Abstract
We have previously reported the role of miR-217 in anti-cancer drug-resistance. miRNA array and miRNA hybridization analysis predicted miR-30a-3p as a target of miR-217. miR-30a-3p and miR-217 formed a negative feedback loop and regulated the expression of each other. Ago1 immunoprecipitation and co-localization analysis revealed a possible interaction between miR-30a-3p and miR-217. miR-30a-3p conferred resistance to anti-cancer drugs and enhanced the invasion, migration, angiogenic, tumorigenic, and metastatic potential of cancer cells in CAGE-dependent manner. CAGE increased the expression of miR-30a-3p by binding to the promoter sequences of miR-30a-3p, suggesting a positive feedback loop between CAGE and miR-30a-3p. miR-30a-3p decreased the expression of p53, which showed the binding to the promoter sequences of miR-30a-3p and CAGE in anti-cancer drug-sensitive cancer cells. Luciferase activity assays showed that p53 serves as a target of miR-30a. Thus, the miR-30a-3p-CAGE-p53 feedback loop serves as a target for overcoming resistance to anti-cancer drugs.
Keywords: anti-cancer drug-resistance, CAGE, feedback loop, miR-30a, p53
INTRODUCTION
CAGE, a cancer/testis antigen, is present in the sera of patients presenting with gastric (Cho et al., 2002), endometrial (Iwata et al., 2005) and hematological malignancies (Liggins et al., 2010). Hypomethylation of CAGE increases its expression (Cho et al., 2003). CAGE displays oncogenic potential and regulates the expression of cyclins (Por et al., 2010). It confers resistance to microtubule-targeting drugs by regulating the expression of p53 through interaction with HDAC2 (Kim et al., 2010). miR-200b forms a negative feedback loop with CAGE and regulates the response to anti-cancer drugs (Kim et al., 2013). Further identification of miRNAs that regulate the expression of CAGE will be necessary for better understanding the mechanisms associated with CAGE-mediated resistance to anti-cancer drugs.
miRNAs regulate anti-cancer drug-resistance, for instance, miR-199a suppresses tumorigenicity and multidrug resistance of ovarian cancer-initiating cells (Cheng et al., 2012). miR-27a reverses the multidrug resistance phenotype by regulating the expression of MDR1 and β-catenin (Chen et al., 2013). Ph (+) leukemia cells acquire resistance to tyrosine kinase inhibitors via down-regulation of miR-217 (Nishioka et al., 2014). miR-326, which is increased in anti-cancer drug-resistant cancer cells, regulates the response to anti-cancer drugs by forming a negative feedback loop with HDAC3 (Kim et al., 2014). miR-217 acts as a tumor suppressor and regulates the resistance to cisplatin in lung cancer cells (Guo et al., 2014). Anti-cancer drug-resistant breast cancer cells spread resistance capacity to sensitive cells by releasing exosomes, and such an effect could be partly attributed to the intercellular transfer of specific miRNAs such as miR-30a (Chen et al., 2014b). miR-30a, miR-382, and miR-136 were shown to be down-regulated in bromocriptine-resistant prolactinomas in comparison with their bromocriptine-sensitive counterparts (Wu et al., 2014). Moreover, miR-136 reverses cisplatin resistance by targeting E2F (Chen et al., 2014a). These reports indicate roles of miRNAs in anti-cancer drug-resistance.
In the present study, we investigated the role of miR-30a, which is increased in anti-cancer drug-resistant cancer cell lines, in anti-cancer drug resistance. We show that miR-30a and CAGE form a positive feedback loop, and that miR-30a regulates the response to anti-cancer drugs in a CAGE-dependent manner. Moreover, we show that miR-30a targets miR-217 to regulate the response to anti-cancer drugs, and suggest a possible interaction between miR-30a and miR-217 based on Ago1 immunoprecipitation and co-localization analysis. Further, we show the binding of p53 to the promoter sequences of CAGE and miR-30a, in addition to the feedback loops among miR-30a, miR-217, CAGE and p53. Our results show that the miR-30a-CAGE-p53 feedback loop serves as a target for overcoming resistance to anti-cancer drugs.
MATERIALS AND METHODS
Materials
Anti mouse and anti rabbit IgG-horse radish peroxidase conjugate antibodies were purchased from Pierce Company. An ECL (enhanced chemiluminiscence) kit was purchased from Amersham. Lipofectamin and Plus™ reagent were purchased from Invitrogen (USA).
miRNA array kit was purchased from Signosis, Inc. miR mimic and miR inhibitor were purchased from Bioneer Company (Korea). miR-30acy3, mutant miR-30acy3 and miR-217cy5 were purchased from Bioneer Company. Anti-CAGE antibody was purchased from AbCam. All other antibodies used in this study were purchased from Santa Cruz Company. Oligonucleotides used in this study were commercially synthesized by Bioneer Company (Korea). Chemicals used in this study were purchased from Sigma Chemical Company.
Cell lines and cell culture
Hepatoma or melanoma cell line made resistant to celstrol, a microtubule-targeting drug (SNU387R or Malme3MR), were established by stepwise addition of celastrol. Cells surviving drug treatment (attached fraction) were obtained and used throughout this study. Malme3MR or SNU387R cells that stably express miR-217 (Malme3MR-miR-217 or SNU387R-miR-217) were also selected by G418 (400 μg/ml).Taxol-resistant cancer cell lines (Malme3MR-Taxol) were made by stepwise addition of taxol. Cancer Cells were grown in Dulbecco’s modified Eagle’s medium containing heat-inactivated fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cultures were maintained in 5% CO2 at 37°C.
Western blot analysis
Western blot analysis and immunoprecipitation were performed according to the standard procedures (Kim et al., 2014). For analysis of proteins from tumor tissues, frozen samples were grounded to a fine powder using a mortar and pestle over liquid nitrogen. Proteins were solubilized in RIPA buffer containing protease inhibitors and insoluble material removed by centrifugation.
Cell viability determination
The cells were assayed for their growth activity using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT; Sigma). Viable cell number counting was carried out by trypan blue exclusion assays.
Caspase-3 activity assays
Caspase-3 activity was measured according to the manufacturer’s instructions (BioVision, USA). Cells were lysed in 0.1 M HEPES buffer, pH 7.4, containing 2 mM dithiothreitol, 0.1% CHAPS, and 1% sucrose. Cell lysates were incubated with a colorimetric substrate, 200 μM Ac-DEVD-p-nitroanilide, for 30 min at 30°C. The fluorescence was measured at 405 nm using a microtiter plate reader.
miRNA array analysis and miRNA hybridization analysis
miRNA analysis was performed according to the standard procedures (Kim et al., 2014). The miRNA array kit was purchased from Koma Biotech (Korea). In order to identify miRNAs that hybridize with miR-217, we employed RNAhybrid and miRanda algorithms.
Chemo invasion assays
The invasive potential of cancer cells was determined by using a transwell chamber system with 8-μm pore polycarbonate filter inserts (CoSTAR, USA). The lower and upper sides of the filter were coated with gelatin and Matrigel, respectively. Trypsinized cells (5 × 103) in the serum-free RPMI 1640 medium containing 0.1% bovine serum albumin were added to each upper chamber of the transwell. RPMI 1640 medium supplemented with 10% fetal bovine serum was placed in the lower chamber and cells were incubated at 37°C for 16 h. The cells were fixed with methanol and the invaded cells were stained and counted. Results were analyzed for statistical significance using the Student’s t test. Differences were considered significant when p < 0.05.
Wound migration
Cells were plated overnight to achieve a confluent layer in 24-well plates. A scratch was made on the cell layer with a micropipette tip and cultures were washed twice with serum-free medium. Cells were then transfected with construct of interest. Wound healing was visualized by comparing photographs taken at the time of transfection and 48 h later.
Chromatin immunoprecipitation (ChIP) assays
Assays were performed according to manufacturer’s instruction (Upstate). For detection of binding of protein of interest to miR-30a promoter sequences, specific primers of miR-30a promoter-1 sequences [5′-AAAAATATCCGCCATAAGAAAAAT-3′ (sense) and 5′-CTGTTTCCATTTGGTTGAACTT-3′ (antisense)], miR-30a promoter-2 sequences [5′-TGAAACTGCAGAAAGGGCAG-3′ (sense) and 5′-TTTAGAGGGCTTCAGCAGGG-3′ (antisense)] were used. For detection of binding of protein of interest to CAGE promoter sequences, specific primers of CAGE promoter-1 sequences [5′-CCTGACAAAGTACTGTATTCACTCCA-3′ (sense) and 5′-TGGCTCAGCTTGAGAGCAAC-3′ (antisense)], CAGE promoter-2 sequences [5′-CGCAGAAGTTAAGGAGG CAG-3′ (sense) and 5′-AAGTTGCCCCAGAAACCAGT-3′ (antisense)] and CAGE promoter-3 sequences [5′-ATGTGACT AGCACCCGGAAA-3′ (sense) and 5′-GGGATAGTGGGAGTA TCGGC-3′ (antisense)] were used.
Immunohistochemistry
Paraffin-embedded tissue sections were immunostained using the Vecta stain ABC Elite Kit (Vector Laboratories).Tissue sections were deparaffinized with xylene and washed in ethanol. Endogenous peroxidase activity is blocked with 3% hydrogen peroxide and H2O for 10min. Slides were then blocked with 5% normal goat serum in TBS containing 0.1%Tween-20 (TBS-T) for 1h. For immunohistochemistry, a primary antibody to CAGE (1:100, Santa Cruz), MDR1 (1:100, Santa Cruz) or IgG (1:100, Santa Cruz) was added and incubation continued at 4°C for 24 h. After washing with TBS-T, slides were treated with biotinylated secondary antibody for 30 min. After washing, slides were incubated in the ABC complex for 30 min, and then stained with diaminobenzidine (DAB, Sigma). For H&E staining, tumor tissue samples were fixed in 10% (v/v) buffered formalin, embedded in paraffin, sectioned at 4 μm, and then stained with hematoxylin and eosin. Sections were mounted using Fixo gum rubber cement (Mercateo, Germany).
Localization of miR-30a and miR-217
Cy3-labeled miR-30a, Cy3-labeled miR-30a-mutant or cy5-labeled miR-217 was transfected by using linear polyethylenimine (PEI, Sigma) according to the manufacturer’s instructions. Briefly, the cells were transfected with 200 nM of cy-labeled miRNAs in serum-free media for 4 h. Medium was then replaced with serum-containing media for additional incubation. Twenty four hours after incubation, the cells were fixed with 4 % paraformaldehyde and imaged using a confocal microscope. Cells were counterstained with DAPI (Sigma).
miRNA target analysis
Genes that contain the miR-binding site(s) in the UTR were obtained using the TargetScan program.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total miRNA was isolated using the mirVana miRNA isolation (Ambion). miRNA was extended by a poly (A) tailing reaction using the A-Plus Poly (A) Polymerase Tailing Kit (Cell Script). CDNA was synthesized from miRNA with poly (A) tail using a poly (T) adaptor primer and qScript™ reverse transcriptase (Quanta Biogenesis). The expression level of miR-30a or other miRNA gene was quantified with SYBR Green qRT-PCR kit (Ambion) using a miRNA-specific forward primer and a universal poly (T) adaptor reverse primer. The expression of miR-30a was defined based on the threshold (Ct), and the relative expression levels were calculates as 2− [(Ct of miR-30a)−(Ct of U6)] after normalization with reference to the expression of U6 small nuclear RNA. For quantitative PCR, SYBR PCR Master Mix (Applied Biosystems) was used in a CFX96 Real-Time System thermocycler (BioRad).
Intravital microscopy
Male BALB/c mice (6–8 week old) were obtained from Daehan Biolink (Korea). In vivo angiogenesis was assessed as follows. The mice were anesthetized with 2.5% avertin (v/v) via intraperitoneal injection (Surgivet, USA), and abdominal wall windows were implanted. Next, a titanium circular mount with eight holes on the edge was inserted between the skin and the abdominal wall. Growth factor-reduced matrigel containing the conditioned medium was applied to the space between the windows, and a circular glass cover slip was placed on top and fixed with a snap ring. After four days, the animals were anesthetized and injected intravenously with 50 μl of 25 ng/ml fluorescein isothiocyanate-labeled dextran (molecular weight, Mr ∼2,000,000) via the tail vein. The mice were then placed on a Zeiss Axiovert 200 M microscope. The epi-illumination microscopy setup included a 100-W mercury lamp and filter set for blue light. Fluorescence images were recorded at random locations of each window using an electron-multiplying charge coupled device camera (Photo Max 512, Princeton Instruments, USA) and digitalized for subsequent analysis using the Metamorph program (Universal Imaging, USA). The assay was scored from 0 (negative) to 5 (most positive) in a double-blinded manner.
Tumorigenic potential
Athymic nude mice (BALB/c nu/ nu, 5-6-week-old females) were obtained from Orient Bio Inc. (Korea) and were maintained in a laminar air-flow cabinet under aseptic conditions. All animal experiments were approved by the Institutional Animal Care and Use Committee of Kangwon National University (KW-140707-1). Cancer cells (1 × 106) were injected subcutaneously into the dorsal flank area of the mice. Tumor volume was determined by direct measurement with calipers and calculated by the following formula: length X width X height X 0.5. To examine whether miR-30a targets CAGE to enhance the tumorigenic potential of cancer cells, Malme3M (106 cells) cells were injected into the dorsal flanks of athymic nude mice. miR-30a mimic (50 μM/kg) and CAGE siRNA (50 μM/kg) were intravenously injected 4 times in a total of 28 days.
In vivo metastasis assay
Female athymic nude mice were used for the studies. To examine whether miR-30a targets CAGE to enhance the meta-static potential of cancer cells, Malme3M (106 cells) cells were intravenously injected into athymic nude mice. The miR-30a mimic (50 μM/kg) and CAGE siRNA (50 μM/kg) were intravenously injected 6 times in a total of 21 days. After 21 days, the mice were sacrificed and analyzed for the lung colonization. The lungs were rinsed with PBS and then fixed and stained with Bouin’s solution. After 24 h, the lungs were rinsed in water to remove excess Bouin’s solution and the extent of lung metastases was quantified.
miR-30a, miR-22, pGL3-3′ UTR-CAGE, pGL3 3′-UTR-p53 and pGL3-miR-217 construct
To generate miR-30a vector, a 335 bp genomic fragment encompassing primary miR-30a gene was PCR amplified and cloned into BamHI/XhoI site of pcDNA3.1 vector. To generate pGL3-miR-217 luciferase construct, 420 bp genomic fragment encompassing primary miR-217 gene was PCR amplified and cloned into XbaI site of pGL3 luciferase plasmid. Luciferase activity assay was performed according to the instruction manual (Promega Company). To generate miR-22 vector, a 438 bp genomic fragment encompassing primary miR-22 gene was PCR amplified and cloned into BamHI/XhoI site of pcDNA3.1 vector. To generate the pGL3 3′-UTR-p53 construct, a 492 bp human p53 gene segment encompassing 3′-UTR was PCR amplified and subcloned into the XbaI site of pGL3 luciferase plasmid. The mutant pGL3 3′-UTR-p53 construct and mutant miR-30a construct were made with the QuikChange site-directed mutagenesis kit (Stratagene). The pGL3 3′-UTR-p53 contains binding sequences for miR-30a. The pGL3 p53 del-3′-UTR construct lacks binding sequences for miR-30a. Luciferase activity assay was performed according to the instruction manual (Promega). Wild type miR-30acy3 sequences used in this study are 5′-CUUUCAGUCGGAUG UU UGCAGC-3′. Mutant miR-30acy3 sequences used in this study are 5′-CUGGACUGAUU AUGUUUGCAGC-3′. Wild type miR-217cy5 sequences used in this study are 5′-UACUGCAUCAGGAA CUGAUUGGA-3′.
Statistical analysis
Statistical differences were determined by using the Student’s t test.
RESULTS
miR-30a forms a negative feedback loop with miR-217
We previously reported the role of the miR-200b-CAGE negative feedback loop in the response to anti-cancer drugs (Kim et al., 2013). HDAC3, which shows an inverse relationship with CAGE, forms a positive feedback loop with miR-217 and miR-200b to confer sensitivity to anti-cancer drugs (Kim et al., 2014). We hypothesized that miRNAs that negatively regulate the expression of miR-217 would confer resistance to anti-cancer drugs. In an effort to further investigate the mechanism of anti-cancer drug-resistance regulated by CAGE, we performed a miRNA array analysis. Our aim was to identify miRNAs that show a higher expression level in Malme3MR cells than in Malme3M cells, and those that show a lower expression in Malme3MR-miR-217 cells than in Malme3MR cells. We subjected Malme3M, Malme3MR, and Malme3MR-miR-217 cells to miRNA array analysis. miR-181d, miR-22, miR-23, miR-24, miR-30a-3p and miR-30a-5p were shown to be regulated by miR-217 (Fig. 1A). RNA hybridization analysis showed that miR-30-3p and miR-22 were predicted to hybridize with miR-217 (Fig. 1B). The miR-30a/Eya2 (Eye absent) axis regulates breast cancer development and progression (Fu et al., 2014). In other words, miR-30a targets Eya2 and suppresses cancer cell proliferation. Moreover, RUNX3 has been shown to suppress gastric cancer cell invasion by activating miR-30a in gastric cancer patients (Liu et al., 2014). However, miRNA array analysis suggests that miR-30a-3p may act as an oncogene in melanoma cells. qRT-PCR analysis showed that the expression level of miR-217 was inversely correlated with miR-30-3p and miR-22 (Fig. 1C). We examined whether miR-30a-3p and miR-22 would directly regulate the expression of miR-217. Specifically, we examined the possibility of negative feedback loop between miR-217 and miR-30a-3p/-22. miR-30a-3p and miR-22 decreased the expression of miR-217 in Malme3M cells (Fig. 1D). miR-30a-3p and miR-22 decreased the luciferase activity associated with 3′-UTR-miR-217 (Fig. 1E). miR-30a-3p and miR-22 increased the expression of CAGE in Malme3M cells (Fig. 1F). Taxol increased the expression of miR-30a-3p in a time-dependent manner (Fig. 1G), suggesting that miR-30a-3p may confer resistance to taxol. Taken together, these results suggest that miR-30a-3p and miR-22 form a negative feedback loop with miR-217 to regulate the expression of CAGE and the response to anti-cancer drugs. Since miR-30a-3p showed stronger binding to miR-217 (Fig. 1B), and a greater effect on the luciferase activity associated with miR-217 (Fig. 1E) than miR-22, we focused our study on miR-30a-3p.
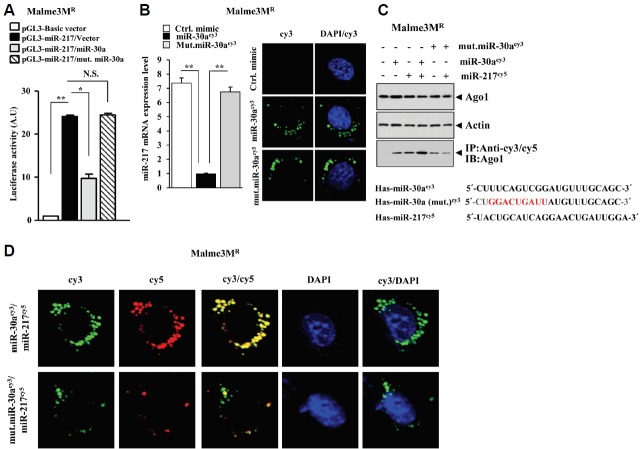
Fig. 1.
miR-30a-3p forms a negative feedback loop with miR-217. (A) miRNA array analysis, employing the indicated cancer cells, was performed as described. The underlined miRNAs are those that are predicted to be regulated by miR-217. (B) The potential duplexes formed between miR-217 and candidate miRNAs. (C) qRT-PCR analysis, employing the indicated cancer cells, was performed as described. *p < 0.05; **p < 0.005. (D) Malme3M cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, cell lysates were prepared and subjected to qRT-PCR analysis. ***p < 0.0005. (E) Malme3MR or SNU387R cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, a luciferase activity assay was performed as described. *p < 0.05; **p < 0.005; ***p < 0.0005. (F) Malme3M cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, cell lysates were prepared and subjected to Western blot analysis. The intensity of the bands was measured using GraphPad Prism 5.04. (G) Malme3M cells were treated with taxol (1 μM) for various time intervals. miRNA isolated at each time point was subjected to qRT-PCR. **p < 0.005; ***p < 0.0005.
miR-30a targets miR-217
Since miR-30a and miR-217 formed a negative feedback loop (Figs.1C and 1D), we examined whether miR-30a directly regulates the expression of miR-217. Wild type miR-30a, but not mutant miR-30a, decreased the luciferase activity associated with miR-217 in Malme3MR cells (Fig. 2A). Point mutations in mutant miR-30a construct do not allow binding of miR-30a to miR-217. Wild type miR-30acy3, but not mutant miR-30acy3, decreased the expression of miR-217 in Malme3MR cells (Fig. 2B) based on qRT-PCR analysis. Confocal image analysis showed the expression and localization of miR-30acy3 and mutant miR-30acy3 (Fig. 2B). These results suggest that miR-217 serves as a direct target of miR-30a. We then examined the possibility of an interaction between miR-30a and miR-217. To this end, Malme3MR cells were transfected with miR-30acy3 or miR-217cy5, or combination of both. Immunoprecipitation with anti-cy3/cy5 antibodies showed that the immunoprecipitates contain Ago1, a component of the RNA induced silencing complex (Fig. 2C), suggesting that miR-30a and miR-217 recruit Ago1. We then examined whether miR-30a and miR-217 would show co-localization. Wild type miR-30a, but not mutant miR-30a, showed co-localization with miR-217 (Fig. 2D). Taken together, these results suggest a potential interaction between miR-30a and miR-217.
Fig. 2.
miR-30a targets miR-217. (A) Malme3MR cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, a luciferase activity assay was performed as described. *p < 0.05; **p < 0.005. (B) Malme3MR cells were transiently transfected with control mimic (10 nM), wild type miR-30acy3 (200 nM) or mutant miR-30acy3 (200 nM). At 48 h after transfection, cell lysates were subjected to qRT-PCR analysis (left panel). Confocal imaging analysis was performed to examine the expression and localization of miR-30a in Malme3MR cells (right panel). **p < 0.005. (C) Malme3MR cells were transiently transfected with the miRNA (each at 200 nM). The 48 h post transfection, cell lyates were subjected to Western blot and immunoprecipitation analysis. (D) Malme3MR cells were transiently transfected with wild type miR-30acy3 (200 nM) or mutant miR-30acy3 (200 nM) along with miR-217cy5 (200 nM). 48 hours post transfection, localization of miR-30a or miR-217 was determined by confocal microscopy.
miR-30a exerts an anti-apoptotic effects and enhances invasion, migration and angiogenic potential of cancer cells
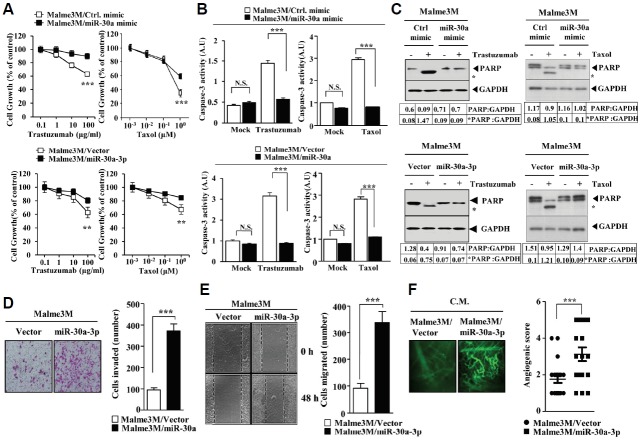
Since miR-30a-3p increased the expression of CAGE (Fig. 1F), we examined whether miR-30a-3p would exert an anti-apoptotic effects. Both the miR-30a-3p mimic and miR-30a-3p conferred resistance to taxol and trastuzumab in Malme3M cells (Fig. 3A), preventing taxol and trastuzumab from increasing caspase-3 activity (Fig. 3B). The miR-30a-3p mimic and miR-30a also prevented taxol and trastuzumab from cleaving PARP in Malme3M cells (Fig. 3C). Over-expression of miR-30a-3p enhanced the invasion (Fig. 3D) and migration potential (Fig. 3E) of Malme3M cells. The conditioned medium of Malme3M cells transfected with miR-30a showed angiogenic potential based on intravital microscopy (Fig. 3F). Taken together, these results suggest that miR-30a-3p exerts an anti-apoptotic effect in a manner associated with its effect on the invasion, migration and angiogenic potential of cancer cells.
Fig. 3.
miR-30a exerts an anti-apoptotic effect and enhances the invasion, migration and angiogenic potential of cancer cells. (A) Malme3M cells were transiently transfected with the indicated mimic (each at 10 nM) or with the indicated construct (1 μg). The following day, cells were treated with various concentrations of taxol or trastzumab for 24 h, followed by the MTT assays. **p < 0.005; ***p < 0.0005. Comparison was made between Malme3M cells transfected with control mimic and Malme3M cells transfected with miR-30 mimic. (B) Malme3M cells were transiently transfected with the indicated mimic (each at 10 nM) or with the indicated construct (each at 1 μg). The following day, cells were treated with taxol (1 μM) or trastuzumab (10 μg/ml) for 24 h, followed by caspase-3 activity assays. ***p < 0.0005. (C) Malme3M cells were transiently transfected with the indicated mimic (each at 10 nM) or with the indicated construct (each at 1 μg). The following, cells were treated with taxol (1 μM) or trastuzumab (10 μg/ml) for 24 h, followed by Western blot analysis. * denotes cleaved PARR. (D) Malme3M cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, a chemoinvasion assay was performed as described. ***p < 0.0005. (E) The indicated cancer cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, cells were subjected to a wound migration assays. ***p < 0.0005. (F) The conditioned medium of Malme3M cells, obtained after transfection with the indicated construct (1 μg), was subjected to intravital microscopy. ***p < 0.0005.
miR-30a requires CAGE to confer resistance to anti-cancer drugs and to regulate the invasion, migration, and angiogenic potential of cancer cells
The down-regulation of CAGE by siRNA prevented the miR-30a mimic from conferring resistance to anti-cancer drugs (Fig. 4A). The miR-30a mimic prevented taxol and trastuzumab from cleaving PARP in Malme3M cells in a CAGE-dependent manner (Fig. 4B), and induced the expression of CAGE in Malme3M cells (Fig. 4B). Malme3M cells showed increased caspase-3 activity in response to taxol and trastuzumab (Fig. 4C). The miR-30a mimic prevented taxol and trastuzumab from increasing caspase-3 activity in a CAGE-dependent manner (Fig. 4C). The miR-30a mimic enhanced the invasion and migration potential of Malme3M cells in a CAGE-dependent manner (Fig. 4D), and also enhanced angiogenic potential of Malme3M cells in a CAGE-dependent manner (Fig. 4E). Taken together, these results suggest that miR-30a-3p requires CAGE to regulate the response to anti-cancer drugs, and the invasion, migration and angiogenic potential of cancer cells.
Fig. 4.
miR-30a requires CAGE to confer resistance to anti-cancer drugs and to regulate the invasion, migration and angiogenic potential of cancer cells. (A) Malme3M cells were transiently transfected with the indicated mimic and siRNA (each at 10 nM). The following day, cells were treated with various concentrations of taxol or trastuzumab, followed by MTT assays. **p, 0.005; ***p < 0.005. Comparison was made between Malme3M cells transfected with control mimic and Malme3M cells transfected with miR-30 mimic. (B) Malme3M cells were transfected with the indicated mimic (each at 10 nM) along with the indicated siRNA (10 nM). Malme3M cells were also transfected with the control mimic. The following day, cells were treated with taxol (1 μM) or trastuzumab (10 μg/ml), followed by Western blot analysis (left panel). Malme3M cells were transfected with the indicated mimic (10 nM) along with the indicated siRNA (10 nM). The 48 h post transfection, cell lysates were isolated and subjected to Western blot analysis (right panel). * denotes cleaved PARP. (C) Malme3M cells were transiently transfected with the indicated mimic (10 nM) along with the indicated siRNA (10 nM). The following day, cells were treated with taxol (1 μM) or trastuzumab (10 μg/ml), followed by caspase-3 activity assays. *p < 0.05; **p < 0.005. (D) Malme3M cells were transiently transfected with the indicated mimic (10 nM) along with the indicated siRNA (10 nM). The 48 h post transfection, chemoinvasion and wound migration assays were performed. **p < 0.005; ***p < 0.0005. (E) Malme3M cells were transiently transfected with the indicated mimic (each at 10 nM) along with the indicated siRNA (each at 10 nM). The conditioned medium, obtained following transfection, was subjected to intravital microscopy. ***p < 0.0005.
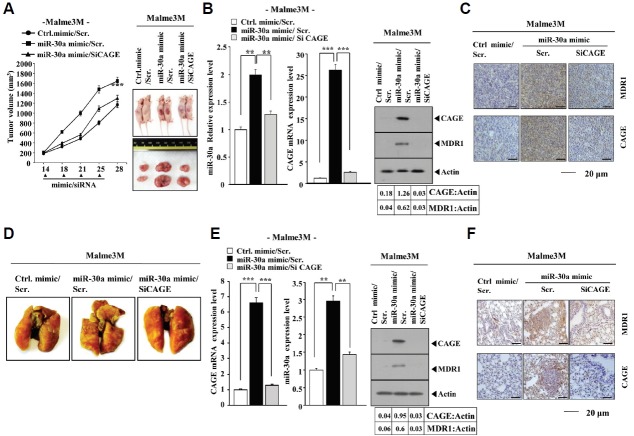
miR-30a requires CAGE to enhance the tumorigenic and metastatic potential of cancer cells
The miR-30a mimic enhanced the tumorigenic potential of Malme3M cells (Fig. 5A). The in vivo down-regulation of CAGE prevented the miR-30a mimic from enhancing the tumorigenic potential of Malme3M cells (Fig. 5A). Western blot analysis of tumor tissue lysates showed that the miR-30a mimic increased the expression of CAGE (Fig. 5B). Immunohistochemistry staining employing tumor tissues showed that the miR-30a mimic increased the expression of CAGE and MDR1 (Fig. 5C), enhanced the metastatic potential of Malme3M cells (Fig. 5D). The in vivo down-regulation of CAGE prevented the miR-30a mimic from enhancing the metastatic potential of Malme3M cells (Fig. 5D). Western blot analysis of tumor tissue lysates showed that the miR-30a mimic increased the expression of CAGE (Fig. 5E). Immunohistochemistry staining employing lung tumor tissue showed that the miR-30 mimic increased the expression of CAGE and MDR1 (Fig. 5F). Taken together, these results suggest that miR-30a-3p requires CAGE to enhance the tumorigenic and metastatic potential of Malme3M cells.
Fig. 5.
miR-30a requires CAGE to enhance the tumorigenic and metastatic potential of cancer cells. (A) Malme3M (106) cells were injected into the dorsal flanks of athymic nude mice. The indicated mimic (50 μM/kg) and siRNA (50 μM/kg) were intravenously injected 4 times in a total of 28 days. The extent of the tumorigenic potential was determined as described. (B) Tumor tissue lysates were subjected to qRT-PCR analysis and Western blot analysis. **p < 0.005; ***p < 0.0005. (C) Immunohistochemistry staining employing tumor tissues was performed as described. (D) Malme3M (106) cells were intravenously injected into athymic nude mice. The indicated mimic (50 μM/kg) and siRNA (50 μM/kg) were intravenously injected 6 times in a total of 21 days. The extent of lung metastases was quantified by staining with Bouin’s solution. (E) Tumor tissue lysates were subjected to qRT-PCR analysis and Western blot analysis. **p < 0.005; ***p < 0.0005. (F) Immunohistochemistry staining employing tumor tissues was performed as described.
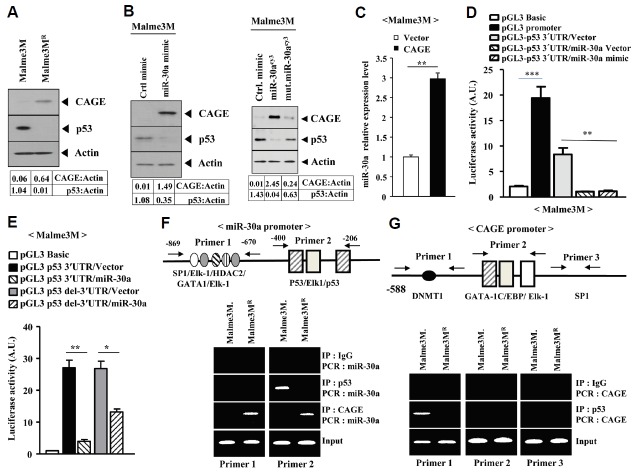
miR-30a forms a feedback loop with CAGE and p53
We have previously reported that CAGE, through the interaction with HDAC2, decreases the expression of p53 and confers resistance to anti-cancer drugs (Kim et al., 2010). miR-30a increased the expression of CAGE in Malem3M cells (Fig. 1F). Malme3MR cells showed a lower expression level of p53, while showing a higher expression of CAGE, than Malme3M cells (Fig. 6A). The miR-30a mimic increased the expression of CAGE and decreased the expression of p53 in Malme3M cells (Fig. 6B). miR-30acy3 increased the expression of CAGE and decreased the expression of p53 in Malme3M cells (Fig. 6B). However, mutant miR-30acy3 did not affect the expression of CAGE or p53 (Fig. 6B). Overexpression of CAGE increased the expression of miR-30a in Malme3M cells (Fig. 6C). TargetScan analysis predicted the binding of the miR-30a to the 3′-UTR of p53. We hypothesized that miR-30a would regulate the expression of p53. The miR-30a vector and miR-30a mimic decreased the luciferase activity associated with the 3′-UTR of p53 in Malme3M cells (Fig. 6D). The miR-30a vector decreased the luciferase activity associated with the 3′-UTR of p53 in Malme3M cells (Fig. 6E). The pGL3 p53 del-3′ UTR construct lacks binding sequences for miR-30a. The miR-30a decreased the luciferase activity associated with p53 del-3′ UTR in Malme3M cells (Fig. 6E). Although miR-30a does not bind to the 3′-UTR of pGL3 p53 del-3′ UTR, miR-30a may regulate the expression of other miRNAs, such as miR-217, to decrease the luciferase activity associated with pGL3 p53 del-3′ UTR. These results suggest that miR-30a exerts both direct and indirect regulation on the expression of p53. TargetScan analysis predicted that the 3′-UTR of p53 contains binding sequences for miR-491-5p, miR485-5p, mi-185, miR-27a and miR-150. It would be interesting to examine the effect of miR-30a on the expression of these miRNAs. The miR-30a promoter contains binding sites for HDAC2 and p53 (Fig. 6F), suggesting the possible binding of CAGE and p53 to the promoter sequences of miR-30a. ChIP assays showed the binding of CAGE to the promoter sequences of miR-30a in Malme3MR cells (Fig. 6F), and the binding of p53 to the promoter sequences of miR-30a in Malme3M cells (Fig. 6F). These results suggest that CAGE and p53 directly regulate the expression of miR-30a. In other words, CAGE and miR-30a form a positive feedback loop, while p53 and miR-30a form a negative feedback loop. ChIP assays showed the binding of p53 to the promoter sequences of CAGE (Fig. 6G). The CAGE promoter contains binding site for DNMT1 (Fig. 6G). The expression of CAGE is under epigenetic regulation (Cho et al., 2003). The expression of DNMT1 is decreased in anti-cancer drug-resistant cancer cells (Kim et al., 2010). Further, it would also be interesting to examine the effect of DNMT 1on the expression of miR-30a. It would be also interesting to examine the potential interaction between p53 and DNMT1 in Malme3M cells. Taken together, these results suggest that miR-30a forms a feedback loop with CAGE and p53 to regulate the response to anti-cancer drugs.
Fig. 6.
miR-30a forms a feedback loops with p53 and CAGE. (A) Cell lysates isolated from the indicated cancer cells were subjected to Western blot analysis. (B) Malme3M cells were transiently transfected with the indicated mimic (each at 10 nM). The 48 h post transfection, cell lysates were subjected to Western blot analysis (left panel). Malme3M cells were transiently transfected with the control mimic (10 nM), miR-30acy3 or mutant miR-30acy3 (200 nM). The 48 h post transfection, cell lysates were subjected to Western blot analysis (right panel). (C) Malme3M cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, cell lysates were subjected to qRT-PCR analysis. **p < 0.005. (D) Malme3M cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, cell lysates were subjected to a luciferase activity assay. **p < 0.005; ***p < 0.0005. (E) Malme3M cells were transiently transfected with the indicated construct (each at 1 μg). The 48 h post transfection, cell lysates were subjected to a luciferase activity assay. *p < 0.05; **p < 0.005. (F) Potential binding sites for transcriptional factors in the promoter sequences of miR-30a (upper panel). Cell lysates isolated from the indicated cancer cells were subjected to ChIP assay. (G) Potential binding sites for transcriptional factors in the promoter sequences of CAGE (upper panel). Cell lysates isolated from the indicated cancer cells were subjected to a ChIP assay (lower panel).
DISCUSSION
We have previously reported that miR-217, which is decreased in anti-cancer drug-resistant cancer cells, forms a positive feedback loop with HDAC3 and confers sensitivity to anti-cancer drugs (Kim et al., 2014). miR-200b, which forms a negative feedback loop with CAGE (Kim et al., 2003), also forms a positive feedback loop with HDAC3 (Kim et al., 2014). HDAC3 shows an inverse relationship with CAGE (Kim et al., 2014). We therefore hypothesized that miRNAs that increase the expression of CAGE would form a negative feedback loop with miR-217. miRNA hybrid analysis predicted that miR-30a-3p and miR-22 form hybrid with miR-217 (Fig. 1B). miR-22 is up-regulated in myelodysplastic syndrome (MDS) and leukemia and its aberrant expression correlates with poor survival (Song et al., 2013). miR-22 is a potent proto-oncogene and aberrations in the miR-22/TET2 regulatory network are common in hematopoietic malignancies (Song et al., 2013). Based on luciferase activity assays (Fig.1E), it is reasonable to suggest that miR-217 forms a negative feedback loop with miR-30a and miR-22. It is necessary to examine the molecular mechanisms associated with the expression regulation of miR-30-3p and miR-22, and to identify transcriptional factor that regulate the expression of miR-30a-3p and miR-22. Since the expression of miR-30a is higher in Malme3MR cells than in Malme3M cells (Figs. 1A and 1C), it is probable that the expression of miR-30a-3p is increased by anti-cancer drugs. CAGE interacts with HDAC2 and negatively regulates the expression of p53 (Kim et al., 2010). miR-30a-3p and miR-22 increase the expression of CAGE (Fig. 1F). It is necessary to examine whether miR-30a-3p and miR-22 would induce an interaction between CAGE and HDAC2.
miR-30a inhibits AECs-II (type II alveolar epithelial cells) apoptosis by repressing the mitochondrial fission dependent on Drp-1, dynamin-related protein-1 (Mao et al., 2014). Our results also show that miR-30a-3p exerts anti-apoptotic effects (Figs. 3B and 3C). Inhibiting miR-30a-3p expression increases HIF2α levels in H1H2 ccRCC cells and promotes cellular proliferation, angiogenesis, and xenograft tumor growth (Mathew et al., 2014). However, our results show that miR-30a-3p promotes the angiogenic potential of Malme3M cells (Fig. 3F). It is necessary to examine the effect of miR-30a-3p on the expression of HIF2α and VEGF in anti-cancer drug-sensitive Malme3M cells. miR-30a-3p is down-regulated in hepatocellular carcinoma and acts as a tumor suppressor (Wang et al., 2013). miRNA array analysis suggests that miR-30a-3p down-regulation results in an overall increased proliferative and angiogenic capacity (Pérez-Rivas et al., 2014). The over-expression of miR-30a increases the metastatic and invasion potential of NPC tumor cells in vivo and in vitro (Wang et al., 2014). E-cadherin serves as a target gene of miR-30a according to computational algorithms (Wang et al., 2014). Our results suggest that miR-30a-3p acts as an oncogene in melanoma cells, enhancing the invasive (Fig. 4D), tumorigenic (Fig. 5A) and metastatic potential (Fig. 5D) of Malme3M cells in a CAGE-dependent manner. It is probable that miR-30a-3p decreases the expression of E-cadherin in a CAGE-dependent manner. miR-30a-5p is a negative regulator of SEPT7 and the oncogenic activity of miR-30a-5p in human gliomas is at least in part due to the repression of SEPT7 (Jia et al., 2013). SEPT7 is a member of the septin family, which is a highly conserved subfamily of GTPases implicated in exocytosis, apoptosis, synaptogenesis, neuro-degeneration and tumorigenesis. It is probable that miR-30a-5p also enhances the invasive, tumorigenic and metastatic potential of Malme3M cells.
Promoter analysis showed that the miR-30a-3p promoter contains binding sites for Sp1, Elk1, GATA and HDAC2 (Fig. 6E). We showed the binding of CAGE to the promoter sequences of miR-30a-3p, and found an interaction between CAGE and Sp1 (unpublished personal observations). It would be interesting to examine the effect of Sp1 on the expression of miR-30a and the response to anti-cancer drugs. miR-200b forms a negative feedback loop with CAGE and regulates the response to anti-cancer drugs (Kim et al., 2013). It is probable that miR-30a-3p and miR-22 negatively regulate the expression of miR-200b. These transcriptional factors may regulate the expression of miR-200b.
In the present study, we showed that the miR-30a mimic confers resistance to trastuzumab, an inhibitor of HER2. We found that Malme3MR cells show a higher expression level of HER2 than Malme3M cells (unpublished personal observations). miR-200b shows an inverse relationship with HER2 in human colorectal and breast cancer cells (Bojmar et al., 2013). It is probable that miR-30a and CAGE increase the expression of HER2. It would be interesting to examine whether CAGE directly regulates the expression of HER2. Promoter analysis showed that the HER2 promoter contains the binding site for p53 (unpublished personal observations). It is probable that p53 exerts a direct regulatory effect on the expression of HER2. Trastuzumab- resistance is associated with the activation of EGFR signaling (Chaudhary et al., 2014; Lee et al., 2014). The EGFR- neutralizing antibody, cetuximab, restores trastuzumab sensitivity to breast cancer BT474-T798M cells and xenografts, suggesting that increased EGFR ligand production is causally associated with trastuzumab resistance (Rexer et al., 2013). Malme3MR cells show higher expression of pEGFRY845 than Malme3M cells (unpublished personal observations). It is probable that miR-30a-CAGE regulates the activation of EGFR.
In conclusion, miR-30a and miR-217 forms a negative feedback loop to regulate the expression of each other and the response to anti-cancer drugs (Fig. 7). miR-30a forms a positive feedback loop with CAGE to regulate the expression of p53 to confer resistance to anti-cancer drugs (Fig. 7). In the present study, we presented evidence that the miR-30a-CAGE-p53 feedback loop serves as a target for the development of anti-cancer drugs in CAGE-expressing cancer patients.
Fig. 7.

Proposed mechanism of the anti-cancer drug-resistance mediated by the miR-30a-CAGE-p53 feedback loop.
Acknowledgments
This work was supported by National Research Foundation Grants (2014R1A2A2A01002448, 2015R1A1A3A04001339, and 2015R1A2A1A15051678), a grant from the BK21 plus Program, and by National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea Grant 1320160. This work was also supported by a grant from Kangwon National University.
REFERENCES
- Bojmar L., Karlsson E., Ellegård S., Olsson H., Björnsson B., Hallböök O., Larsson M., Stål O., Sandström P. The role of microRNA-200 in progression of human colorectal and breast cancer. PLoS One. 2013;8:e84815. doi: 10.1371/journal.pone.0084815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary P., Thamake S.I., Shetty P., Vishwanatha J.K. Inhibition of triple-negative and Herceptin-resistant breast cancer cell proliferation and migration by Annexin A2 antibodies. Br. J. Cancer. 2014;111:2328–2341. doi: 10.1038/bjc.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Ma T., Huang C., Zhang L., Lv X., Xu T., Hu T., Li J. miR-27a modulates the MDR1/P-glycoprotein expression by inhibiting FZD7/β-catenin pathway in hepatocellular carcinoma cells. Cell. Signal. 2013;25:2693–2701. doi: 10.1016/j.cellsig.2013.08.032. [DOI] [PubMed] [Google Scholar]
- Chen W., Yang Y., Chen B., Lu P., Zhan L., Yu Q., Cao K., Li Q. MiR-136 targets E2F1 to reverse cisplatin chemo-sensitivity in glioma cells. J. Neurooncol. 2014a;120:43–53. doi: 10.1007/s11060-014-1535-x. [DOI] [PubMed] [Google Scholar]
- Chen W.X., Liu X.M., Lv. M.M., Chen L., Zhao J.H., Zhong S.L., Ji M.H., Hu Q., Luo Z., Wu J.Z., et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 2014b;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W., Liu T., Wan X., Gao Y., Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279:2047–2059. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- Cho B., Lim Y., Lee D.Y., Park S.Y., Lee H., Kim W.H., Yang H., Bang Y.J., Jeoung D.I. Identification and characterization of a novel cancer/testis antigen gene CAGE. Biochem. Biophys. Res. Commun. 2002;292:715–726. doi: 10.1006/bbrc.2002.6701. [DOI] [PubMed] [Google Scholar]
- Cho B., Lee H., Jeong S., Bang Y.J., Lee H.J., Hwang K.S., Kim H.Y., Lee Y.S., Kang G.H., Jeoung D.I. Promoter hypomethylation of a novel cancer/testis antigen gene CAGE is correlated with its aberrant expression and is seen in premalignant stage of gastric carcinoma. Biochem. Biophys. Res. Commun. 2003;307:52–63. doi: 10.1016/s0006-291x(03)01121-5. [DOI] [PubMed] [Google Scholar]
- Fu J., Xu X., Kang L., Zhou L., Wang S., Lu J., Cheng L., Fan Z., Yuan B., Tian P., et al. miR-30a suppresses breast cancer cell proliferation and migration by targeting Eya2. Biochem. Biophys. Res. Commun. 2014;445:314–319. doi: 10.1016/j.bbrc.2014.01.174. [DOI] [PubMed] [Google Scholar]
- Guo J., Feng Z., Huang Z., Wang H., Lu W. Micro RNA-217 functions as a tumor suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Mol. Cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T., Fujita T., Hirao N., Matsuzaki Y., Okada T., Mochimaru H., Susumu N., Matsumoto E., Sugano K., Yamashita N., et al. Frequent immune responses to a cancer/testis antigen, CAGE, in patients with microsatellite instability-positive endometrial cancer. Clin. Cancer Res. 2005;11:3949–3957. doi: 10.1158/1078-0432.CCR-04-1702. [DOI] [PubMed] [Google Scholar]
- Jia Z., Wang K., Wang G., Zhang A., Pu P. MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth by targeting SEPT7. PLoS One. 2013;8:e55008. doi: 10.1371/journal.pone.0055008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim Y., Park H., Park D., Lee Y.S., Choe J., Hahn J.H., Lee H., Kim Y.M., Jeoung D. Cancer/testis antigen CAGE exerts negative regulation on p53 expression through HDAC2 and confers resistance to anti-cancer drugs. J. Biol. Chem. 2010;285:25957–25968. doi: 10.1074/jbc.M109.095950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Park D., Kim H., Choi M., Lee H., Lee Y.S., Choe J., Kim Y.M., Jeoung D. miR-200b and cancer/testis antigen CAGE form a feedback loop to regulate the invasion and tumorigenic and angiogenic responses of a cancer cell line to microtubule-targeting drugs. J. Biol. Chem. 2013;288:36502–36518. doi: 10.1074/jbc.M113.502047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kim H., Park H., Park D., Lee H., Lee Y.S., Choe J., Kim Y.M., Jeoung D. miR-326-histone deacetylase-3 feedback loop regulates the invasion and tumorigenic and angiogenic response to anti-cancer drugs. J. Biol. Chem. 2014;289:28019–28039. doi: 10.1074/jbc.M114.578229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.M., Nam K., Oh S., Lim J., Kim Y.P., Lee J.W., Yu J.H., Ahn S.H., Kim S.B., Noh D.Y., et al. Extracellular matrix protein 1 regulates cell proliferation and trastuzumab resistance through activation of epidermal growth factor signaling. Breast Cancer Res. 2014;16:479. doi: 10.1186/s13058-014-0479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggins A.P., Lim S.H., Soilleux E.J., Pulford K., Banham A. H. A panel of cancer-testis genes exhibiting broad-spectrum expression in haematological malignancies. Cancer Immun. 2010;10:8. [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chen L., Zhang X., Xu X., Xing H., Zhang Y., Li W., Yu H., Zeng J., Jia J. RUNX3 regulates vimentin expression via miR-30a during epithelial-mesenchymal transition in gastric cancer cells. J. Cell. Mol. Med. 2014;18:610–623. doi: 10.1111/jcmm.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Zhang J., Lin S., Jing L., Xiang J., Wang M., Wang B., Xu P., Liu W., Song X., et al. MiRNA-30a inhibits AECs-II apoptosis by blocking mitochondrial fission dependent on Drp-1. J. Cell. Mol. Med. 2014;18:2404–2416. doi: 10.1111/jcmm.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew L.K., Lee S.S., Skuli N., Rao S., Keith B., Nathanson K.L., Lal P., Simon M.C. Restricted expression of miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas enhances HIF2α activity. Cancer Discov. 2014;4:53–60. doi: 10.1158/2159-8290.CD-13-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka C., Ikezoe T., Yang J., Nobumoto A., Tsuda M., Yokoyama A. Downregulation of miR-217 correlates with resistance of Ph (+). leukemia cells to ABL tyrosine kinase inhibitors. Cancer Sci. 2014;105:297–307. doi: 10.1111/cas.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rivas L.G., Jerez J.M., Carmona R., de Luque V., Vicioso L., Claros M.G., Viguera E., Pajares B., Sánchez A., Ribelles N., et al. A microRNA signature associated with early recurrence in breast cancer. PLoS One. 2014;9:e91884. doi: 10.1371/journal.pone.0091884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Por E., Byun H.J., Lee E.J., Lim J.H., Jung S.Y., Park I., Kim Y.M., Jeoung D.I., Lee H. The cancer/testis antigen CAGE with oncogenic potential stimulates cell proliferation by up-regulating cyclins D1 and E in an AP-1- and E2F-dependent manner. J. Biol. Chem. 2010;285:14475–14485. doi: 10.1074/jbc.M109.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer B.N., Ghosh R., Narasanna A., Estrada M.V., Chakrabarty A., Song Y., Engelman J.A., Arteaga C.L. Human breast cancer cells harboring a gatekeeper T798M mutation in HER2 overexpress EGFR ligands and are sensitive to dual inhibition of EGFR and HER2. Clin. Cancer Res. 2013;19:5390–5401. doi: 10.1158/1078-0432.CCR-13-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.J., Ito K., Ala U., Kats L., Webster K., Sun S.M., Jongen-Lavrencic M., Manova-Todorova K., Teruya-Feldstein J., Avigan D.E., et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin H., Zhou L., Zhu Q., Gao S., Xie H., Liu Z., Xu Z., Wei J., Huang X., et al. MicroRNA-30a-3p inhibits tumor proliferation, invasiveness and metastasis and is downregulated in hepatocellular carcinoma. Eur. J. Surg. Oncol. 2013;40:1586–1594. doi: 10.1016/j.ejso.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Li Y.Y., Fu S., Wang X.P., Huang M.Y., Zhang X., Shao Q., Deng L., Zeng M.S., Zeng Y.X., et al. MicroRNA-30a promotes invasiveness and metastasis in vitro and in vivo through epithelial-mesenchymal transition and results in poor survival of nasopharyngeal carcinoma patients. Exp. Biol. Med. (Maywood). 2014;239:891–898. doi: 10.1177/1535370214532758. [DOI] [PubMed] [Google Scholar]
- Wu Z.B., Li W.Q., Lin S.J., Wang C.D., Cai L., Lu J.L., Chen Y.X., Su Z.P., Shang H.B., Yang W.L., et al. MicroRNA expression profile of bromocriptine-resistant prolactinomas. Mol. Cell. Endocrinol. 2014;395:10–18. doi: 10.1016/j.mce.2014.07.014. [DOI] [PubMed] [Google Scholar]