplification. PCR cycling conditions which were followed by predenaturation process (95°C, 5 min) were as follows: denaturation process (95°C, 30 s); annealing process (52°C, 30 s); extension process (72°C, 40 s) × 36 cycles; and final extension process (72°C, 5 min). The amplified PCR product was separated from other gradients using 1.5% agarose gel electrophoresis after the staining by the addition of Safe-white™ (abm, Canada). Amplified products were analyzed using MyImage (Seoulin Biotechnology, Korea).
2.4. Determination of DNA Sequence of PCR Product
PCR product separated from agarose gel was cloned using MG™ TOPcloner TA kit (Macrogen, Korea) and DNA sequence of cloned PCR product was determined through the interpretation performed by Macrogen (Korea).
2.5. Analysis of DNA Sequence and Preparation of Dendrogram
DNA sequence was analyzed using ClustalW multiple sequence alignment (Bioedit, v7.0.9; available at http://www.mbio.ncsu.edu/BioEdit/page2.html) and the phylogenetic tree was created by using DNADist (Bioedit). To study the relationship of Atractylodes, the nucleotide sequences of the genera Atractylis and Carlina deposited in NCBI GenBank were used. The genera Brachylaena, Cardopatium, Cirsium, Echinops, Phonus, and Tarchonanthus were also used as reference for phylogenetic relationship. Magnolia heptapeta, Zanthoxylum rhoifolium, and Lilium cernuum were used as out-groups in the phylogenetic analyses, based on previous studies [28, 29]. ITS sequences of these taxa were collected from GeneBank in NCBI (the accession numbers were shown in Table 3).
Table 3.
The GenBank accession number of species that are used in phylogenetic tree analysis.
| Species | GenBank accession number |
|---|---|
| Atractylis cancellata | AY826231.1 |
| Atractylis arabica | KF850563.1 |
| Atractylis carduus | AY826232.1 |
| Atractylis arbuscula | KF301215.1 |
| Carlina falcata | AY826243.1 |
| Carlina vulgaris | KF301217.1 |
| Carlina acanthifolia | KF301216.1 |
| Echinops viscosus | AY826283.1 |
| Phonus riphaeus | AY826310.1 |
| Cirsium palustre | EU143268.1 |
| Cardopatium corymbosum | AY826238.1 |
| Tarchonanthus camphoratus | AY826340.1 |
| Brachylaena discolor | AY826236.1 |
| Magnolia heptapeta | AY858638.1 |
| Zanthoxylum rhoifolium | KC502933.1 |
| Lilium cernuum | HQ686064.1 |
2.6. Preparation of Samples for HPLC Analysis
The dried powder of the rhizomes (100 mg) was weighed and then extracted by sonication with 2 mL of ethanol (HPLC grade; Phillipsburg, NJ, USA) for 40 min. The extract was filtered through a 0.45 μm membrane filter (Adventec, Tokyo, Japan) and concentrated in vacuum oven at 45°C. Concentrated extract was dissolved with methanol at the concentration of 5000 μg/mL and filtered through a 0.45 μm membrane filter, prior to HPLC injection.
2.7. HPLC Conditions for Chromatographic Fingerprinting
An Agilent 1260 liquid chromatography system (Agilent Technologies, Palo Alto, CA, USA) equipped with an autosampler, degasser, quaternary solvent pump, and diode array detector (DAD) was used for chromatographic fingerprinting. The data were processed by using Chemstation software (Agilent Technologies Inc., USA). The separation of compounds was carried out on a Capcell Pak Mg II C18 column (4.6 mm × 250 mm, 5 μm; Shiseido, Tokyo, Japan) with Mg II C18 guard cartridge (4.0 mm × 10 mm, S-5; Shiseido, Tokyo, Japan) at 35°C. The flow rate was 1 mL/min and the injection volume was 10 μL. The mobile phase consisted of solvent A (HPLC-grade water; Phillipsburg, NJ, USA) and solvent B (HPLC-grade acetonitrile; Phillipsburg, NJ, USA), with the following gradient elution, 20% (B) over 0–2 min, 20–55% (B) over 2–10 min, 55% (B) over 10–13 min, 55–60% (B) over 13–35 min, 60% (B) over 35–38 min, and 60–75% (B) over 38–50 min, held for 2 min, and then reequilibrated to 20% until the end of the analysis. Detection was performed using a UV detector at the wavelengths of 255, 275, 295, 315, and 340 nm.
2.8. Chemometric Statistical Analysis
The 34 samples that were genetically identified and recoded were used for PCA, HCA, and Pearson's correlation analysis. Total 27 peaks were selected as “reference peaks,” and their absolute area was calculated by peak area integration. The 27 reference peaks for the chromatographic fingerprinting, which were representative and >1.0% of total peak area, were chosen at their optimal UV absorption. The absolute area of chosen peak was calculated for chromatographic fingerprinting. A matrix composed of the rows (Atractylodes sample) and columns (absolute area of each reference peak) was used for construction of PCA plot, HCA dendrogram, and Pearson's correlation analysis, which were conducted using open-source software R (v.3.1.1).
3. Results
3.1. ITS Genotype and Genetic Identification of Atractylodes Rhizomes
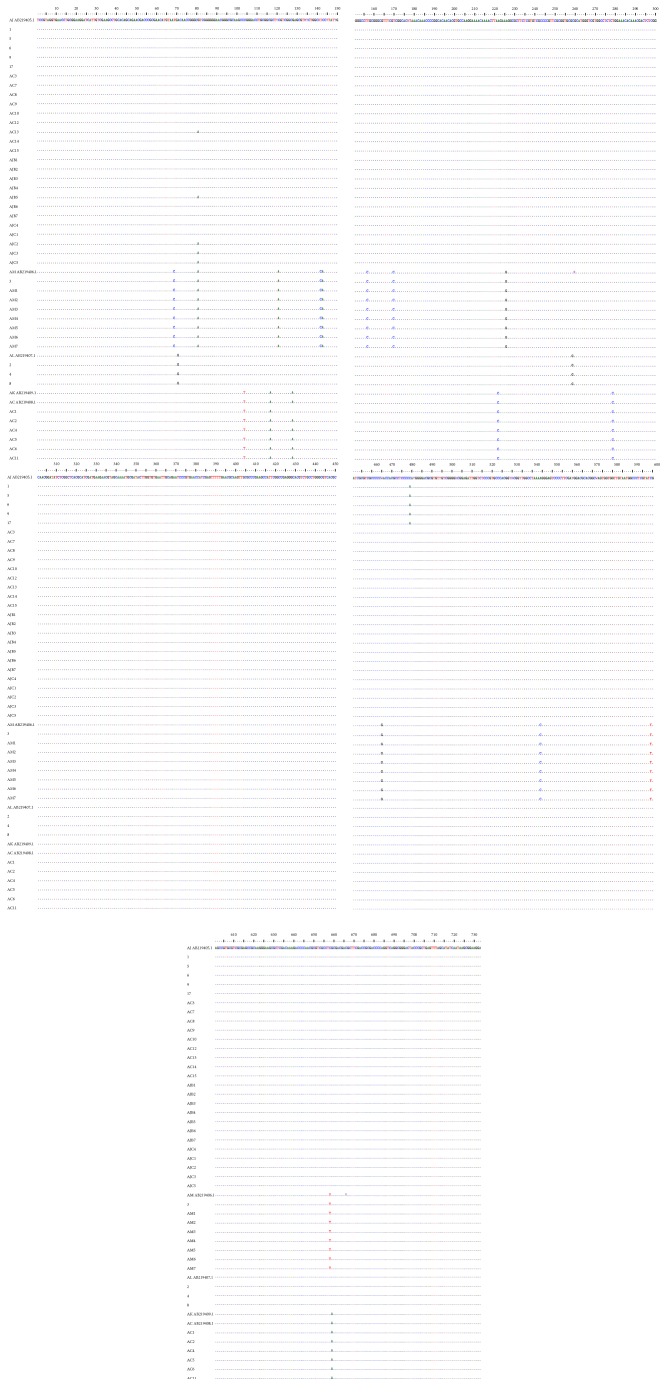
The amplification of internal transcribed spacer (ITS) region produced overall 733 bp of nucleotide sequences from 34 samples listed in Table 1 and nine dried-voucher specimens (Figure 1). The determined ITS nucleotide sequence of samples were confirmed by using DNA sequence registered in NCBI GenBank as well as previous paper [14] with comparison of the accession numbers: A. japonica (AB219405), A. macrocephala (AB219406), A. lancea (AB219407), A. chinensis (AB219408), and A. koreana (AB219409). As presented in Table 4, there was nucleotide substitutions observed on 37 sites on the ITS regions of Atractylodes samples. Type 1, ITS sequence of A. japonica, showed multiple sequences comparing to other species, while types 2 and 3 were the genotypes of A. macrocephala and A. lancea, respectively. Type 4, the genotype of A. chinensis, was identical to type 5, the genotype of A. koreana.
Figure 1.
Multiple alignments of ITS nucleotide sequence among the sample listed in Table 1. The dots indicate the consensus nucleotide, and the dashes represent the gaps. AB219405.1 accession numbers of NCBI GenBank for the nucleotide sequences of the ITS for Atractylodes japonica; AB219409.1 and AB210407.1 accession numbers of NCBI GenBank for the nucleotide sequences of the ITS for A. koreana and A. lancea, respectively; AB219406.1 accession number of NCBI GenBank for the nucleotide sequence of ITS for A. macrocephala; and AB219408.1 accession number of NCBI GenBank for the nucleotide sequence of ITS for A. chinensis. The samples with Arabic number were dried-voucher specimens deposited in the Korea Institute of Oriental Medicine.
Table 4.
Genotypes of ITS nucleotide sequences deposited in the NCBI GenBank for recognition in the Atractylodes species.
| Nucleotide position | 69 | 71 | 81 | 104 | 117 | 120 | 121 | 128 | 134 | 135 | 141 | 142 | 143 | 145 | 149 | 157 | 170 | 177 | 222 | 226 | 259 | 260 | 279 | 465 | 481 | 494 | 502 | 529 | 530 | 534 | 544 | 560 | 572 | 599 | 658 | 659 | 666 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | |||||||||||||||||||||||||||||||||||||
| Type 1 | T | A | G | C | G | Y | C | G | M | Y | R | T | C | Y | Y | T | T | R | A | A | C | G | T | W | Y | Y | R | Y | R | Y | A | Y | W | C | C | G | C |
| Type 2 | C | A | A | C | G | C | A | G | C | C | G | C | A | C | C | C | C | G | A | G | C | R | T | G | C | C | G | C | G | C | C | C | T | T | T | G | Y |
| Type 3 | T | G | G | C | G | C | C | G | C | C | G | T | C | C | C | T | T | A | A | A | G | G | T | A | C | C | G | T | G | C | A | C | A | C | C | G | C |
| Type 4 | T | A | G | T | A | C | C | A | C | C | G | T | C | C | T | T | T | G | C | A | C | G | C | T | C | C | G | C | G | C | A | C | T | C | C | A | C |
| Type 5 | T | A | G | T | A | C | C | A | C | C | G | T | C | C | T | T | T | G | C | A | C | G | C | T | C | C | G | C | G | C | A | C | T | C | C | A | C |
∗Bold characters indicate nucleotide additives, M = A and C; R = A and G; W = A and T; Y = C and T. Type 1 genotype resulted from deposited ITS nucleotide sequences in NCBI Genbank of Atractylodes japonica. Type 2 and type 3 represent A. macrocephala and A. lancea. Type 4 and type 5 represent A. chinensis and A. koreana.
All 7 samples labelled as “AM” were determined as A. macrocephala and the difference of DNA sequence between samples was not observed. Among 15 samples labelled as “AC,” AC-1, AC-2, AC-4, AC-5, AC-6, and AC-11 were determined as A. chinensis, whereas the rest were A. japonica. The sequence of AC-1 was different from that of type 4 by 1 bp (A → G) at nucleotide position, 128 bp, which indicates intraspecific variation. All 7 samples labelled as “AJB” as well as 5 samples labelled as “AJC” were determined as A. japonica. AC-13 and AJB-5 were different form that of type 1 by 1 bp (G → A), at nucleotide position, 80 bp. AJC-2, AJC-3, and AJC-5 have 2 different nucleotide positions at 80 bp (G → A) and 481 bp (C → T), showing intraspecific variation.
3.2. Genetic Relationship of the Atractylodes
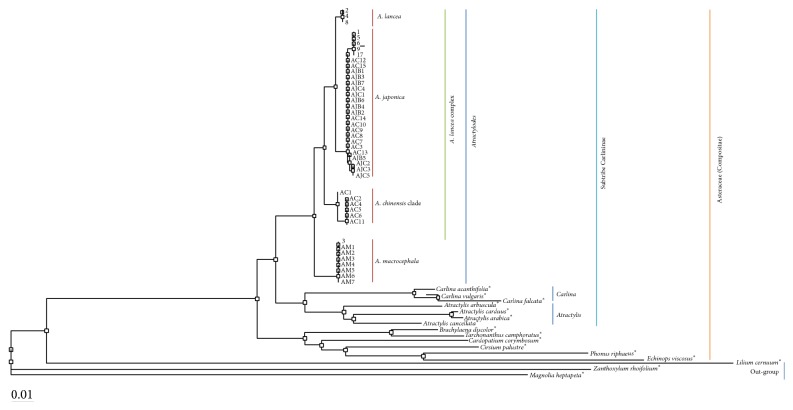
Nine dried-voucher specimens were confirmed as A. japonica, A. lancea, and A. macrocephala. Thirty-four samples of Atractylodes rhizomes were identified as A. chinensis, A. japonica, and A. macrocephala. Phylogenetic classification based on ITS region was constructed and the inferred evolutionary relationships among Atractylodes rhizomes were represented on phylogenetic tree. The genus Atractylodes is well separated from other close genera and out-groups. The samples of A. japonica, A. lancea, and A. chinensis formed A. lancea complex, whereas those of A. macrocephala formed their own complex, namely, A. macrocephala complex. Five AC samples, AC-2, AC-4, AC-5, AC-6, and AC-11, were involved in A. chinensis group. AJB, AJC samples, and the rest AC samples were contained in A. japonica group (Figure 2).
Figure 2.
Phylogenic tree from DNADist (Neighbor phylogenetic tree) analysis of the ITS nucleotide sequences. The ITS sequences of taxa with “∗” such as Atactylis, Carlina, and out-group were downloaded from Genbank in NCBI. The samples with Arabic number were dried-voucher specimens deposited in the Korea Institute of Oriental Medicine.
Comparing the genotypes listed in Figures 1 and 2 and Table 4, the species of 34 samples in Table 1 of Atractylodes rhizomes were recoded as shown in Table 5.
Table 5.
Genetic identification of original species of Atractylodes rhizomes listed in Table 1.
| Code | Genetically identified species | Recode | Code | Genetically identified species | Recode |
|---|---|---|---|---|---|
| AC-1 | A. chinensis | AC-1 | AJB-3 | A. japonica | AJB-3 |
| AC-2 | A. chinensis | AC-2 | AJB-4 | A. japonica | AJB-4 |
| AC-3 | A. japonica | AJ-1 | AJB-5 | A. japonica | AJB-5 |
| AC-4 | A. chinensis | AC-3 | AJB-6 | A. japonica | AJB-6 |
| AC-5 | A. chinensis | AC-4 | AJB-7 | A. japonica | AJB-7 |
| AC-6 | A. chinensis | AC-5 | AJC-1 | A. japonica | AJC-1 |
| AC-7 | A. japonica | AJ-2 | AJC-2 | A. japonica | AJC-2 |
| AC-8 | A. japonica | AJ-3 | AJC-3 | A. japonica | AJC-3 |
| AC-9 | A. japonica | AJ-4 | AJC-4 | A. japonica | AJC-4 |
| AC-10 | A. japonica | AJ-5 | AJC-5 | A. japonica | AJC-5 |
| AC-11 | A. chinensis | AC-6 | AM-1 | A. macrocephala | AM-1 |
| AC-12 | A. japonica | AJ-6 | AM-2 | A. macrocephala | AM-2 |
| AC-13 | A. japonica | AJ-7 | AM-3 | A. macrocephala | AM-3 |
| AC-14 | A. japonica | AJ-8 | AM-4 | A. macrocephala | AM-4 |
| AC-15 | A. japonica | AJ-9 | AM-5 | A. macrocephala | AM-5 |
| AJB-1 | A. japonica | AJB-1 | AM-6 | A. macrocephala | AM-6 |
| AJB-2 | A. japonica | AJB-2 | AM-7 | A. macrocephala | AM-7 |
3.3. HPLC Fingerprinting of Atractylodes Samples
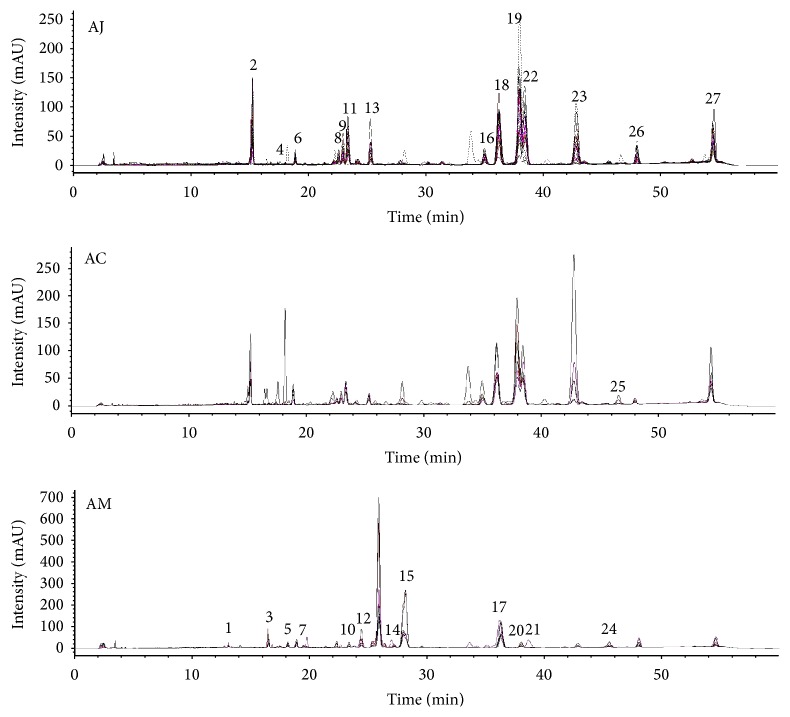
Through macroscopic observation of the results of HPLC fingerprinting, the chromatograms of A. japonica samples (AJBs + AJCs + AJs) and A. chinensis samples (ACs) showed peak patterns similar to each other, whereas the chromatograms of A. macrocephala samples (AMs) represented different peak patterns compared with those of A. japonica and A. chinensis samples. Moreover, chromatographic difference between Baekchul samples (AJBs) and Changchul samples (AJCs) was not apparently distinguished (Figures 3 and 4).
Figure 3.
Representative chromatograms of the methanol extracts of AC, AJ, and AM at 255 nm. AC: A. chinensis Koidz.; AJ: A. japonica Koidz.; AM: A. macrocephala Koidz.
Figure 4.
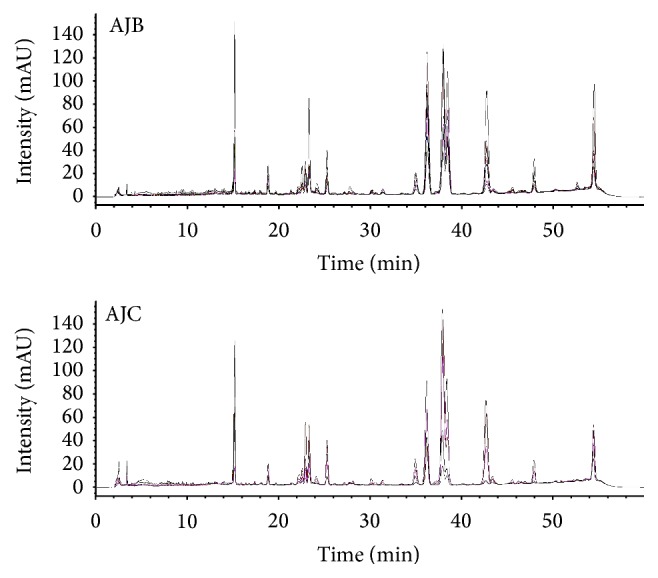
Representative chromatograms of the methanol extracts of AJB and AJC at 255 nm. AJB and AJC: A. japonica Koidz.
3.4. Chemometric Statistical Analysis of Atractylodes Samples
The results of the chromatographic fingerprinting were further analyzed using principle component analysis (PCA), hierarchical clustering analysis (HCA), and Pearson's correlation analysis to evaluate the correlations between the samples whose code names were redetermined by genetic identification as listed in Table 5.
Principle component 1 (PC1) score divided the samples of “A. japonica + A. chinensis” group in the positive plot with exception of AC-6, AJ-9, and AJC-5, while “A. macrocephala” group was in the negative plot. PC2 score further differentiated the samples having positive PC1 scores into positive PC2 plot (AC-1, AC-3, AC-5, AJ-1, AJ-2, AJ-6, AJ-7, AJ-8, AJB-2, AJB-3, AJB-7, AJC-1, and AJC-2) and negative PC2 plot (AC-2, AC-4, AJ-3, AJ-4, AJ-5, AJB-1, AJB-4, AJB-5, AJB-6, AJC-3, and AJC-4) and samples having negative PC1 scores into positive PC2 (AM-1, AM-5, AM-6, and AM-7) and negative PC2 (AM-2, AM-3, and AM-4) (Figure 5).
Figure 5.
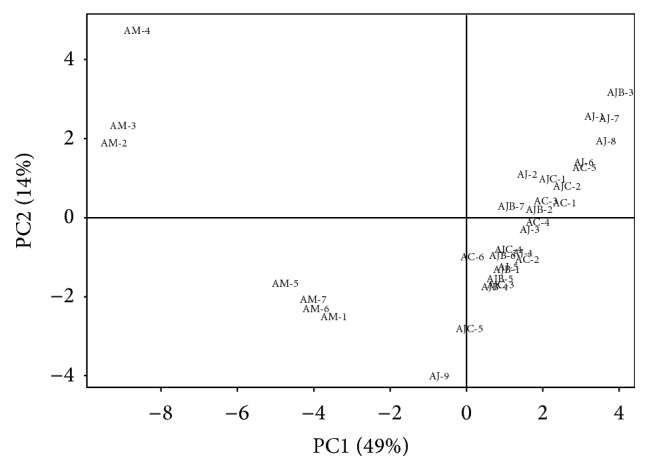
Score plot of principal components (PC1 versus PC2) on the variables (absolute area of reference peaks) with Atractylodes samples. PC1 and PC2 represent 49% and 14% of the total variance, respectively. AC: A. chinensis Koidz.; AJ, AJB, and AJC: A. japonica Koidz.; AM: A. macrocephala Koidz.
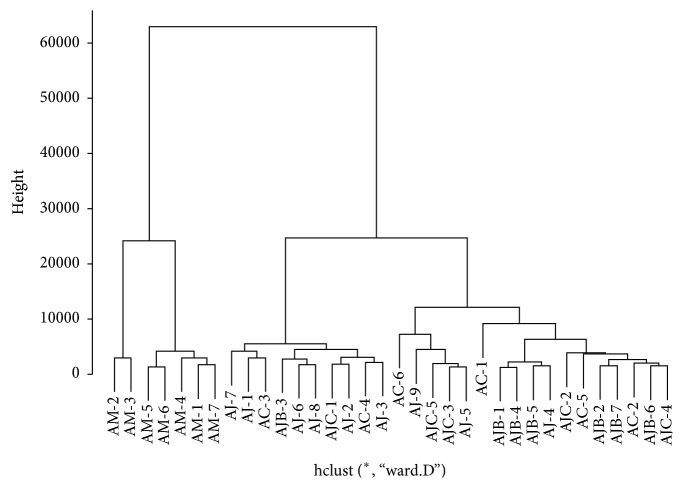
HCA results also showed similar result obtained from PCA. Below the height of 30000, while the samples of A. macrocephala formed their own clustering, those of A. japonica and A. chinensis were gathered together, where A. japonica samples and A. chinensis samples were mixed. Therefore, apparent discrimination between A. japonica samples and A. chinensis samples was not observed. Moreover, as seen in PCA score plot, AJB and AJC samples were not clearly distinguished in HCA dendrogram, as they were grouped undistinguishably within the same levels (Figure 6).
Figure 6.
Hierarchical clustering analysis of Atractylodes samples. AC: A. chinensis Koidz.; AJ, AJB, and AJC: A. japonica Koidz.; AM: A. macrocephala Koidz.
Box plot of average Pearson's correlation coefficient (r) showed two distinct sample groups, the group of A. macrocephala samples and the group of A. japonica and A. chinensis samples. The average coefficients of A. macrocephala samples ranged from −0.2 to 0.0, whereas those of A. japonica and A. chinensis samples ranged from 0.5 to 1.0, except for AJ-9 showing the average coefficient between 0.0 and 0.2. Furthermore, AJB and AJC samples showed similar values of average Pearson's coefficients (Figure 7).
Figure 7.
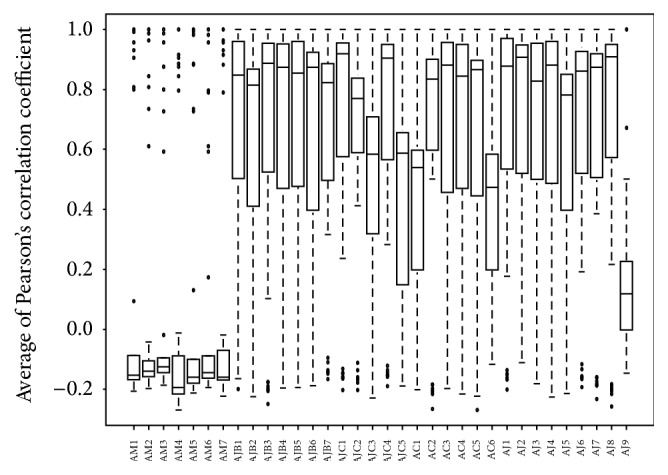
Average coefficients of Pearson's correlation of 34 Atractylodes samples. AC: A. chinensis Koidz.; AJ, AJB, and AJC: A. japonica Koidz.; AM: A. macrocephala Koidz.
4. Discussion
Discrimination of herbal medicines according to their therapeutic effect is most important in using herbal medicines as therapeutic agents. To achieve such purpose, correct classification of herbal medicines should be preceded at their species levels. The rhizome of A. japonica has been at the center of controversy as it is differently classified in many countries; Korea and Japan have used that as “Baekchul,” whereas China has used that as “Changchul.” Furthermore, the rhizome of A. japonica is further divided into “Baekchul” and “Changchul” by the age; tuberous part grown for less than a year is used as Baekchul, while fibrous part grown for more than two years is used as Changchul. Hence, we classified Atractylodes samples which were genetically identified by ITS genotypes using analytical tools combined with chemometric statistics, in order to propose correct categorization of Atractylodes rhizomes, especially A. japonica rhizomes.
4.1. Genetic Identification of Atractylodes Samples
Atractylodes, Atractylis, and Carlina belong to subtribe Carlininae [28]. ITS sequence of A. japonica showing multiple sequences indicates that there are diverse intraspecific variations in A. japonica samples. The genotype of A. chinensis was identical to that of A. koreana. As the distribution of A. koreana is limited in Shandong and Liaoning area, A. koreana has been possibly derived from A. chinensis [14, 17]. The samples of A. japonica, A. lancea, and A. chinensis formed A. lancea complex, in accordance with the previous research [17]. A. japonica is most closely related to A. lancea and next related to A. chinensis, as suggested in previous researches [17, 30, 31]. The samples identified as A. japonica were obviously separated from those identified as A. macrocephala by DNA sequencing analysis [14, 16, 32]. According to the results, there was difference in classification between morphological identification and genetic identification which might be caused by morphological similarity.
4.2. Classification of Atractylodes Samples Using Chemometric Analysis
Principal components analysis (PCA) was performed for the clustering of the samples and for investigating the relationships among the samples using principal components on the PCA plot. Principal component 1 (PC1) explains most of the variance, and PC2, which is orthogonal to PC1, represents most of the variance not explained by PC1 [33]. HCA is another clustering method which classifies similar objects mathematically into the same group and the classifications were represented as tree diagram called dendrogram [34]. Pearson's correlation coefficient of each sample was calculated to evaluate the correlations between samples (−1 ≤ r ≤ +1; the closer the r is to 1, the more correlated the two fingerprints are) [35].
From the results of PCA, HCA, and Pearson's correlation analysis, the A. japonica samples were obviously distinct from A. macrocephala samples but distributed close to the A. chinensis samples without apparent separation of AJBs and AJCs samples. These results indicate that A. japonica is chemically different from A. macrocephala; however, the crude drugs derived from A. japonica and A. chinensis were not clearly distinguishable by their chemical components. These results indicate that the therapeutic effects of A. japonica and A. chinensis whose effects can be exerted by the chemical components are thought to be presumably analogous [36], although they are genetically different. Moreover, the separation of A. japonica rhizomes as “Baekchul” and “Changchul” is not recommended because they had the closest PC scores, equal levels of hierarchies, and similar Pearson's coefficients [37]; therefore, AJBs and AJCs can be assumed to be the same medicinal parts.
4.3. Assured Medicinal Categorization of Atractylodes japonica Rhizomes
The categorization of A. japonica as Baekchul in Korea and Japan, therefore, should be removed, because its relationship to A. macrocephala, the only species regarded as Baekchul in China, is not close, which supports the results from previous studies [14, 17, 32, 38]. Moreover, we do not recommend the segregation of A. japonica into Baekchul and Changchul, because the segregated crude drugs of A. japonica are not distinguishable by their chemical and genetic characteristics. This suggests that there is the lack of chemical or genetic reason to categorize the rhizome of A. japonica as Changchul and Baekchul based on plant age.
Therefore, we propose that A. japonica should be separated from A. macrocephala and should be used as “Changchul” as the Chinese literatures define it, regardless of its age. Further biological and clinical experiments are necessary to confirm the results of the present study.
In conclusion, the 34 crude drugs derived from Atractylodes rhizomes were identified using ITS sequencing analysis and classified using HPLC fingerprinting and statistical tools such as PCA, HCA, and Pearson's correlation analysis. The macroscopically authenticated Atractylodes samples were genetically identified as A. japonica, A. chinensis, and A. macrocephala by ITS DNA sequencing. The hyphenated HPLC fingerprinting and statistical analyses showed that the chemical components of A. japonica were not related to those of A. macrocephala but related to those of A. chinensis. Moreover, the rhizome of A. japonica could not be segregated as Baekchul and Changchul by its age. The results from these chemical and genetic analyses demonstrate that A. japonica should be classified as “Changchul” and should not be classified by plant age.
Acknowledgments
This study was supported by the Convergence of Conventional Medicine and Traditional Korean Medicine R&D program funded by the Ministry of Health & Welfare through the Korea Health Industry Development Institute (KHIDI) (HI14C0750).
Disclosure
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the paper.
Competing Interests
The authors declare that they have no competing interests.
Authors' Contributions
J.-H. Kim and E.-J. Doh contributed equally to this work.
References
- 1.Bensky D., Clavey S., Stöger E. Chinese Herbal Medicine: Materia Medica. 3rd. Seattle, Wash, USA: Eastland Press; 2004. [Google Scholar]
- 2.The Korea Food and Drug Administration. The Korean Pharmacopeia. 10th. The Korea Food and Drug Administration; 2013. [Google Scholar]
- 3.The Pharmacopoeia Commission of the Ministry of Health of the People's Republic of China. The Pharmacopoeia of the People's Republic of China. 10th. Beijing, China: China Science and Technology Press; 2010. [Google Scholar]
- 4.Society of Japanese Pharmacopoeia. Japanese Pharmacopeia. Vol. 65. The Ministry of Health; 2011. (Labor and Welfare Ministerial Notification). [Google Scholar]
- 5.Li J., You W. Analysis of nuclear types in chromosome of Japanese Atractylodes. Journal of Shenyang Agricultural University. 1987;18:75–77. [Google Scholar]
- 6.Kang E. M., Jeong C. H., Shim K. H. Functional properties of Korean Atractylodes japonica Koidz. Korean Journal of Post-Harvest Science and Technology of Agricultural Products. 2001;8:86–91. [Google Scholar]
- 7.Wu Z. Y. Flora of China. Vol. 20. Beijing, China: Science Press; 2011. [Google Scholar]
- 8.Park J. M., Jang K. H., Lee S. T., et al. Growth characteristics of Atractylodes japonica Koidz. In its native habitat. Korean Journal of Medicinal Crop Science. 2000;8:327–333. [Google Scholar]
- 9.Ma X. Q., Duan J. A., Zhu D. Y., Dong T. T. X., Tsim K. W. K. Species identification of Radix astragali (Huangqi) by DNA sequence of its 5S-rRNA spacer domain. Phytochemistry. 2000;54(4):363–368. doi: 10.1016/s0031-9422(00)00111-4. [DOI] [PubMed] [Google Scholar]
- 10.Choo B. K., Moon B. C., Ji Y., et al. Development of SCAR markers for the discrimination of three species of medicinal plants, Angelica decursiva (Peucedanum decursivum), Peucedanum praeruptorum and Anthricus sylvestris, based on the internal transcribed spacer (ITS) sequence and random amplified polymorphic DNA (RAPD) Biological and Pharmaceutical Bulletin. 2009;32(1):24–30. doi: 10.1248/bpb.32.24. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y.-Q., Wang N., Qu L.-H., Li T.-H., Zhang W.-M. Determination of the anamorph of Cordyceps sinensis inferred from the analysis of the ribosomal DNA internal transcribed spacers and 5.8S rDNA. Biochemical Systematics and Ecology. 2001;29(6):597–607. doi: 10.1016/s0305-1978(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 12.Takamiya T., Wongsawad P., Tajima N., et al. Identification of Dendrobium species used for herbal medicines based on ribosomal DNA Internal transcribed spacer sequence. Biological and Pharmaceutical Bulletin. 2011;34(5):779–782. doi: 10.1248/bpb.34.779. [DOI] [PubMed] [Google Scholar]
- 13.Wu C.-T., Hsieh C.-C., Lin W.-C., et al. Internal transcribed spacer sequence-based identification and phylogenic relationship of I-Tiao-Gung originating from Flemingia and Glycine (Leguminosae) in Taiwan. Journal of Food and Drug Analysis. 2013;21(4):356–362. doi: 10.1016/j.jfda.2013.08.002. [DOI] [Google Scholar]
- 14.Shiba M., Kondo K., Miki E., et al. Identification of medicinal Atractylodes based on ITS sequences of nrDNA. Biological and Pharmaceutical Bulletin. 2006;29(2):315–320. doi: 10.1248/bpb.29.315. [DOI] [PubMed] [Google Scholar]
- 15.Kondo K., Shiba M., Yotsuyanagi Y., Nishimura N., Maruyama T., Goda Y. Discrimination between Atractylodes rhizome (Byaku-jutsu) and Atractylodes lancea rhizome (So-jutsu) by the PCR-RFLP analysis of ITS region on nrDNA nrDNA. Journal of Japanese Botany. 2009;84(6):356–359. [Google Scholar]
- 16.Ge Y.-F., Hang Y.-Y., Xia B., Wei Y.-L. Sequencing of trnL-F and analysis of interspecific genetic relationship of five medicinal species in Atractylodes DC. Journal of Plant Resources and Environment. 2007;16(2):12–16. [Google Scholar]
- 17.Peng H.-S., Yuan Q.-J., Li Q.-Q., Huang L.-Q. Molecular systematics of genus atractylodes (Compositae, Cardueae): evidence from internal transcribed spacer (ITS) and trnL-F sequences. International Journal of Molecular Sciences. 2012;13(11):14623–14633. doi: 10.3390/ijms131114623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D.-Z., An Y.-Q., Jiang X.-L., et al. Development of a novel method combining HPLC fingerprint and multi-ingredients quantitative analysis for quality evaluation of traditional Chinese medicine preparation. Talanta. 2011;85(2):885–890. doi: 10.1016/j.talanta.2011.04.059. [DOI] [PubMed] [Google Scholar]
- 19.Kang J., Zhou L., Sun J., Han J., Guo D.-A. Chromatographic fingerprint analysis and characterization of furocoumarins in the roots of Angelica dahurica by HPLC/DAD/ESI-MSn technique. Journal of Pharmaceutical and Biomedical Analysis. 2008;47(4-5):778–785. doi: 10.1016/j.jpba.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Xie B., Gong T., Tang M., et al. An approach based on HPLC-fingerprint and chemometrics to quality consistency evaluation of Liuwei Dihuang Pills produced by different manufacturers. Journal of Pharmaceutical and Biomedical Analysis. 2008;48(4):1261–1266. doi: 10.1016/j.jpba.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Xu X., Jiang J., Liang Y., Yi L., Cheng J. Chemical fingerprint analysis for quality control of Fructus aurantii immaturus based on HPLC-DAD combined with chemometric methods. Analytical Methods. 2010;2(12):2002–2010. doi: 10.1039/c0ay00455c. [DOI] [Google Scholar]
- 22.Sajewicz M., Staszek D., Wróbel M. S., et al. The HPLC/DAD fingerprints and chemometric analysis of flavonoid extracts from the selected Sage (Salvia) species. Chromatography Research International. 2012;2012:8. doi: 10.1155/2012/230903.230903 [DOI] [Google Scholar]
- 23.Zhang Q.-F., Cheung H.-Y., Zeng L.-B. Development of HPLC fingerprint for species differentiation and quality assessment of Rhizoma Smilacis Glabrae . Journal of Natural Medicines. 2013;67(1):207–211. doi: 10.1007/s11418-012-0648-9. [DOI] [PubMed] [Google Scholar]
- 24.Tao W., Duan J., Zhao R., et al. Comparison of three officinal Chinese pharmacopoeia species of Glycyrrhiza based on separation and quantification of triterpene saponins and chemometrics analysis. Food Chemistry. 2013;141(3):1681–1689. doi: 10.1016/j.foodchem.2013.05.073. [DOI] [PubMed] [Google Scholar]
- 25.Nakai Y., Yano K., Shiba M., et al. Chemical characterization of rhizomes of Atractylodes lancea and A. chinensis identified by ITS sequences of nrDNA. Journal of Japanese Botany. 2006;81(2):63–74. [Google Scholar]
- 26.Xia Y.-G., Yang B.-Y., Wang Q.-H., Liang J., Wang D., Kuang H.-X. Species classification and quality assessment of Cangzhu (Atractylodis rhizoma) by high-performance liquid chromatography and chemometric methods. Journal of Analytical Methods in Chemistry. 2013;2013:7. doi: 10.1155/2013/497532.497532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White T. J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., editors. PCR Protocols: A Guide to Methods and Applications. New York, NY, USA: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 28.Garcia-Jacas N., Garnatje T., Susanna A., Vilatersana R. Tribal and subtribal delimitation and phylogeny of the cardueae (Asteraceae): a combined nuclear and chloroplast DNA analysis. Molecular Phylogenetics and Evolution. 2002;22(1):51–64. doi: 10.1006/mpev.2001.1038. [DOI] [PubMed] [Google Scholar]
- 29.Susanna A., Garcia-Jacas N., Hidalgo O., Vilatersana R., Garnatje T. The Cardueae (Compositae) revisited: insights from ITS, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Annals of the Missouri Botanical Garden. 2006;93(1):150–171. doi: 10.3417/0026-6493(2006)93[150:tccrif]2.0.co;2. [DOI] [Google Scholar]
- 30.Mizukami H., Shimizu R., Kohjyouma M., Kohda H., Kawanishi F., Hiraoka N. Phylogenetic analysis of Atractylodes plants based on chloroplast trnK sequence. Biological and Pharmaceutical Bulletin. 1998;21(5):474–478. doi: 10.1248/bpb.21.474. [DOI] [PubMed] [Google Scholar]
- 31.Guo L. P., Huang L. Q., Wang M., Feng X. F., Fu G. F., Yan Y. N. A preliminary study on relationship between Atractylodes lancea and A. chinensis as analyzed by RAPD. Zhongguo Zhong Yao Za Zhi. 2001;26(3):156–158. [PubMed] [Google Scholar]
- 32.Huh M. K., Bang K. H. Identification of Atractylodes japonica and A. macrocephala by RAPD analysis and SCAR markers. Silvae Genetica. 2006;55(3):101–105. [Google Scholar]
- 33.Lee D.-Y., Cho J.-G., Lee M.-K., et al. Discrimination of Panax ginseng roots cultivated in different areas in Korea using HPLC-ELSD and principal component analysis. Journal of Ginseng Research. 2011;35(1):31–38. doi: 10.5142/jgr.2011.35.1.031. [DOI] [Google Scholar]
- 34.Kumooka Y. Hierarchical cluster analysis as a tool for preliminary discrimination of ATR-FT-IR spectra of OPP acrylic and rubber-based adhesives. Forensic Science International. 2009;189(1–3):104–110. doi: 10.1016/j.forsciint.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 35.Goodarzi M., Russell P. J., Heyden Y. V. Similarity analyses of chromatographic herbal fingerprints: a review. Analytica Chimica Acta. 2013;804:16–28. doi: 10.1016/j.aca.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z.-J. Therapeutic effects of herbal extracts and constituents in animal models of psychiatric disorders. Life Sciences. 2004;75(14):1659–1699. doi: 10.1016/j.lfs.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Lu J., Wang J.-S., Kong L.-Y. Anti-inflammatory effects of Huang-Lian-Jie-Du decoction, its two fractions and four typical compounds. Journal of Ethnopharmacology. 2011;134(3):911–918. doi: 10.1016/j.jep.2011.01.049. [DOI] [PubMed] [Google Scholar]
- 38.Tian L., Bi K.-S., Sun W.-J., Zhao S.-C., Wu G.-F., Lu Y. Chemical pattern recognition in the Rhizoma of Atractylodes macrocephala . China Journal of Chinese Materia Medica. 2003;28(2):143–146. [PubMed] [Google Scholar]