Abstract
Almost all of eukaryotic mRNAs are subjected to polyadenylation during mRNA processing. Recent discoveries showed that many of these mRNAs contain more than one polyadenylation sites in their 3′ untranslated regions (UTR) and that alternative polyadenylation (APA) is prevalent among these genes. Many biological processes such as differentiation, proliferation, and tumorigenesis have been correlated to global APA events in the 3′ UTR of mRNAs, suggesting that these APA events are tightly regulated and may play important physiological roles. In this review, recent discoveries in the physiological roles of APA events, as well as the known and proposed mechanisms are summarized. Perspective for future directions is also discussed.
Keywords: 3′ UTR, 3′-end processing, alternative polyadenylation, mTOR, post-transcriptional gene regulation
INTRODUCTION
Polyadenylation
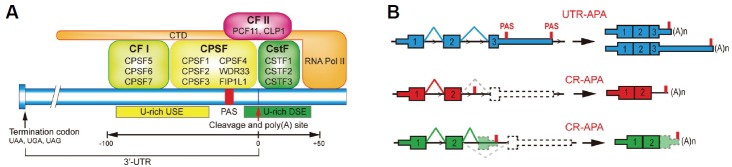
The maturation of most of the eukaryotic mRNAs involve three vital processes: capping, splicing, and polyadenylation. In the process of polyadenylation, a long stretch of untemplated adenosines, the poly(A) tail, is added to the 3′ end of mRNAs. Many studies have unveiled the molecular mechanisms of mRNA 3′ end processing machinery, as well as the roles of polyadenylation in the metabolism of mRNAs (Proudfoot, 2011). Briefly, the maturation of 3′ end of an mRNA involves a two-step reaction: the endonucleolytic cleavage of the pre-mRNA and the addition of poly(A) tail at the cleavage site. This two-step reaction requires the coordinated actions of various protein factors as well as specific sequence elements in pre-mRNA, which can guide the trans-acting factors to form 3′ end processing complex at the cleavage site (Fig. 1A). The core players in the plethora of these protein factors that carry out the cleavage and polyadenylation include multi-subunit protein complexes CPSF (cleavage and polyadenylation specificity factor), CSTF (cleavage stimulation factor), CFI and CFII (cleavage factors I and II), as well as the poly(A) polymerase (PAP). As for the essential sequence elements that signal 3′ end formation, they center on a hexanucleotide poly(A) signal (PAS), whose canonical sequence is AAUAAA. It is often flanked by auxiliary upstream elements (USE, U-rich or UGUA elements) and downstream elements (DSE, U-rich or GU-rich elements). Both USE and DSE have been shown to have a role in facilitating the usage of the PAS, and the “strength” of the PAS can be influenced by the relative positions and sequence variations of these elements (Beaudoing et al., 2000; Danckwardt et al., 2007b; Proudfoot, 2011). Together they form a USE-PAS-DSE pattern for signaling 3′ end processing; this pattern is largely conserved in eukaryotes (Graber et al., 1999). CPSF is the complex that recognizes and binds to the PAS. This binding is assisted by CSTF complex, which binds to DSE, as well as many other factors not mentioned here. The endonuclease CPSF3 then cleaves the pre-mRNA at the cleavage site, which is usually after a CA dinucleotide 15–30 nucleotide downstream of the PAS and 0–20 nucleotide upstream of DSE. Finally, PAP is recruited to the cleavage site to catalyze the addition of the poly(A) tail (Colgan and Manley, 1997). The poly(A) tail has been shown to be essential for many aspects of mRNA metabolism, including translation, stability, localization, etc. (Zhang et al., 2010).
Fig. 1.
(A) 3′ end processing machinery assembly at poly(A) site. The endonucleolytic cleavage at poly(A) site is mediated by the binding of 3′ end processing factors including multi-subunit protein complexes CPSF, CSFT, CFI, and CFII to the USE-PAS-DSE sequence elements. (B) Schematic illustration of UTR-APA and CR-APA of mRNA. Exons are represented by rectangular squares with coding DNA sequences being thicker than UTRs. Introns are represented by thin lines. For UTR-APA, alternative PAS resides in 3′ UTR. Therefore, UTR-APA can generate transcripts with varying UTR lengths without changing the coding sequences. There are two major types of CR-APA, as illustrated. Both types of CR-APA may yield transcripts with truncated coding sequences.
Alternative polyadenylation
With the advent of modern sequencing technologies, recent studies revealed that most of the human genes contain more than one poly(A) sites, suggesting the prevalence of alternative polyadenlylation (APA). In fact, possibly more than 70% of human genes are capable of producing mRNA isoforms by APA (Elkon et al., 2013). There are two major types of APA: UTR-APA (untranslated region alternative polyadenylation) and CR-APA (coding region alternative polyadenylation) (Fig. 1B) (Di Giammartino et al., 2011; Tian and Manley, 2013). In the case of UTR-APA, the alternative poly(A) sites are located in the 3′ UTR of the mRNA, with the majority them residing closer to stop codons (proximal) compared to canonical poly(A) sites (distal). Therefore, usage of the alternative poly(A) sites will result in 3′ UTR shortening in most cases, without changing the coding capacity of the mRNA. Since the 3′ UTR of mRNA often contains many binding sites for regulatory RNA-binding proteins and microRNAs, UTR-APA may lead to altered mRNA stability, localization, or protein translation efficiency due to variation of 3′ UTR length (Fabian et al., 2010). For CR-APA, the alternative poly(A) sites reside in the upstream introns of genes. Unlike UTR-APA, CR-APA can produce different protein isoforms; these protein isoforms lack a chunk of polypeptides at the C-termini, rendering the truncated proteins the potential to behave differently from the full-length counterparts (Hoque et al., 2013).
Alternative poly(A) sites often possess non-canonical poly(A) signals such as AUUAAA, AGUAAA, or UAUAAA, etc. and are considered “weaker”. However, these non-canonical PAS can be favored over the canonical PAS under certain physiological conditions and/or when the levels of trans-acting factors vary, leading to APA (Beaudoing et al., 2000). APA has gained much attention recently as it is considered a new layer of general mechanism for gene expression regulation.
GLOBAL UTR-APA EVENTS IN VARYING BIOLOGICAL PROCESSES
In recent years, advanced sequencing technologies have allowed us to study APA at a transcriptome-wide scale. Many tissue-specific and biological process-specific global APA events have been reported. For example, it has been shown that mRNA transcripts in brain and nervous tissues are prone to usage of distal poly(A) sites, leading to 3′ UTR lengthening, while transcripts in tissues a such as retina and placenta tend to use proximal poly(A) sites (Licatalosi et al., 2008; Zhang et al., 2005). In another study, widespread 3′ UTR lengthening is observed during mouse embryonic development. This progressive 3′ UTR lengthening can also be recapitulated during the differentiation of mouse myoblast cells C2C12 (Ji et al., 2009). On the contrary, intriguingly, 3′ UTR shortening is observed when generating induced pluripotent stem cells from differentiated tissues (Ji and Tian, 2009). Moreover, during B cell and T cell activation, global 3′ UTR shortening is also observed (Sandberg et al., 2008b). Together, these studies suggest that global UTR-APA events are tissue-specific, and that 3′ UTR shortening seem to correlate positively to cell proliferation and negatively to cellular differentiation. Indeed, Sandberg et al. (2008b) showed that, by comparing among different tissues and samples, there is a good correlation between the “proliferative-ness” of the cells and the degree of global 3′ UTR shortening.
Global 3′ UTR shortening is also linked to cancer development. It has been found that transcripts with short 3′ UTRs are widespread in the transcriptomes of various transformed cell lines and cancer samples (Mayr and Bartel, 2009; Singh et al., 2009). Moreover, global 3′ UTR shortening is reported in human colorectal carcinomas, breast, and lung cancers (Lembo et al., 2012; Morris et al., 2012); surprisingly, in these studies, the APA profiles are shown to correlate with the degree of cancer development and prognosis, suggesting the potential utility of APA as a diagnostic marker and treatment target. Nevertheless, whether the APA is driven by the proliferation or transformation of the cancer cells is still unclear (Tian and Manley, 2013).
Despite the substantial progress in our observations of global UTR-APA events and their correlation with biological processes, their biological importance and functional roles in various biological processes are still elusive. Nevertheless, in recent years, a number of specific UTR-APA events are studied more extensively, which can help reveal the physiological roles of the widespread UTR-APA observed in various cellular contexts.
PHYSIOLOGICAL ROLES OF UTR-APA
Since more than 50% of the conserved microRNA target sites reside downstream of proximal poly(A) sites in mammalian genes (Sandberg et al., 2008a), UTR-APA can enable the transcripts to evade microRNA regulation, and can have significant biological effects especially if the mRNAs targeted by UTR-APA are coding for genes that play regulatory roles in cellular signaling. One interesting example is one of the master regulators of myogenic differentiation Pax3, whose transcript has a miR-206 target site in the 3′ UTR. It has been shown that in different muscle types, muscle stem cells express varying ratios of long and short 3′ UTR containing Pax3 transcripts, leading to varying degrees of negative regulation by miR-206, and thus varying translation efficiencies, which may partially explain the different differentiation patterns in different muscle types (Boutet et al., 2012).
3′ UTRs also serve as binding platforms for many different RNA-binding proteins (RBPs). The binding of RBPs to these elements can control various characteristics of the bound mRNA transcripts, such as their stability and localization (Di Giammartino et al., 2011; Tian and Manley, 2013). Therefore, UTR-APA can allow a transcript to avoid the regulation of RBPs, resulting in altered characteristics. For example, RNA-binding protein CELF1 binds to GU-rich elements (GRE) in the 3′ UTR of select transcripts and target them for rapid decay. However, during T cell activation, a number of transcripts involved in cell division undergo UTR-APA and exclude the GRE in their 3′ UTRs, allowing them to evade the regulation by CELF1 and produce more protein products (Beisang et al., 2014). This is a potential mechanism of cellular proliferation regulation. Another example is the RNA-binding protein Mbnl1. It has been shown that many mRNA transcripts that contain Mbnl1 binding sites in their 3′ UTR display differing localization between the long and short 3′ UTR containing isoforms, with the long 3′ UTR transcripts preferentially localizing to insoluble compartments compared to short 3′ UTR transcripts (Wang et al., 2012). Furthermore, in a study on RNA granule formation, it has been observed that transcripts with longer 3′ UTRs tend to precipitate in RNA granules (Han et al., 2012), while in another study, transcripts with short 3′ UTRs tend to form polysomes more efficiently, leading to a more robust protein translation (Chang et al., 2015).
These examples demonstrate that UTR-APA is another layer of gene expression regulation. The modulations of the 3′ UTR length of a transcript can determine when, where, and how much a transcript is to be translated.
The fact that various physiological conditions correlate with the varying profiles of global and specific APA events, as presented above, indicates that APA events are actively regulated. In the past decade, many mechanistic studies on APA have been performed, providing much insight in the mechanistic details of the APA regulation. Below, we will briefly discuss what has been known and proposed regarding the mechanisms of UTR-APA.
MECHANISMS OF UTR-APA
It has been known that the process of polyadenylation is regulated by the strength and availability of the cis-acting sequence elements, as well as the concentration and activity of the trans-acting protein factors (Barabino and Keller, 1999; Danckwardt et al., 2007a). These factors indeed also constitute the regulation of APA events (Di Giammartino et al., 2011; Tian and Manley, 2013):
One of the first mechanistic insights in APA regulation is the differential expression of CSTF2 during B cell activation (Takagaki et al., 1996). When B cells are activated, the level of CSTF2 is upregulated, leading to a preferential usage of the weaker proximal poly(A) sites of some genes (Chuvpilo et al., 1999). Later on, a transcriptome-wide study showed that the level of CSTF2 is indeed positively correlated to the tendency of global 3′ UTR shortening (Yao et al., 2012). The proposed mechanism for CSTF2 regulated APA is the following: when CSTF2 is limited, distal poly(A) site with strong canonical PAS is preferred. However, when CSTF2 is upregulated, it can bind to the DSE near proximal poly(A) site more efficiently to facilitate its usage, albeit having a weaker PAS sequence. Moreover, according the first-come-first-served model (Danckwardt et al., 2007a), which is well-accepted in the field, another advantage the proximal poly(A) site has is that with the facilitation of enhanced CSFT2 binding, it has a higher chance to be utilized before the distal poly(A) site is even transcribed and can become a competition for the recognition of 3′ end processing factors. Apart from CSTF2, factors belonging to CFI, CPSF5 and CPSF6, have also been implicated in influencing poly(A) site choices. It has been reported that the loss-of-function and knockdown of CPSF5 and CPSF6 lead to global usage of proximal poly(A) site (Martin et al., 2012; Masamha et al., 2014; Yang et al., 2010). The proposed mechanism is that since distal poly(A) sites tend to possess stronger CPSF5/6 binding sites in their USEs compared to proximal poly(A) sites, down-regulation of CPSF5 and CPSF6 will render the preferential recruitment of core 3′ end processing factors to distal sites by CPSF5/6 less prominent, leading to more usage of proximal poly(A) sites (Martin et al., 2012). Interestingly, in a subset of glioblastoma tumors, CPSF5 is downregulated. And the downregulation of CPSF5 results in enhanced tumorigenesis in tumor cells, possibly via its APA modulations (Masamha et al., 2014).
Many other 3′ end processing factors and different RBPs have also been found to be capable of regulating APA. For example, PABPN1, whose function was thought to control poly(A) tail length (Kühn et al., 2009), has recently been shown to have a role in modulating 3′ UTR length, as well (de Klerk et al., 2012; Jenal et al., 2012). These studies showed that loss-of-function and knockdown of PABPN1 can promote widespread 3′ UTR shortening in the transcriptome. Moreover, they proposed that PABPN1 has a role in suppressing the usage of proximal poly(A) sites via direct interaction with non-canonical PAS. Splicing factors have also been implicated in APA modulation. U2AF2, the splicing factor binding to polypyrimiding tracts, has been shown to be capable of interacting with and recruiting CFI to facilitate 3′ end formation near polypyrimidine tracts (Millevoi et al., 2006). Other RBPs, such as CPEB1 and Hu, have also been reported to be capable of modulating 3′ UTR lengths, possibly through similar mechanisms (Bava et al., 2013; Zhu et al., 2007).
RECENT DEVELOPMENTS IN THE MECHANISM AND PHYSIOLOGICAL RELEVANCE OF UTR-APA
In an effort to study the mechanism(s) of APA regulation systematically, Li et al. (2015) performed siRNA knockdown and deep sequencing on various 3′ end processing factors, splicing factors, and some RNA binding proteins, and examined their effect on global APA events. Indeed, factors previously known to be capable of regulating global APA events such as CFI, CFII, PABPN1, etc. are shown to affect UTR-APA events across the transcriptome upon knockdown. Moreover, they were able to deduce several regulatory rules in APA modulation.
Although this type of study can reveal the general mechanistic actions of APA regulation, it still comes up short in terms of linking to physiological contexts. For instance, Li et al. (2015) performed their experiments in C2C12 cell line. The APA events and regulations they observe in this particular cell line may not be translatable to other physiological conditions. Moreover, while showing that differential expression of trans-acting factors can modulate APA events is crucial to understandin the mechanism of APA, the mechanisms through which the differential expressions of these factors are achieved in various physiological contexts are still lacking.
One of the puzzle pieces that can potentially fill in this missing link is the transcription regulation by E2F transcription factors. It has been shown that many 3′ end processing factors tend to possess proliferation-related sequence elements in their promoters, such as E2F binding sites, and their transcriptions are indeed affected by E2F knockdown (Elkon et al., 2012). This may provide a partial mechanistic explanation for the correlation between proliferation and upregulation of polyadenylation factors, which leads to 3′ UTR shortening.
Furthermore, we have recently shown that mTORC1 activation also drives global 3′ UTR shortening in the transcriptome (Chang et al., 2015). By using various genetic and chemical means to modulate mTORC1 activity, we observed that ∼16.4% of the UTR-APA-eligible transcripts can be actively targeted for 3′ UTR shortening. To understand the physiological role of the mTORC1-activated 3′ UTR shortening events, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on the genes that show significant 3′ UTR shortening upon mTORC1 activation. Intriguingly, in our analysis, ubiquitin-mediated proteolysis pathway is the pathway most targeted by mTORC1-mediated 3′ UTR shortening. When comparing the differential expression data from quantitative mass spectrometry and RNA deep sequencing, we observed that genes belonging to ubiquitin-mediated proteolysis pathway show increased protein expressions without changing mRNA levels significantly upon mTORC1 activation, suggesting that the upregulation in protein level is mediated by 3′ UTR shortening. Indeed, in the polysome profiling experiment, we found that shorter 3′ UTR-containing transcripts of these genes tend to form polysomes more efficiently than their longer 3′ UTR-containing counterparts, indicating elevated translation efficiency. Overall, we demonstrated that mTORC1 can activate ubiquitin-mediated proteolysis pathway by selectively targeting their mRNA transcripts for 3′ UTR shortening (Fig. 2).
Fig. 2.
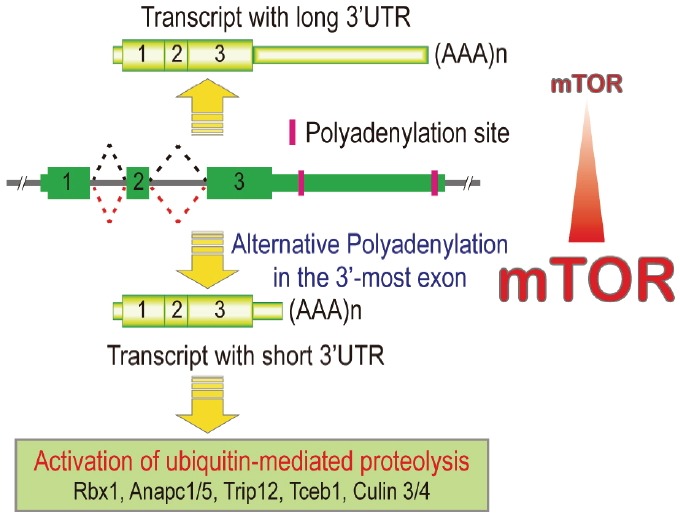
mTORC1 activation mediates global 3′ UTR shortening in the transcriptome. Genes belonging to the ubiquitin-mediated proteolysis pathway are shown to be preferentially targeted by mTOR-mediated 3′ UTR shortening.
As mentioned above, widespread 3′ UTR shortening is a characteristic of cancer transcriptomes. Our study may provide a cellular mechanistic explanation for these global 3′ UTR shortening events as mTORC1 pathway is often hyperactivated in various cancer types (Laplante and Sabatini, 2012). Moreover, novel physiological roles of 3′ UTR shortening and mTORC1 pathway in cancer biology are implicated as we have preliminary data demonstrating that tumor suppressors such as Arf and Securin are targeted for faster turnover by the upregulated ubiquitin-mediated proteolysis pathway.
As for the mechanism of how mTORC1 mediates global 3′ UTR shortening, we have observed that many of the known regulators of UTR-APA display altered expression levels upon mTORC1 activation. We believe that the mTORC1-mediated global 3′ UTR shortening events are the result of the combinatorial actions by the differential expressions of these trans-acting factors. Nonetheless, more detailed mechanistic research is needed to obtain a clearer picture in this regard.
CONCLUSION
UTR-APA is a widespread post-transcriptional regulatory mechanism that can modulate the behavior of mRNA transcripts. As demonstrated by the above mentioned individual examples of the physiological roles of UTR-APA, the global UTR-APA events coupled with various biological processes can most likely play significant roles in regulating cellular biology. However, the physiological roles of most of the biological process-coupled global UTR-APA events are still missing. To this end, technologies and experiments capable of examining the differential protein expression efficiency, stability, and localization of UTR-APA isoforms at a transcriptome-wide scale need to be employed in various relevant biological contexts. Moreover, these global UTR-APA events have been shown to be actively regulated. Current understandings of the mechanisms of APA regulation tend to be limited to the direct actions of various trans-acting factors. However, how are the actions of these trans-acting factors are regulated by cellular signaling pathways in various cellular contexts is still largely unknown. To better understand the functional role and significance of global UTR-APA events in their respective biological processes, and perhaps to be able to better manipulate these APA events as therapeutic means, more research needs to be done to link cellular signaling pathways to global APA events.
Acknowledgments
This work was partly supported by the start-up funds from the University of Minnesota Twin Cities and the grant from National Institute of Health 1 R01 GM113952-01A1.
REFERENCES
- Barabino S.M.L., Keller W. Last but not least: fegulated Poly(A) tail formation. Cell. 1999;99:9–11. doi: 10.1016/s0092-8674(00)80057-4. [DOI] [PubMed] [Google Scholar]
- Bava F., Eliscovich C., Ferreira P.G., Minana B., Ben-Dov C., Guigo R., Valcarcel J., Mendez R. CPEB1 coordinates alternative 3prime]-UTR formation with translational regulation. Nature. 2013;495:121–125. doi: 10.1038/nature11901. [DOI] [PubMed] [Google Scholar]
- Beaudoing E., Freier S., Wyatt J.R., Claverie J., Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisang D., Reilly C., Bohjanen P.R. Alternative polyadenylation regulates CELF1/CUGBP1 target transcripts following T cell activation. Gene. 2014;550:93–100. doi: 10.1016/j.gene.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S., Cheung T., Quach N., Liu L., Prescott S.L., Edalati A., Iori K., Rando T. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Zhang W., Yeh H., de Jong E.P., Jun S., Kim K., Bae S.S., Beckman K., Hwang T.H., Kim K., et al. mRNA 3prime]-UTR shortening is a molecular signature of mTORC1 activation. Nat. Commun. 2015;6:7218. doi: 10.1038/ncomms8218. [DOI] [PubMed] [Google Scholar]
- Chuvpilo S., Zimmer M., Kerstan A., Glöckner J., Avots A., Escher C., Fischer C., Inashkina I., Jankevics E., Berberich-Siebelt F., et al. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity. 1999;10:261–269. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Danckwardt S., Hentze M.W., Kulozik A.E. 3â€2 end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2007a;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S., Kaufmann I., Gentzel M., Foerstner K.U., Gantzert A., Gehring N.H., Neu-Yilik G., Bork P., Keller W., Wilm M., et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007b;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk E., Venema A., Anvar S.Y., Goeman J.J., Hu O., Trollet C., Dickson G., den Dunnen J.T., van der Maarel S.M., Raz V., et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 2012;40:9089–9101. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino D.C., Nishida K., Manley J.L. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R., Drost J., van Haaften G., Jenal M., Schrier M., Oude Vrielink J., Agami R. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R., Ugalde A.P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T., Kato M., Xie S., Wu L., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B. Analysis of alternative cleavage and polyadenylation by 3prime] region extraction and deep sequencing. Nat. Meth. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M., Elkon R., Loayza-Puch F., van Haaften G., Kühn U., Menzies F., Vrielink J.F., Bos A., Drost J., Rooijers K., et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Ji Z., Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Lee J.Y., Pan Z., Jiang B., Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U., Gündel M., Knoth A., Kerwitz Y., Rüdel S., Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo A., Di Cunto F., Provero P. Shortening of 3â€2 UTRs correlates with poor prognosis in breast and lung cancer. PLoS One. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., Park J.Y., Gunderson S.I., Kalsotra A., Manley J.L., et al. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 2015;11:e1005166. doi: 10.1371/journal.pgen.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Gruber A., Keller W., Zavolan M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Masamha C.P., Xia Z., Yang J., Albrecht T.R., Li M., Shyu A., Li W., Wagner E.J. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C., Bartel D.P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S., Loulergue C., Dettwiler S., Karaa S.Z., Keller W., Antoniou M., Vagner S. An interaction between U2AF 65 and CF Im links the splicing and 3â€2 end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.R., Bos A., Diosdado B., Rooijers K., Elkon R., Bolijn A.S., Carvalho B., Meijer G.A., Agami R. Alternative cleavage and polyadenylation during colorectal cancer development. Clin. Cancer Res. 2012;18:5256–5266. doi: 10.1158/1078-0432.CCR-12-0543. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J., Sarma A., Sharp P., Burge C. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science (New York, N.Y.) 2008a;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008b;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Alley T.L., Wright S.M., Kamdar S., Schott W., Wilpan R.Y., Mills K.D., Graber J.H. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y., Seipelt R.L., Peterson M.L., Manley J.L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Tian B., Manley J.L. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Cody N.L., Jog S., Biancolella M., Wang T., Treacy D., Luo S., Schroth G., Housman D., Reddy S., et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Gilmartin G.M., Doublié S. Structural basis of UGUA recognition by the Nudix protein CFIm25 and implications for a regulatory role in mRNA 3′ processing. Proc. Natl. Acad. Sci. USA. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Biesinger J., Wan J., Weng L., Xing Y., Xie X., Shi Y. Transcriptome-wide analyses of CstF64–RNA interactions in global regulation of mRNA alternative polyadenylation. Proc. Natl. Acad. Sci. USA. 2012;109:18773–18778. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lee J., Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Virtanen A., Kleiman F.E. To polyadenylate or to deadenylate: That is the question. Cell Cycle. 2010;9:4437–4449. doi: 10.4161/cc.9.22.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhou H., Hasman R.A., Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]