Abstract
Nitrification directly contributes to the ammonia removal in sponges, and it plays an indispensable role in sponge-mediated nitrogen cycle. Previous studies have demonstrated genomic evidences of nitrifying lineages in the sponge Theonella swinhoei. However, little is known about the transcriptional activity of nitrifying community in this sponge. In this study, combined DNA- and transcript-based analyses were performed to reveal the composition and transcriptional activity of the nitrifiers in T. swinhoei from the South China Sea. Transcriptional activity of ammonia-oxidizing archaea (AOA) and nitrite-oxidizing bacteria (NOB) in this sponge were confirmed by targeting their nitrifying genes,16S rRNA genes and their transcripts. Phylogenetic analysis coupled with RDP rRNA classification indicated that archaeal 16S rRNA genes, amoA (the subunit of ammonia monooxygenase) genes and their transcripts were closely related to Nitrosopumilus-like AOA; whereas nitrifying bacterial 16S rRNA genes, nxrB (the subunit of nitrite oxidoreductase) genes and their transcripts were closely related to Nitrospira NOB. Quantitative assessment demonstrated relative higher abundances of nitrifying genes and transcripts of Nitrosopumilus-like AOA than those of Nitrospira NOB in this sponge. This study illustrated the transcriptional potentials of Nitrosopumilus-like archaea and Nitrospira bacteria that would predominantly contribute to the nitrification functionality in the South China Sea T. swinhoei.
The marine nitrogen cycle controls the availability of nitrogenous nutrients and the biological productivity in oceanic systems1. The nitrogen cycle consists of multiple transformations of nitrogenous compounds, primarily driven by microbes. One key link of this cycle is the nitrification process, which occurs in many marine ecosystems, such as seawaters, sediments, hydrothermal vents and in invertebrates like sponges2,3,4,5.
Nitrification describes the oxidation of ammonia to nitrite and subsequently to nitrate for energy purposes6. Ammonia-oxidizing archaea (AOA) and bacteria (AOB) perform the first, often rate-determining step of nitrification from ammonia to nitrite, while nitrite-oxidizing bacteria (NOB) are responsible for the oxidation of nitrite to nitrate. Nearly all the reported AOA are ascribed into the phylum Thaumarchaeota, and AOB are affiliated with some genera of the phylum Proteobacteria (e.g. Nitrosomonas, Nitrosococcus, Nitrosospira, Nitrosovibrio); while all known NOB belong to some genera of the phyla Proteobacteria (e.g. Nitrobacter, Nitrococcus, Nitrotoga), Nitrospinae (e.g. Nitrospina), Nitrospirae (e.g. Nitrospira) and Chloroflexi (e.g. Nitrolancea)7. In view of their phylogenetic diversity, the 16S rRNA gene, amoA gene (encoding the subunit of the ammonia monooxygenase in AOA and AOB) and nxrA or nxrB gene (encoding the subunit of the nitrite oxidoreductase in NOB) have served as useful phylogenetic markers for investigating nitrifying communities in various ecosystems8,9,10,11,12,13,14,15.
Sponges are sessile invertebrates that grow widely in virtually all aquatic ecosystems with high filtration rates16. Many sponges harbor phylogenetically complex microbial assemblages including bacteria, archaea, fungi and microalgae17. For example, surveys of sponge symbiotic communities have revealed no less than 47 bacterial phyla and two archaeal phyla so far18. However, compared with the knowledge about the diversity of sponge microbial symbionts, our understanding of their functions is still limited19. Given the overall importance of ecological functions of symbionts to the sponge holobionts, nitrogen metabolism in sponges is continuing to receive considerable attention, since nitrogen is generally a limiting factor in the oligotrophic habitat of sponges.
Like many other marine invertebrates, sponges excrete ammonia as a metabolic waste20. Nitrification process that contributes to ammonia removal in sponges has been suggested by using incubation experiments20,21. In sponges, nitrifiers have been corroborated by the isolation of individual species22,23, and by the identification of 16S rRNA genes or functional genes of nitrifying microbes from sponge microbiomes by using molecular-based approaches. For example, diverse Nitrosospira AOB and Planctomycetes anaerobic bacteria in the sponges Ircinia strobilina and Mycale laxissima were confirmed by amoA gene and 16SrRNA gene assays24. Meanwhile, vertical transmission of AOA from mother to offspring was suggested in a diverse range of sponges from different biogeographic areas by 16S rRNA gene and amoA gene detection results5. Moreover, the increasing gene-targeted and shotgun high-throughput sequencing technologies have strongly illustrated the ecological potentials of sponge microbes, although many limitations persist25. For example, 16S rRNA-targeted pyrosequencing analysis has revealed the Nitrosopumilus lineage in the sponge holobionts26. Meanwhile, shotgun metagenomic analysis of the sponge Neamphius huxleyi has demonstrated the genomic potentials, including Nitrosopumilus ammonia oxidation, at the functional-population level27. Recent shotgun metagenomic sequencing analysis of the sponges Didiscus oxeata and Scopalina ruetzleri has uncovered the symbionts’ functions at the whole-community level, including the nitrification process that might be performed by a diverse set of archaeal and bacterial nitrifying lineages, including Nitrosopumilus, Nitrosococcus, Nitrosomonas and Nitrosospira28, suggesting the presence of various nitrifying microbes, e.g. AOA and AOB in sponges.
In many niches, such as sponges, the presence of functional genes is not the direct evidence of their corresponding metabolic activity in situ, whereas RNA-based strategies would provide valuable hints on the activity of microbial assemblage29, albeit imprecisely in particular cases30. To date, a number of RNA-based analyses have been performed in revealing the activity of microbial communities in sponges in their natural physiological state or under certain environmental stresses31,32, and in conjecturing the active microbial metabolic processes, such as nitrogen fixation, ammonia oxidation or urea hydrolysis in sponges33,34,35. Moreover, metatranscriptomic analysis has been performed to give a whole-community-level exploration of the in situ active functions, such as archaeal ammonia oxidation and photosynthetic carbon fixation, carried out by sponge symbionts36,37,38. However, functional gene-targeted high-throughput sequencing of interested functional genes, such as amoA and nxrA/nxrB genes, has not been performed in sponge metagenome/metatranscriptome so far.
The sponge Theonella swinhoei (class Demospongiae, order Lithistida, family Theonellidae) is widely distributed in tropical and subtropical oceans39. Previous DNA-based investigations have demonstrated the nitrifying potential of the Nitrosopumilus, Nitrosospira and Nitrospira lineages in T. swinhoei near Caroline Islands, Palau (Palauan T. swinhoei)21,40. However, little is known about the transcriptional activity of nitrifiers in this sponge. Hence, the present study aims to reveal the community structure and transcriptional activity of nitrifiers in T. swinhoei from the South China Sea using a DNA and RNA combined approach targeting the nitrifying genes, 16S rRNA genes and their transcripts.
Results
Amplification of 16S rRNA, amoA and nxrA/nxrB
The archaea and bacteria were detected by PCR amplification using the16S rRNA primers (Table 1) and nitrifiers were verified using the primer pairs (Table 1) targeting amoA, nxrA and nxrB genes from T. swinhoei DNA and cDNA templates. Cloning and sequencing results showed that archaeal and bacterial 16S rRNA gene, archaeal amoA gene and Nitrospira nxrB gene were positively amplified from both DNA and cDNA templates; while bacterial amoA gene, Nitrobacter nxrA gene, Nitrococcus nxrA gene, Nitrolancea nxrA gene and Nitrospina nxrB gene were not detected from either DNA or cDNA templates (data not shown). After quality check and trimming, archaeal 16S rRNA fragments of 657 ± 3-bp length, bacterial 16S rRNA fragments of 694 ± 14-bp length, archaeal amoA fragments of 532-bp length and Nitrospira nxrB fragments of 469-bp length were used for further analysis. Rarefaction curve analysis showed that the amount of sequenced clones in each clone library, shown in Table 1, reached saturation at 95% or 97% sequence similarity (Supplementary Fig. S1).
Table 1. Primers for the PCR-screen of nitrifying functional genes and 16S rRNA sequences and their amplifying results.
| Primer | Sequence (5′-3′) | Target gene | Ta (°C) | Approach | No. of DNA clones | No. of transcript clones | Reference |
|---|---|---|---|---|---|---|---|
| 340F | CCCTAYGGGGYGCASCAG | Archaea 16S rRNA | 57 | PCR | 69 | 65 | 9 |
| 1000R | GGCCATGCACYWCYTCTC | ||||||
| 356F | ACWCCTACGGGWGGCWGC | Bacteria 16S rRNA | 60 | PCR | 204 | 187 | 15 |
| 1064R | AYCTCACGRCACGAGCTGAC | ||||||
| amo111F | TTYTAYACHGAYTGGGCHTGGACATC | Archaea amoA | 55 | PCR | 73 | 68 | 8 |
| amo643R | TCCCACTTWGACCARGCGGCCATCCA | ||||||
| archamoAqF | CCRGTCTGGTTRCCDTCAGG | Archaea amoA | 57 | qPCR | – | – | This study |
| archamoAqR | CTTGAAYGCVGTYTCAAGCG | ||||||
| amoA1F | GGGGTTTCTACTGGTGGT | Bacteria amoA | 50 | PCR | N.A. | N.A. | 12 |
| amoA2R | CCCCTCKGSAAAGCCTTCTTC | ||||||
| amoA3F | GGTGAGTGGGYTAACMG | Bacteria amoA-amoB | 45 | PCR | N.A. | N.A. | 11 |
| amoB4R | GCTAGCCACTTTCTGG | ||||||
| nxrA1F | CAGACCGACGTGTGCGAAAG | Nitrobacter/Nitrococcus nxrA | 55 | PCR | N.A. | N.A. | 14 |
| nxrA2R | TCYACAAGGAACGGAAGGTC | ||||||
| nxrA0038F | GCCAGTGGGAAGAGTTCTATA | Nitrolancea nxrA | 68 | PCR | N.A. | N.A. | 13 |
| nxrA3595R | GCCACGTGCGTGTCCCGSGT | ||||||
| NpnnxrBF | TAYATGTGGTGGAACAAYGTRG | Nitrospina nxrB | 54 | PCR | N.A. | N.A. | This study |
| NpnnxrBR | GCCTTKCGMGGACAMGCCGC | ||||||
| nxrB169F | TACATGTGGTGGAACA | Nitrospira nxrB | 55 | PCR | 16 | 27 | 10 |
| nxrB638R | CGGTTCTGGTCRATCA | ||||||
| nxrBqF | TGTGGTGGAACAACGTGGAA | Nitrospira nxrB | 56 | qPCR | – | – | This study |
| nxrBqR | CCCGGCATCGAAAATGGTCA |
N.A., PCR detection was not available; “–”, clone sequencing was not performed.
Active archaea and AOA in the South China Sea T. swinhoei
Composition and activity of archaea in the South China Sea T. swinhoei was detected by 16S rRNA gene and transcript amplification. The 69 16S rRNA gene amplicons and 65 16S rRNA transcript amplicons were grouped into five OTUs (accession no. KM010247–KM010250 and KT380844) belonging to the Nitrosopumilus cluster, according to phylogenetic analysis and RDP Classifier identification (Fig. 1A; Supplementary Table S1). Four of these OTUs were shared at DNA/transcript levels, while the left one was unique at the DNA level (Supplementary Table S3). All the OTUs showed 94–99% similarity to the sequences from the sponge Luffariella sp. (accession no. EU049816 and EU049817) and T. swinhoei (accession no. AF186424); meanwhile, these OTUs shared 93–94% identity with the sequences of Nitrosopumilus spp. (accession no. KR737579 and HQ331116) and 90–92% identity with the sequences of Cenarchaeum spp. (accession no. AF083071 and AF083072). Each OTU took different proportions of the sequences in the corresponding clone library ranging from 9–48% (Fig. 1B). Thus, our findings suggested that Nitrosopumilus species dominated the archaeal community in the South China Sea T. swinhoei, and a large fraction of archaea had transcriptional activity in this sponge host.
Figure 1.
Maximum-likelihood phylogenetic analysis of archaeal 16S rRNA genes and transcripts (A) and the percentage of each OTU (97% sequence similarity) in the corresponding clone library (B), and maximum-likelihood phylogenetic analysis of archaeal amoA genes and transcripts (C) and the percentage of each OTU (95% sequence similarity) in the corresponding clone library (D) from the South China Sea T. swinhoei. OTU representatives are marked. Scale bar represents 5% (for 16S rRNA) or 10% (for amoA) nucleotide sequence divergence per homologues position. Bootstrap values more than 50% of 1000 replicates are shown on the trees. The outgroup of the archaeal 16S rRNA tree is a bacterial 16S rRNA sequence of candidatus Poribacteria sp. WGA-4CII (accession no. KC713966) and the outgroup of the archaeal amoA tree is a bacterial amoA sequence of Nitrosospira multiformis (accession no. AF042171).
Furthermore, PCR assays of archaeal amoA genes and transcripts verified the presence of active ammonia-oxidizing population in the South China Sea T. swinhoei. All the amplicons of archaeal amoA genes and transcripts were assorted to five shared OTUs at DNA/transcript levels (accession no. KM010251–KM010255) (Fig. 1C). Nucleic acid sequences rather than deduced amino acid sequences were used for phylogeny reconstruction, as the former shows higher phylogenetic resolution. Each amoA OTU accounted for 5–41% of the sequences in the respective clone library (Fig. 1D). These OTUs were closely (93–99% sequence identity) related to the sequences from the sponges Luffariella sp. (accession no. EU049831- EU049833) and Xestospongia muta (accession no. GQ485791). Meanwhile, these OTUs showed 79–81% similarity with the amoA sequences of Nitrosopumilus spp. (accession no. CP000866, CP003843, HM345611 and HM345609) and 77–79% similarity with the amoA sequences of Cenarchaeum spp. (accession no. DQ397569 and DQ397580) (Supplementary Table S2). On the peptide level, these OTUs shared 94–96% similarity with the AmoA sequences of Nitrosopumilus spp. (accession no. CP000866, CP003843, HM345611 and HM345609) and 92–93% similarity with the AmoA sequences of Cenarchaeum spp. (accession no. DQ397569 and DQ397580), which indicated that they were more likely the Nitrosopumilus-like amoA sequences.
Altogether, the phylogeny of archaeal 16S rRNA genes and transcripts strongly demonstrated that the active archaeal community was dominated by Nitrosopumilus species in the South China Sea T. swinhoei; particularly, Nitrosopumilus-like AOA with transcriptional activity might predominantly contribute to the aerobic ammonia oxidation in this sponge.
Active bacteria and NOB in the South China Sea T. swinhoei
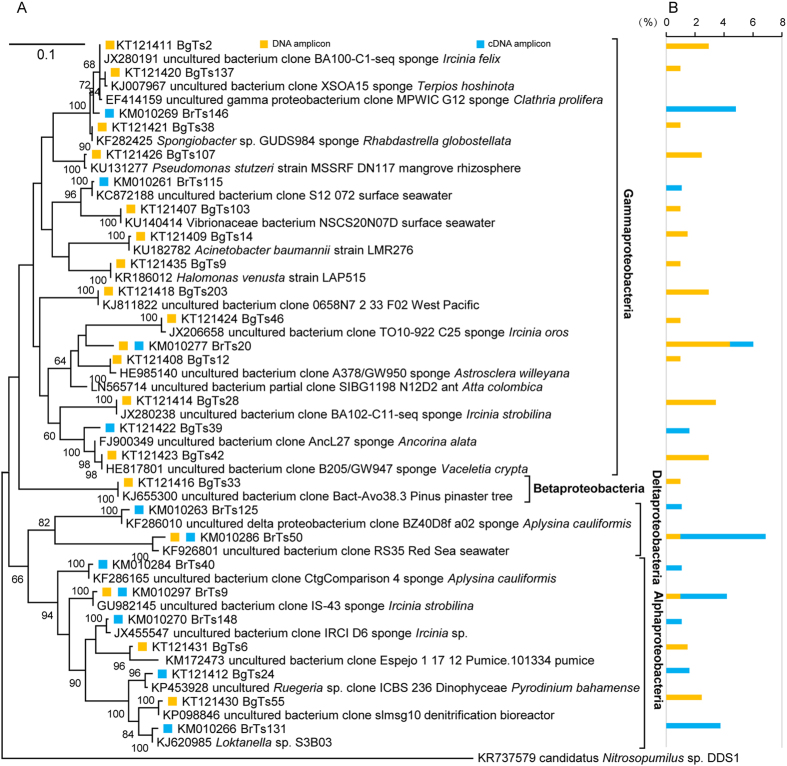
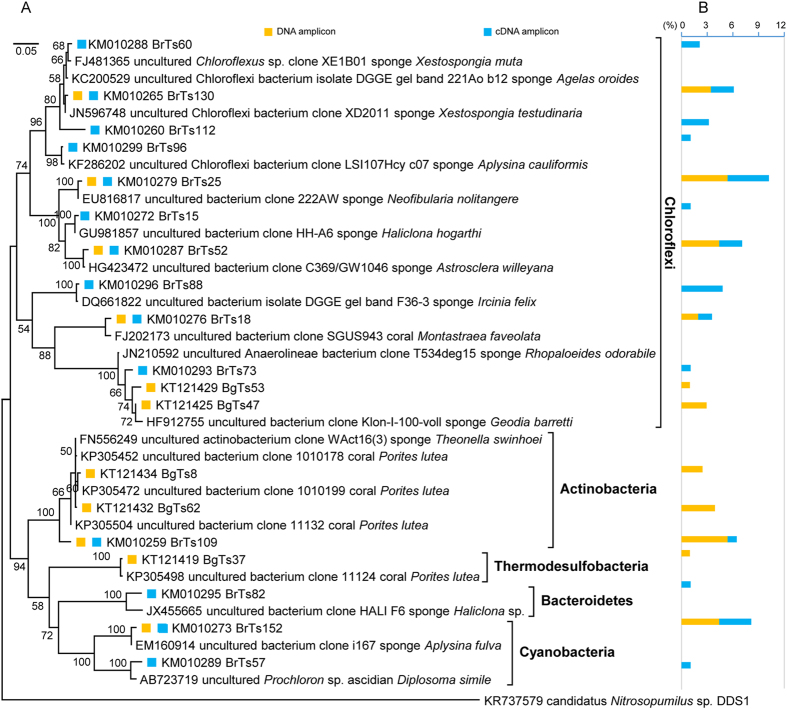
Bacteria showed higher diversity than archaea in the South China Sea T. swinhoei, since 66 bacterial 16S rRNA OTUs were revealed (Figs 2A,3A and 4A; Supplementary Table S3). These OTUs interspersed among 12 reported phyla according to the RDP Classifier identification (Supplementary Table S1) and phylogenetic analysis (Figs 2A,3A and 4A; Supplementary Fig. S2A). Each of these OTUs accounted for 0.5–7.8% of the sequences in the corresponding clone library (Figs 2B,3B and 4B; Supplementary Fig. S2B). In detail, 14 OTUs interspersing among Acidobacteria, Actinobacteria, Chloroflexi, Cyanobacteria, Firmicutes, Nitrospirae, Poribacteria and Proteobacteria were detected at DNA/transcript levels (Supplementary Table S3). Twenty-six OTUs belonging to Acidobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Poribacteria, Proteobacteria and Spirochaetes were uniquely revealed at the transcript level, and 28 OTUs falling into Acidobacteria, Actinobacteria, Chloroflexi, Nitrospirae, Proteobacteria, Spirochaetes and Thermodesulfobacteria were uniquely identified by their 16S rRNA genes (Supplementary Table S3). Overall, 40 of 66 OTUs were revealed at the transcript level and fell into 12 reported phyla, indicating diverse bacteria with transcriptional activity in this sponge.
Figure 2.
Maximum-likelihood phylogenetic analysis of Proteobacteria 16S rRNA genes and transcripts (A) and the percentage of each OTU (97% sequence similarity) in the corresponding clone library (B) from the South China Sea T. swinhoei. OTU representatives are marked. Scale bar represents 10% nucleotide sequence divergence per homologues position. Bootstrap values more than 50% of 1000 replicates are shown on the tree. The outgroup is an archaeal 16S rRNA sequence of candidatus Nitrosopumilus sp. DDS1 (accession no. KR737579).
Figure 3.
Maximum-likelihood phylogenetic analysis of Chloroflexi, Actinobacteria, Thermodesulfobacteria, Bacteroidetes and Cyanobacteria 16S rRNA genes and transcripts (A) and the percentage of each OTU (97% sequence similarity) in the corresponding clone library (B) from the South China Sea T. swinhoei. OTU representatives are marked. Scale bar represents 5% nucleotide sequence divergence per homologues position. Bootstrap values more than 50% of 1000 replicates are shown on the tree. The outgroup is an archaeal 16S rRNA sequence of candidatus Nitrosopumilus sp. DDS1 (accession no. KR737579).
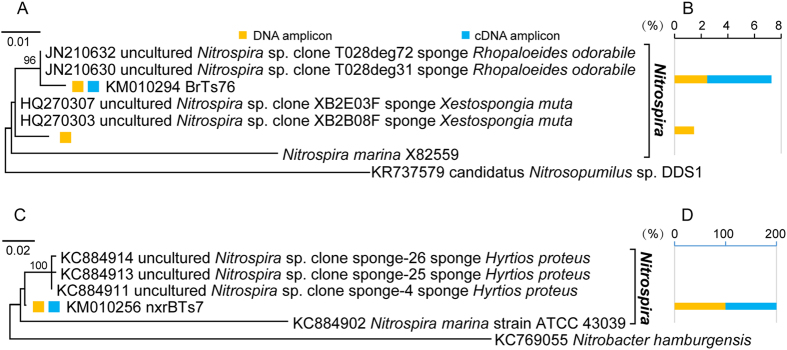
Figure 4.
Maximum-likelihood phylogenetic analysis of Nitrospira 16S rRNA genes and transcripts (A) and the percentage of each OTU (97% sequence similarity) in the corresponding clone library (B), and maximum-likelihood phylogenetic analysis of Nitrospira nxrB genes and transcripts (C) and the percentage of this OTU (95% sequence similarity) in the corresponding clone library (D) from the South China Sea T. swinhoei. OTU representatives are marked. Scale bar represents 1% (for 16S rRNA) or 2% (for nxrB) nucleotide sequence divergence per homologues position. Bootstrap values more than 50% of 1000 replicates are shown on the tree. The outgroup of Nitrospira 16S rRNA tree is an archaeal 16S rRNA sequence of candidatus Nitrosopumilus sp. DDS1 (accession no. KR737579) and the outgroup of Nitrospira nxrB tree is a nxrB sequence of Nitrobacter hamburgensis (accession no. KC769055).
Particularly, one shared 16S rRNA OTU at DNA/transcript levels (accession no. KM010294) and one unique 16S rRNA OTU at the DNA level (accession no. KT121415) fell into the Nitrospira genus of Nitrospirae (Fig. 4A; Supplementary Table S1). These OTUs were most similar to the sequences (99% sequence identity) from the sponges Rhopaloeides odorabile (accession no. JN210630 and JN210632) and Xestospongia muta (accession no. HQ270303 and HQ270307). Meanwhile, none of the Proteobacteria and Chloroflexi 16S rRNA OTUs was ascribed into the AOB genera Nitrosomonas, Nitrosococcus, Nitrosospira or Nitrosovibrio, or the NOB genera Nitrobacter, Nitrospina, Nitrococcus, Nitrospina, Nitrotoga or Nitrolancea, according to the phylogenetic analysis and RDP classification (Figs 2A and 3A; Supplementary Table S1). This result indicated that the Nitrospira species might dominate the nitrifying bacterial community in the South China Sea T. swinhoei.
The presence and activity of NOB in the South China Sea T. swinhoei was verified by positive PCR detection of Nitrospira nxrB genes and their transcripts. All the Nitrospira nxrB gene and transcript amplicons were clustered into one shared OTU at DNA/transcript levels (accession no. KM010256) belonging to the Nitrospira cluster (Fig. 4C,D). No other NOB genera were revealed by nxrA or nxrB PCR assays. This OTU was most similar (98% sequence identity) to the sequences from the sponge Hyrtios proteus (accession no. KC884911), and it shared 85% identity to the sequence of Nitrospira marina strain ATCC 43039 (accession no. KC884902) (Supplementary Table S2).
Collectively, our results on 16S rRNA genes and transcripts suggested that a fraction of bacteria interspersing among 12 reported phyla had transcriptional activity in the South China Sea T. swinhoei. Particularly, Nitrospira species dominated the nitrifying bacterial community in this sponge. The revealed Nitrospira nxrB genes/transcripts and Nitrospira16S rRNA genes/transcripts demonstrated that the Nitrospira NOB species had transcriptional activity, and they were predominantly responsible for nitrite oxidation in the South China Sea T. swinhoei.
Quantification of Nitrosopumilus-like amoA and Nitrospira nxrB genes and transcripts
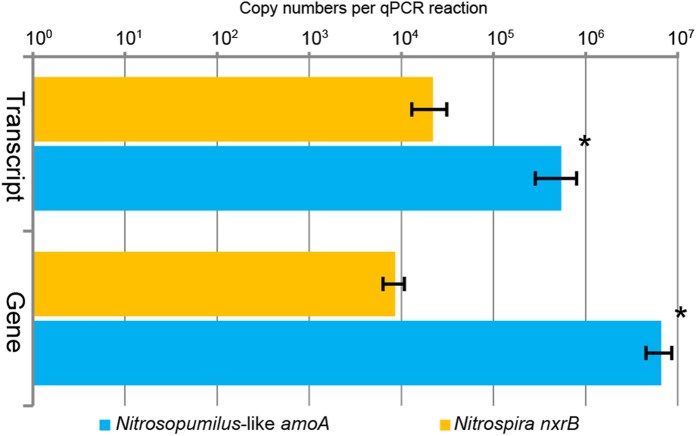
qPCR assays were employed to quantify the abundance of Nitrosopumilus-like amoA and Nitrospira nxrB genes and their transcripts in the South China Sea T. swinhoei. Copy numbers of Nitrosopumilus-like amoA genes and their transcripts were (6.59 ± 1.21) × 106 and (5.41 ± 2.57) × 105 per reaction, respectively; while copy numbers of Nitrospira nxrB genes and their transcripts were (8.55 ± 2.24) × 103 and (2.20 ± 0.91) × 104 per reaction, respectively (Supplementary Table S4). At a significance level of α = 0.05, the abundance of Nitrosopumilus-like amoA genes was higher than that of Nitrospira nxrB genes (P < 0.001, independent-sample t-test) in T. swinhoei, which was similar to the transcript-level revelation of this sponge that the abundance of Nitrosopumilus-like amoA transcripts was higher than that of Nitrospira nxrB transcripts (P < 0.001, independent-sample t-test) (Fig. 5). This finding suggested the higher abundances of nitrifying genes and transcripts of Nitrosopumilus-like AOA than those of Nitrospira NOB in the South China Sea T. swinhoei.
Figure 5.
Abundance of Nitrosopumilus-like amoA genes/transcripts and Nitrospira nxrB genes/transcripts estimated by qPCR for the South China Sea T. swinhoei. The Mean and standard deviation (SD) of amoA and nxrB abundances were calculated in Microsoft Excel 2007. T-test was performed using the t-test command of Microsoft Excel 2007 at a significance level of α = 0.05. A statistically significant difference (P < 0.001) between copy numbers of Nitrosopumilus-like amoA genes and Nitrospira nxrB genes, and Nitrosopumilus-like amoA transcripts and Nitrospira nxrB transcripts is indicated with an asterisk.
Discussion
Although some lineages, like AOB, retain an appreciable abundance of rRNAs even in the dormant period when their activity is expected to be minimal41, it is widely accepted that transcriptional expressions of rRNA genes are typically correlated with cellular growth rate and activity in microbes42. The RNA-based analysis of rRNA or functional genes has been successfully used to investigate the activity of sponge symbiotic communities. For example, active microbial community including Nitrosopumilus, Nitrosococcus and Nitrospira nitrifiers, in the sponge Rhopaloeides odorabile, showed significant shifts under changed thermal stresses by 16S rRNA assays43. Meanwhile, PCR assays have uncovered diverse Nitrosospira AOB amoA genes but no transcript counterparts in the sponges Ircinia strobilina and Mycale laxissima24. Thus, RNA-based identification of rRNAs or functional genes is a useful means to reveal the metabolically active microbial populations in sponges.
The shared OTUs at DNA/transcript levels, including five amoA OTUs, one nxrB OTU, four archaeal 16S rRNA OTUs and 14 bacterial 16S rRNA OTUs, indicated the existence of community-wide transcriptionally active prokaryotic populations, especially the nitrifying microbes in the South China Sea T. swinhoei. The unique OTUs at the DNA level, including one archaeal 16S rRNA OTUs and 28 bacterial 16S rRNA OTUs, implied that the microbes represented by these OTUs might be during their dormant period or be transcribed at low level which was below the PCR sensitivity threshold. Meanwhile, 26 bacterial 16S rRNA OTUs were uniquely revealed at the transcript level, indicating that these bacterial members may have very low genomic abundance but high transcriptional activity in the South China Sea T. swinhoei. Similar DNA- and transcript-level discrepancies of rRNA or functional genes have also been discovered in various sponge species24,32,35, indicating the common existence of selective activity of microbial community in their hosts. Consequently, a large fraction of active archaea and bacteria were suggested by rRNA-based findings, while the active nitrifying community dominated by Nitrosopumilus-like AOA and Nitrospira NOB were revealed by functional transcript-based assays. This study provides direct molecular evidences for the inhabitancy of nitrifying community with transcriptional activity in the South China Sea T. swinhoei.
In contrast to Palauan T. swinhoei21, no AOB was detected in the South China Sea T. swinhoei in this study, suggesting that archaeal not bacterial ammonia oxidizers dominate the first step in nitrification in this sponge, which is consistent with the previous reports on the sponge Rhopaloeides odorabile adults and larvae, and other sponges5,43. The Nitrospira bacteria, known to be able to catalyze the nitrite oxidation step, were active in the South China Sea T. swinhoei, which is consistent with the revelation of the sponges Stylissa carteri and Geodia barretti37,38. No other types of bacteria with recognized capability to oxidize nitrite were detected by 16S rRNA or nxrA/nxrB targeting, indicating the strict nitrifying bacteria in the South China Sea T. swinhoei.
Quantitative assays on nitrifying genes or transcripts have reflected the abundance and transcriptional activity of nitrifying communities in sponges. For example, quantification of amoA genes/transcripts has indicated that AOA greatly outnumbered AOB in the sponges Halisarca caerulea, Higginsia thielei and Nodastrella nodastrella34. Meanwhile, the abundance of AOA amoA transcripts would be significantly affected by the physiological status of sponge Xestospongia muta44. In this study, qPCR assays revealed higher abundances of nitrifying genes and transcripts of Nitrosopumilus-like AOA than those of Nitrospira NOB in the South China Sea T. swinhoei. Within the nitrification process, the oxidation of ammonia to nitrite is the rate-limiting step, and archaeal ammonia oxidation rates have been reported to be correlated with AOA abundance in the ocean45. Therefore, higher abundance of AOA would contribute to the timely removal of the excreted ammonia from sponge to keep the host healthy. Meanwhile, the fact of nitrite oxidation rates exceeding ammonia oxidation rates in the ocean has been verified in the former case46. Therefore, higher abundance of AOA than NOB would be necessary in the nitrification process in the investigated T. swinhoei samples.
As a ubiquitous sponge species in the Indo-Pacific Ocean39, T. swinhoei-mediated nitrogen cycle has been partially predicted by the genomic information of the bacterial isolates from T. swinhoei. For example, genomic analyses of T. swinhoei-derived candidatus Synechococcus spongiarum and candidatus Entotheonella sp. have revealed the genes related with nitrogen fixation, nitrate reduction, ammonia/nitrite/urea assimilation and ammonia remineralization47,48. Meanwhile, genomic potentials of ammonia oxidation of Nitrosopumilus and Nitrosospira, and that of nitrite oxidation of Nitrospira have been suggested for Palauan T. swinhoei by amoA and 16S rRNA genes assays21,40. In this study, by comparing amoA/nxrB/16S rRNA genes with their transcripts, we are able to provide a community-wide profile of the in situ active microbes, including Nitrosopumilus-like AOA and Nitrospira NOB in the South China Sea T. swinhoei, thus gaining a picture of the active nitrification potentials in this sponge.
Archaea and bacteria have been previously revealed by 16S rRNA gene targeting for Palauan T. swinhoei21,40. Comparison of archaeal 16S rRNA sequences between Palauan T. swinhoei and the South China Sea T. swinhoei indicated that, archaeal community in these allopatric T. swinhoei were dominated by the Nitrosopumilus genus. ClustalW sequence alignment analysis (http://www.genome.jp/tools/clustalw/) indicated that, not all of the archaeal 16S rRNA sequences from these allopatric T. swinhoei were overlapped, as multiple alignment similarity of these archaeal 16S rRNA fragments is 95.6% (coverage region is archaeal 16S rRNA V3-V5 region, coverage value is 100%) (Supplementary Fig. S3A). Meanwhile, eight bacterial phyla, including Chloroflexi, Proteobacteria, Cyanobacteria, Actinobacteria, Acidobacteria, Nitrospirae, Bacteroidetes and Spirochaetes, were detected in these allopatric T. swinhoei; while four phyla, including Firmicutes, Thermodesulfobacteria, Gemmatimonadetes and Poribacteria were only identified from the South China Sea T. swinhoei. Particularly, ClustalW sequences alignment analysis indicated that, the Nitrospira 16S rRNA fragments from these allopatric T. swinhoei showed significant distinction, as multiple alignment similarity of these Nitrospira 16S rRNA sequences is 98.6% (coverage region is bacterial 16S rRNA V3 region, coverage value is 100%) (Supplementary Fig. S3B). These revelations indicated the different structures of archaeal and bacterial communities, including AOA and NOB lineages, between these allopatric T. swinhoei. Since samples of the South China Sea T. swinhoei and Palauan T. swinhoei were collected from separate biogeographic regions, different environmental factors, such as temperature, substrate concentration or dissolved oxygen, might lead to the composition shifts of archaeal and bacterial communities between these allopatric T. swinhoei. Meanwhile, some microbes that colonize their sponge host could be filtered in from local surrounding seawater, which could also affect the community structure of sponge symbionts49.
In this study, PCR cloning and Sanger sequencing methods have been used to reveal the putatively active nitrifying community in the South China Sea T. swinhoei. These methods are acceptable in conducting censuses of environmental microbes at the current time. For example, PCR and qPCR analyses of archaeal amoA gene have revealed the AOA diversity and abundance in Qinghai Lake50. Similar assays have revealed the composition and abundance shifts of archaeal amoA transcript in the deep sea51. Meanwhile, applications of high-throughput sequencing technologies have expanded the information about community memberships and diversity in various ecosystems. The majority of microbial ecology studies apply high-throughput sequencing by focusing on either gene-targeted or shotgun sequencing25. Gene-targeted high-throughput sequencing can provide functional gene information from microbial communities; however, there are several challenges associated with this approach. First, widespread lack of sequence conservation across functionally homologous genes can make PCR primer design difficult, leading to lack of detection of relevant functional genes in the environment. Second, even though fairly conserved primers can be designed for some functional genes of interest, the success of amplification is habitat/ecosystem dependent, most likely due to variations in the quality of extracted DNA, community complexity, sequence divergence, and target gene abundance25. Shotgun high-throughput sequencing can avoid many of the biases encountered in amplicon sequencing because it does not require amplification prior to sequencing. While it often fails to provide sufficient sequence depth to assemble the genomes of individual species, especially in complex microbial communities25. Another obstacle to adequate sequence coverage is contaminant DNA or RNA, particularly in host-associated microbiome studies, where sequence data may be predominantly from the host38. Shotgun-based approaches can also be impaired by dominant populations in the sample, which may be excessively oversampled38. In metatranscriptomic studies, this issue can be compounded by high rRNA abundance37. Data analysis can be challenging for the high-throughput sequencing technologies, particularly the shotgun sequencing data, as the analysis and assembly of large sequencing data sets are often computationally demanding52. In addition, statistical analysis of large short read datasets depends on the length of the reads and the availability of representative reference genomes25. Therefore, in this study, PCR-targeted cloning and Sanger sequencing approaches are reliable in specifically elucidating certain ecological function, i.e. the nitrification process in the South China Sea T. swinhoei.
Conclusions
In summary, the composition and activity of nitrifying community in the South China Sea T. swinhoei were revealed by targeting 16S rRNA genes, amoA and nxrB genes and their transcripts. Our investigation demonstrated the inhabitancy and transcriptional activity of Nitrosopumilus-like AOA and Nitrospira NOB that dominate the nitrifying community in this sponge host. Meanwhile, quantitative assessments indicated relative higher abundances of nitrifying gene and transcript of Nitrosopumilus-like AOA than those of Nitrospira NOB in this sponge. These results replenished an growing data pool of associations between sponge and nitrifiers, and further contributed to the understanding of the ecological functions of so far uncultivated functional community in sponges.
Materials and Methods
Sample collection
T. swinhoei was collected by Scuba diving near Xisha Archipelago (16°50′N; 112°20′E) in the South China Sea at a depth of ca. 20 m in May, 2009. The sponge species was identified by morphological characterization and 28S rRNA Sequencing. The 28S rRNA gene from this sponge showed 99% similarity to T. swinhoei voucher NCI218 28S rRNA gene (accession no. KC884844) and was submitted to GenBank under the accession number JF506040. Samples from three different individuals were cut into 15 mm thick slices, then immediately preserved in RNAfixer (YuanPingHao, Beijing, China) and stored at −70 °C before DNA or RNA extraction.
RNA, DNA extraction and cDNA synthesis
T. swinhoei RNA was extracted using the RNApure Plant Kit (CoWin Biotech, Beijing, China), and DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), respectively, according to the manufacturer’s instructions. RNA and DNA were extracted from the samples of three individuals, respectively; for each individual, several specimens were homogenized to minimize the sampling bias. RNA was treated with DNase (CoWin Biotech) at 37 °C for 30 min and purified with RNA Cleanup Kit (CoWin Biotech). RNA and DNA were accurately quantified by Qubit 2.0 Fluorometer using Qubit RNA BR Assay Kit (Invitrogen, Darmstadt, Germany) and Qubitds DNA BR Assay Kit (Invitrogen), respectively. For reverse-transcription, purified RNA was converted into single strand cDNA with random hexamers primer using the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocol. DNA and cDNA products were stored at −70 °C for subsequent experiments.
Clone library construction
Reported primers (Table 1) targeting amoA, nxrA, nxrB and 16S rRNA gene fragments were selected for PCR-screening from T. swinhoei DNA and cDNA templates. A novel primer pair NpnnxrBF/NpnnxrBFR specially targeting the Nitrospina nxrB gene was designed using the CODEHOP tool53 in this research. nxrB sequences of Nitrospina gracilis3/211 (accession no. KC262217), Nitrospina sp. AB-629-B18 (accession no. A3QC_RS0100660) and an uncultured Nitrospina sp. clone (accession no. KJ571877) were selected for primer design to cover a 425 bp region. Forward primer NpnnxrBF and reverse primer NpnnxrBR target the 685–707 and the 1089–1109 nucleotide regions of nxrB2 gene in Nitrospina gracilis3/2, respectively. Specificity of this primer pair targeting Nitrospina nxrB was tested by BLASTn (www.ncbi.nlm.nih.gov/blast/) searches against the GenBank database. PCR mixture (40 μl) consists of 2 μl cDNA or 4 ng DNA templates, 0.1 μM of each primer and 20 μl TaqMaster Mix (CoWin Biotech). PCR amplifications were carried out on a Thermocycler (Eppendorf, Hamburg, Germany) according to the following procedures: 95 °C for 5 min; followed by 30 cycles consisting of 95 °C for 40 s, annealing (temperature referring to Table 1) for 1min and 72 °C for 1min; and finally 72 °C for 15 min. For negative control, similar procedure was carried out using purified RNA to ensure that there was no genomic DNA contamination. Triplicate PCR products were pooled to reduce amplification bias and determined by 1.5% agarose gel electrophoresis. PCR products were gel-purified with MinElute Gel Extraction Kit (Qiagen), then ligated into pUC-T vectors (CoWin Biotech) and transformed into Escherichia coli DH5α competent cells (CoWin Biotech) according to the standardized protocols. For each gene marker, individual triplicates-derived clones were screened based on ampicillin resistance; while inserts were identified by PCR with vector-specific M13 primers and read on an ABI 3100 capillary sequencer (Sangon Corp., Sequencing Service, Shanghai, China).
Phylogenetic analysis
The obtained 16S rRNA sequences were checked for chimeras by DECIPHER’s Find Chimeras web tool54. All the obtained nucleotide sequences were trimmed manually by using ClustalW implemented in MEGA 6 with default settings. The trimmed sequences were used to perform BLASTn searches against available sequences in the GenBank. Operational taxonomic unit (OTU) was defined as sequence group in which nucleotide sequences differed by 3% for 16S rRNA genes and by 5% for functional genes55, respectively, by using the Mothur package56. Individual triplicates-derived clones of each gene marker were simultaneously sequenced to make one rarefaction curve and sequencing of clones would be sufficient when the rarefaction curves reach saturation at 3% or 5% nucleotide cutoff value using Mothur package56. The 16S rRNA sequences were assigned into different taxa using the RDP Classifier tool57. One representative sequence from each OTU and its closest sequence(s) retrieved from GenBank were aligned by using ClustalW implemented in the MEGA 6 for constructing phylogenic trees. Maximum-likelihood tree was constructed by using the MEGA 6 with the Kimura-2 parameter model according to a published guideline58. Bootstrap analysis was used to estimate the reliability of phylogenetic reconstructions (1000 replicates). Representative of each OTU was submitted to GenBank under the accession numbers: KM010251 - KM010255 for amoA sequences, KM010256 for nxrB sequence, KM010247 - KM010250, KM010257 - KM010299, KM121407 - KM121435 and KT380844 for 16S rRNA sequences.
qPCR assays
qPCR were performed using an ABI 7500 Fast Real-time PCR platform (Applied Biosystems, Foster, CA, USA), according to the procedures used by Radax et al.33. Three independent biological replicates were performed for each sample with three technical replicates each. For Nitrosopumilus-like amoA gene, Nitrospira nxrB gene, or their transcripts, the PCR was performed in a total volume of 25 μl containing 12.5 μl of SYBR Premix Ex Taq™ II (Takara, Dalian, China), 2ng of DNA template or 1μl of cDNA template and 0.1 μM of each primer (Table 1). The qPCR thermocycling steps were set as follows: 95 °C for 5 min and 40 cycles at 95 °C for 45 s, 57 °C (for amoA) or 56 °C (for nxrB) for 45 s, and 72 °C for 45 s. Cycling was followed by a final elongation step at 72 °C for 10 min. Standard curves (log-linear R2 > 0. 99) were generated using purified and quantified plasmids containing amoA sequence (168bp) or nxrB sequence (179bp) in a dilution series that spanned from 101 to 107 gene copies per reaction. Plasmid DNA was extracted with the PurePlasmid 96 Kit (CoWin Biotech), and the plasmid concentration was measured with a Qubit 2.0 Fluorometer using Qubitds DNA BR Assay Kit (Invitrogen). Since the sequences of the vector and PCR insert are known, we calculated the copy numbers of amoA or nxrB directly from the concentration of extracted plasmid DNA according to the reported formula59: copy numbers μl−1 = (A × 6.022 × 1023) × (660 × B)−1, where A is the plasmid concentration (g μl−1), B is the recombinant plasmid length (bp) containing the amoA or nxrB sequence, 6.022 × 1023 is the Avogadro’s number and 660 is the average molecular weight of one bp. For negative control, similar procedure was carried out using purified RNA to ensure that there was no genomic DNA contamination. After real-time PCR assay, the specificity of amplification was verified by generation of melting curves and agarose gel electrophoresis. Data acquisition and analysis of the Real-time PCR assay were performed using the 7500 System SDS Software Version 1.2 (Applied Biosystems). Data statistics and independent-sample t-test were performed using corresponding commands in Microsoft Excel 2007.
Additional Information
How to cite this article: Feng, G. et al. Inhabitancy of active Nitrosopumilus-like ammonia-oxidizing archaea and Nitrospira nitrite-oxidizing bacteria in the sponge Theonella swinhoei. Sci. Rep. 6, 24966; doi: 10.1038/srep24966 (2016).
Supplementary Material
Acknowledgments
Financial supports from National Natural Science Foundation of China (NSFC) (41076077, 41176127) and Minhang Leading Talent Project (Shanghai, China) are greatly acknowledged.
Footnotes
Author Contributions G.F. and Z.L. conceived the study and designed experiments; G.F., W.S. and F.Z. performed the fieldwork and molecular analysis; G.F. and Z.L. analyzed the data and G.F., L.K. and Z.L. wrote the paper.
References
- Zehr J. P. & Kudela R. M. Nitrogen cycle of the open ocean, from genes to ecosystems. Ann. Rev. Mar. Sci. 3, 197–225 (2011). [DOI] [PubMed] [Google Scholar]
- Byrne N. et al. Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J. 3, 117–123 (2009). [DOI] [PubMed] [Google Scholar]
- Cleary D. F., de Voogd N. J., Polónia A. R., Freitas R. & Gomes N. C. Composition and predictive functional analysis of bacterial communities in seawater, sediment and sponges in the Spermonde Archipelago, Indonesia. Microb. Ecol. 70, 889–903 (2015). [DOI] [PubMed] [Google Scholar]
- Francis C. A., Roberts K. J., Beman J. M., Santoro A. E. & Oakley B. B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102, 14683–14688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger D. et al. Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ. Microbiol. 10, 1087–1094 (2008). [DOI] [PubMed] [Google Scholar]
- Kowalchuk G. A. & Stephen J. R. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55, 485–529 (2001). [DOI] [PubMed] [Google Scholar]
- Pfister C. A., Gilbert J. A. & Gibbons S. M. The role of macrobiota in structuring microbial communities along rocky shores. Peer J. 2, e631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle-Yarwood S. A., Bottomley P. J. & Myrold D. D. Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ. Microbiol. 10, 2956–2965 (2008). [DOI] [PubMed] [Google Scholar]
- Gantner S., Andersson A. F., Alonso-Sáez L. & Bertilsson S. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 84, 12–18 (2011). [DOI] [PubMed] [Google Scholar]
- Pester M. et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 16, 3055–3071 (2014). [DOI] [PubMed] [Google Scholar]
- Purkhold U. et al. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis, implications for molecular diversity surveys. Appl. Environ. Microbiol. 66, 5368–5382 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthauwe J. H., Witzel K. P. & Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker, molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J. Exploring the distribution and activity of novel nitrite oxidizers in their natural and environmental habitats. Masterarbeit, Universität Wien (2013). [Google Scholar]
- Wertz S., Poly F., Le Roux X. & Degrange V. Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil. FEMS Microbiol. Ecol. 63, 261–271 (2008). [DOI] [PubMed] [Google Scholar]
- Winsley T., Van Dorst J. M., Brown M. V. & Ferrari B. C. Capturing greater 16S rRNA gene sequence diversity within the domain bacteria. Appl. Environ. Microbiol. 78, 5938–5941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antcliffe J. B., Callow R. H. & Brasier M. D. Giving the early fossil record of sponges a squeeze. Biol. Rev. Camb. Philos. Soc. 89, 972–1004 (2014). [DOI] [PubMed] [Google Scholar]
- Rodríguez-Marconi S. et al. Characterization of bacterial, archaeal and eukaryote symbionts from Antarctic sponges reveals a high diversity at a three-domain level and a particular signature for this ecosystem. PloS One 10, e0138837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveillaud J. et al. Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8, 1198–1209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U., Piel J., Degnan S. M. & Taylor M. W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10, 641–654 (2012). [DOI] [PubMed] [Google Scholar]
- Diaz M. C. & Ward B. B. Sponge-mediated nitrification in tropical benthic communities. Mar. Ecol. Prog. Ser. 156, 97–107 (1997). [Google Scholar]
- Bayer K., Schmitt S. & Hentschel U. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ. Microbiol. 10, 2942–2955 (2008). [DOI] [PubMed] [Google Scholar]
- Hallam S. J. et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. USA 103, 18296–18301 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Off S., Alawi M. & Spieck E. Enrichment and physiological characterization of a novel Nitrospira-like bacterium obtained from a marine sponge. Appl. Environ. Microbiol. 76, 4640–4646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed N. M., Saito K., Tal Y. & Hill R. T. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J. 4, 38–48 (2010). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. MBio 6, e02288–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polónia A. R., Cleary D. F., Freitas R., Voogd N. J. & Gomes N. The putative functional ecology and distribution of archaeal communities in sponges, sediment and seawater in a coral reef environment. Mol. Ecol. 24, 409–423 (2015). [DOI] [PubMed] [Google Scholar]
- Li Z. Y., Wang Y. Z., He L. M. & Zheng H. J. Metabolic profiles of prokaryotic and eukaryotic communities in deep-sea sponge Neamphius huxleyi indicated by metagenomics. Sci. Rep. 4, 3895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua C. P. et al. Potential metabolic strategies of widely distributed holobionts in the oceanic archipelago of St Peter and St Paul (Brazil). FEMS Microbiol. Ecol. 91, fiv043 (2015). [DOI] [PubMed] [Google Scholar]
- Klindworth A. et al. Diversity and activity of marine bacterioplankton during a diatom bloom in the North Sea assessed by total RNA and pyrotag sequencing. Mar. Genomics 18, 185–192 (2014). [DOI] [PubMed] [Google Scholar]
- Blazewicz S. J., Barnard R. L., Daly R. A. & Firestone M. K. Evaluating rRNA as an indicator of microbial activity in environmental communities, limitations and uses. ISME J. 7, 2061–2068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Liu M., Simister R., Webster N. S. & Thomas T. Marine microbial symbiosis heats up, the phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 7, 991–1002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamke J., Taylor M. W. & Schmitt S. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 4, 498–508 (2010). [DOI] [PubMed] [Google Scholar]
- Radax R., Hoffmann F., Rapp H. T., Leininger S. & Schleper C. Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ. Microbiol. 14, 909–923 (2012). [DOI] [PubMed] [Google Scholar]
- Mohamed N. M., Colman A. S., Tal Y. & Hill R. T. Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ. Microbiol. 10, 2910–2921 (2008). [DOI] [PubMed] [Google Scholar]
- Su J. et al. Phylogenetically diverse ureC genes and their expression suggest the urea utilization by bacterial symbionts in marine sponge Xestospongia testudinaria. PLoS One 8, e64848 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore C. L., Labrie M., Jarett J. K. & Lesser M. P. Transcriptional activity of the giant barrel sponge, Xestospongia muta Holobiont: molecular evidence for metabolic interchange. Front. Microbiol. 6, 364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitinho-Silva L. et al. Revealing microbial functional activities in the Red Sea sponge Stylissa carteri by metatranscriptomics. Environ. Microbiol. 16, 3683–3698 (2014). [DOI] [PubMed] [Google Scholar]
- Radax R. et al. Metatranscriptomics of the marine sponge Geodia barretti, tackling phylogeny and function of its microbial community. Environ. Microbiol. 14, 1308–1324 (2012). [DOI] [PubMed] [Google Scholar]
- Bewley C. A., Holland N. D. & Faulkner D. J. Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia 52, 716–722 (1996). [DOI] [PubMed] [Google Scholar]
- Hentschel U. et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68, 4431–4440 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenroth E. et al. Effect of long-term idle periods on the performance of sequencing batch reactors. Water Sci. Technol. 41, 105–113 (2000). [Google Scholar]
- Poulsen L. K., Ballard G. & Stahl D. A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59, 1354–1360 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister R. et al. Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environ. Microbiol. 14, 3232–3246 (2012). [DOI] [PubMed] [Google Scholar]
- López-Legentil S., Erwin P. M., Pawlik J. R. & Song B. Effects of sponge bleaching on ammonia-oxidizing Archaea, distribution and relative expression of ammonia monooxygenase genes associated with the barrel sponge Xestospongia muta. Microb. Ecol. 60, 561–571 (2010). [DOI] [PubMed] [Google Scholar]
- Beman J. M., Popp B. N. & Alford S. E. Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of California and eastern tropical North Pacific Ocean. Limnol. Oceanogr. 57, 711–726 (2012). [Google Scholar]
- Füssel J. et al. Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 6, 1200–1209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin P. M. & Thacker R. W. Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Mol. Ecol. 17, 2937–2947 (2008). [DOI] [PubMed] [Google Scholar]
- Wilson M. C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014). [DOI] [PubMed] [Google Scholar]
- Sipkema D. et al. Similar sponge-associated bacteria can be acquired via both vertical and horizontal transmission. Environ. Microbiol. 17, 3807–3821 (2015). [DOI] [PubMed] [Google Scholar]
- Yang J. et al. Sedimentary archaeal amoA gene abundance reflects historic nutrient level and salinity fluctuations in Qinghai Lake, Tibetan Plateau. Sci. Rep. 5, 18071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cono V. et al. Shifts in the meso- and bathypelagic archaea communities composition during recovery and short-term handling of decompressed deep-sea samples. Environ. Microbiol. Rep. 7, 450–459 (2015). [DOI] [PubMed] [Google Scholar]
- Nagarajan N. & Pop M. Sequence assembly demystified. Nat. Rev. Genet. 14, 157–167 (2013). [DOI] [PubMed] [Google Scholar]
- Rose T. M., Henikoff J. G. & Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31, 3763–3766 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. S., Yilmaz L. S. & Noguera D. R. DECIPHER, a Search-Based Approach to Chimera Identification for 16S rRNA Sequences. Appl. Environ. Microbiol. 78, 717–725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. & Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and species richness. Appl. Environ. Microbiol. 71, 1501–1506 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naïve bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 6, 1229–1235 (2013). [DOI] [PubMed] [Google Scholar]
- Perini F. et al. New approach using the real-time PCR method for estimation of the toxic marine dinoflagellate Ostreopsis cf. ovata in marine environment. PLoS One 6, e17699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.