Abstract
Bacterial adaptation requires large-scale regulation of gene expression. We have performed a genome-wide analysis of the Csr system, which regulates many important cellular functions. The Csr system is involved in post-transcriptional regulation, but a role in transcriptional regulation has also been suggested. Two proteins, an RNA-binding protein CsrA and an atypical signaling protein CsrD, participate in the Csr system. Genome-wide transcript stabilities and levels were compared in wildtype E. coli (MG1655) and isogenic mutant strains deficient in CsrA or CsrD activity demonstrating for the first time that CsrA and CsrD are global negative and positive regulators of transcription, respectively. The role of CsrA in transcription regulation may be indirect due to the 4.6-fold increase in csrD mRNA concentration in the CsrA deficient strain. Transcriptional action of CsrA and CsrD on a few genes was validated by transcriptional fusions. In addition to an effect on transcription, CsrA stabilizes thousands of mRNAs. This is the first demonstration that CsrA is a global positive regulator of mRNA stability. For one hundred genes, we predict that direct control of mRNA stability by CsrA might contribute to metabolic adaptation by regulating expression of genes involved in carbon metabolism and transport independently of transcriptional regulation.
Organisms have developed multiple mechanisms of gene expression regulation to adapt their physiology and metabolism to changing environment in order to compete with other species. At any time in the adaptation process, the intracellular level of an mRNA is the direct result of gene expression regulation. For each gene, the balance of two independent cellular processes, synthesis and degradation of mRNA determines mRNA quantity. These two levels were previously demonstrated to contribute to adaptation processes in various organisms1,2,3,4.
At the genome-wide scale, global regulators allow coordinated expression control of a large set of genes after sensing environmental changes. Most of the known global regulators in Bacteria are transcriptional regulators (CRP, Fis, etc…) with well described mechanisms of action. They activate or repress initiation of mRNA transcription by the RNA polymerase, and control global metabolism such as carbon uptake and respiration5,6. Only a few global regulators act at the post-transcriptional level on mRNA stability. One example is the RNA binding protein Hfq, homolog of the Sm and Lsm proteins that form the core of splicing and mRNA degradation complexes in eukaryotes. Hfq facilitates base-pairing interactions of regulatory noncoding small RNAs (sRNAs) on multiple target mRNAs7 to destabilize mRNAs and regulate key bacterial metabolic pathways8,9.
The Csr/Rsm (carbon storage regulator/repressor of secondary metabolites) system is a multi-component global regulatory system, well conserved in Bacteria, that controls gene expression of many important cellular functions. The Csr system represses glycogen metabolism, gluconeogenesis, biofilm formation and quorum sensing while it activates glycolysis, cell motility, virulence and pathogenesis as demonstrated in γ-Proteobacteria such as Escherichia, Salmonella, Erwinia, Pseudomonas or Vibrio10. It is composed of two proteins, CsrA, an RNA binding protein, and CsrD, a putative signaling protein, and two regulatory sRNAs, CsrB and CsrC11. The Csr system is mainly known for its post-transcriptional role in mRNA stability. In E. coli K-12 strains, mRNA binding of CsrA has been demonstrated to destabilize four transcripts (glgCAP, pgaABCD, ydeH and ycdT)12,13,14,15 and to stabilize one (flhDC)16. The CsrD protein stimulates the degradation of CsrB and CsrC17. An originality of the Csr system is to combine multi-level regulation acting at the post-translational level but also at the transcriptional level. Besides its role in mRNA stability, the Csr system is indeed involved in post-translational regulation through the CsrA-dependent activation of enzyme activity18. The CsrA activity is itself regulated via sequestration by the sRNA CsrB, CsrC and McaS19,20,21,22. At the transcriptional level, CsrA interacts with the global stringent response23 and the BarA/UvrY two-component signal transduction system24,25, two major actors in regulation of E. coli gene expression. The role of CsrD at the transcriptional level is not yet fully understood. Although CsrD contains GGDEF/EAL domains, CsrD is not directly involved in c-di-GMP metabolism17,26. However, CsrD was demonstrated to regulate expression of a transcriptional regulator CsgD via mechanisms not yet fully elucidated26.
How the Csr system combines transcriptional and post-transcriptional regulation at the genome-wide scale to control major physiological functions is not yet determined. There are a limited number of genomic studies of the Csr system, and they all focused on CsrA. Lists of CsrA targeted genes were provided by mRNA-protein interaction analysis23, bioinformatics tools27,28 and with transcriptomic analysis15. When changes in mRNA amounts were identified by transcriptomic analysis in response to csrA overexpression15, measurement of mRNA stability was not simultaneously performed impeding to discriminate between transcriptional and post-transcriptional regulations.
Here we have elucidated the regulation networks of CsrA and CsrD at both the transcriptional and post-transcriptional levels. We determined and compared genome-wide data of mRNA quantity and mRNA stability in a wild type MG1655 strain, in a strain containing a partial deletion of csrA, MG1655(csrA51), and in a strain deleted of csrD, MG1655(ΔcsrD). Using an integrative approach, we show that in our experimental condition CsrA and CsrD are both involved in massive transcriptional regulation whereas genome-wide regulation of mRNA stability is only dependent on CsrA. We discuss these results in terms of direct and indirect effects, and show how the direct control of mRNA stability by CsrA contributes to metabolic adaptation.
Results
Construction and growth of the MG1655(csrA51) and MG1655(ΔcsrD) mutant strains
The csrA gene is essential for cell viability in E. coli K-12 strains grown on glycolytic sources29,30. A hypomorphic mutant corresponding to a transposon insertion into the csrA gene is viable and has been used in numerous studies of the role of CsrA31,32,33,34. CsrA is a dimeric 61 amino acid RNA binding protein35. The transposon insertion results in the production of a 50 amino acid protein deleted of the C-terminal dimerization domain36. In order to create isogenic strains in the MG1655 background, we engineered the MG1655(csrA51) strain by inserting a stop codon at position 51. In this construct, the endogenous transcription termination signal downstream of the stop codon is conserved. As expected36, the MG1655(csrA51) strain produced much more glycogen than the MG1655 strain and this phenotype is abolished by complementation with an ectopic copy of the wild type csrA (Supplementary Fig. S1). The mutant strain also displayed significantly less motility and more biofilm formation (Supplementary Fig. S1) as expected14,16. Measurement by Northern Blot of CsrB and CsrC levels in the MG1655(csrA51) strain showed the absence of expression of the two sRNAs (Supplementary Fig. S1) confirming the role of CsrA in transcriptional activation of CsrB and CsrC24. All these results confirmed that the activity of the C-terminal truncated variant of CsrA was drastically diminished in our MG1655(csrA51) strain10.
The MG1655(ΔcsrD) strain was constructed by total deletion of the csrD gene in E. coli MG1655. When cultured in batch in M9 minimal medium supplemented with glucose, MG1655(csrA51) cells exhibited a two-fold reduced maximal growth rate (μmax = 0.31 ± 0.01 h−1) compared to wild type (μmax = 0.63 ± 0.01 h−1) (Fig. 1); normal growth was restored by complementation with an ectopic copy of the wild type csrA gene (Fig. 1). In contrast, the maximal growth rate was only slightly affected in MG1655(ΔcsrD) (0.52 ± 0.08 h−1) compared to the MG1655 strain (Fig. 1). To eliminate the growth rate effects on mRNA half-lives and levels3 and to permit comparison between strains, the MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains were cultivated in continuous culture at an equivalent growth rate of 0.10 h−1. The csrA51 or ΔcsrD mutations did not affect the macro kinetics behavior of the strains (Table 1). Glucose (3 g/l) was totally consumed and no acetate was produced. A slightly higher biomass yield was measured in the MG1655(csrA51) strain. The csrA51 mutation was associated with a 5-fold higher intracellular glycogen content compared to the MG1655 strain (Table 1).
Figure 1. Maximal growth rates (μmax) of the MG1655, MG1655(csrA51), MG1655(csrA51) complemented with a plasmid copy of the wild type csrA gene (csrA+), and MG1655(ΔcsrD) strain in M9 minimal medium supplemented with glucose.
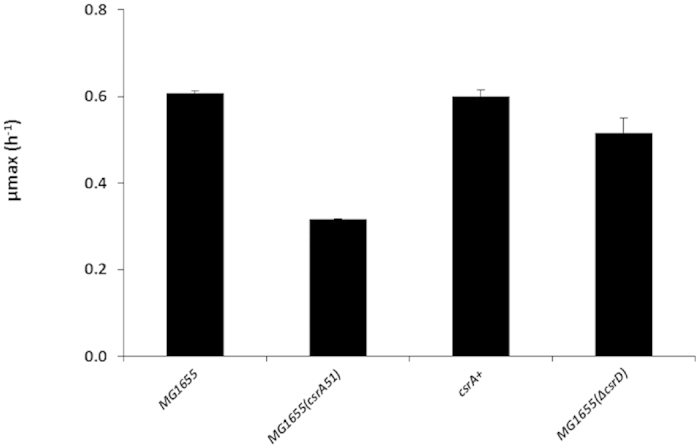
Table 1. Macro kinetics behavior and intracellular glycogen content of the strains MG1655, MG1655(csrA51) and MG1655(ΔcsrD) in continuous cultures at 0.1 h−1 in M9 minimal medium supplemented with 3 g/l glucose at 37 °C.
| Strain | Residual glucose (mM) | Produced acetate (mM) | qS (g glucose/g of dry cell weight h−1) | Yx/s (g of dry cell weight/g glucose) | Intracellular glycogen content (g glucose/g of dry cell weight) |
|---|---|---|---|---|---|
| MG1655 | ND | ND | 0.29 ± 0.03 | 0.36 ± 0.01 | 0.034 ± 0.006 |
| MG1655(csrA51) | ND | ND | 0.26 ± 0.01 | 0.41 ± 0.02 | 0.194 ± 0.006 |
| MG1655(ΔcsrD) | ND | ND | 0.29 ± 0.01 | 0.35 ± 0.01 | 0.040 ± 0.007 |
ND: not detected.
Effect of the csrA51 and ΔcsrD mutations on mRNA stability
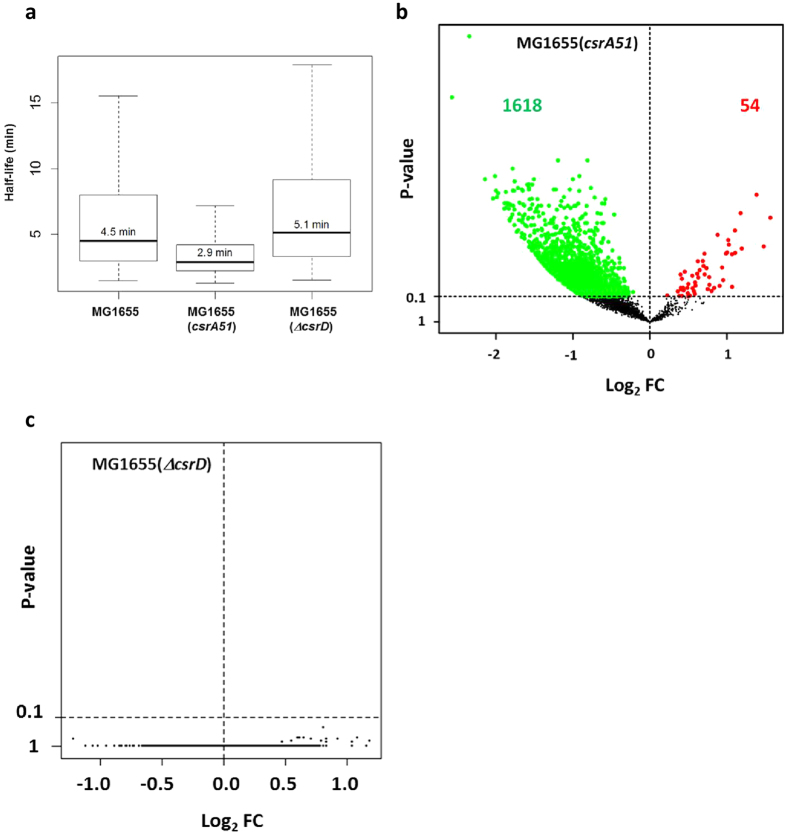
CsrA has been demonstrated to regulate the stability of a few mRNAs11. However, many more putative targets are suggested by interaction study23 and bioinformatics analysis27,28. CsrD is involved in the turn-over of the two sRNA CsrB and CsrC17 but its involvement in mRNA stability regulation has not yet been investigated. To explore CsrA and CsrD involvement in mRNA stability regulation, we compared genome-wide measurements of mRNA stability in the MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains grown in continuous culture at 0.1 h−1 (Supplementary Table S1). Transcripts were globally less stable in MG1655(csrA51) than in the MG1655 strain, the median half-life decreasing from 4.5 min in the MG1655 to 2.9 min in the mutant strain (Fig. 2a). More precisely, a statistical test (P-value ≤ 0.1) identified 1672 messengers with differential half-life between the MG1655(csrA51) and MG1655 strains: 1618 were significantly destabilized in the mutant strain whereas 54 were significantly stabilized (Fig. 2b). Interestingly, these results show a genome-wide involvement of CsrA in mRNA stability regulation and more particularly a massive destabilization of mRNA in the MG1655(csrA51) mutant strain.
Figure 2. Effect of the csrA51 and ΔcsrD mutations on mRNA half-life.
(a) Box plots of transcript half-life for the MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains (n = 3351 mRNAs). Values are separated into four quartiles by horizontal bars. The central bar (in the middle of the rectangle) represents the median value, which is given above the bar. VolcanoPlot of the log2 fold change (Log2 FC) of mRNA half-lives (b) between the MG1655(csrA51) and MG1655 strains (n = 3028 mRNAs) and (c) between the MG1655(ΔcsrD) and MG1655 strains (n = 3333 mRNAs). A P-value ≤ 0.1 was required for fold change significance (above the horizontal dashed line). The significantly stabilized mRNAs in the mutant strain compared to the MG1655 strain are colored in red whereas the significantly destabilized mRNAs are colored in green.
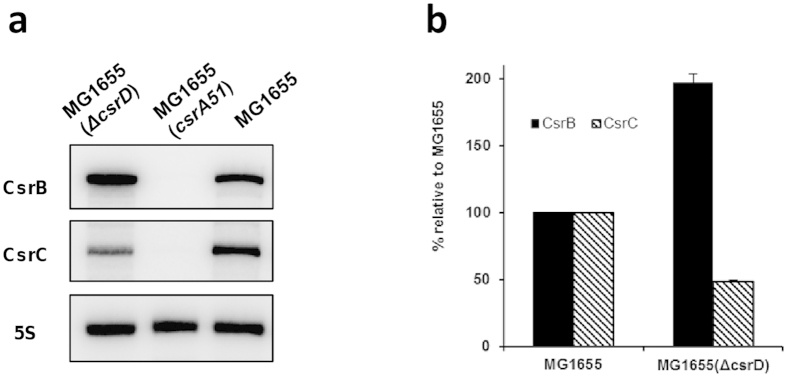
The median mRNA half-life in MG1655(ΔcsrD) was only slightly higher than in the MG1655 strain (Fig. 2a). However, applying the same statistical test used for the MG1655(csrA51) strain (P-value ≤ 0.1), no mRNA with a significant variation of stability was found between the MG1655(ΔcsrD) and MG1655 strains (Fig. 2c) suggesting a small systematic increase in global mRNA stability. This observation is quite surprising since according to the established regulatory network of the Csr system, CsrD is believed to positively regulate CsrA activity via CsrB and CsrC destabilization17. Consequently, a destabilization as already observed for MG1655(csrA51) strain was expected in the MG1655(ΔcsrD) strain. In order to understand better this phenotype, we measured by Northern Blot CsrB and CsrC half-lives and levels. As previously shown17, CsrB and CsrC sRNAs were strongly stabilized in the MG1655(ΔcsrD) strain (Supplementary Fig. S2) but their levels were not modified accordingly, suggesting a transcriptional retro-control: indeed the CsrB level was only increased around two fold in our experimental condition whereas the CsrC level was even decreased around two fold (Fig. 3). Since CsrD regulation of CsrB and CsrC turnover depends on the physiological status of the cells37, the CsrB and CsrC sRNA stabilization shows that CsrD was active in our growth condition. The opposite regulation of CsrB and CsrC levels is expected to only slightly modify the amount of CsrA sequestrated by the two sRNAs in the MG1655(ΔcsrD) strain compared to the MG1655 strain. Therefore, CsrA should exhibit a rather similar activity in the two strains. This conclusion was supported at the phenotypic level by the absence of a CsrA-related glycogen response in the MG1655(ΔcsrD) strain compared to the MG1655 strain. We measured the glycogen content in the MG1655(ΔcsrD) strain and we did not find an increase of the glycogen content (Table 1). In our growth conditions, the presence/absence of CsrD did not affect mRNA stability neither via CsrD direct targeting nor via CsrA activity.
Figure 3. Level of CsrB and CsrC in the MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains in continuous culture.
(a) Northern blots of total RNA probed for CsrB and CsrC. (b) Hybridization signals quantified on a PhosphorImager. The signals for MG1655 were set to 100%.
Effect of the csrA51 and ΔcsrD mutations on mRNA levels
To assess the transcriptional influence of CsrA and CsrD, we first measured the transcriptomic response in MG1655(csrA51) and MG1655(ΔcsrD) mutant strains (Supplementary Table S2). In contrast to the massive destabilization of mRNA in the MG1655(csrA51) strain, transcriptomic analyses showed a global up-regulation of mRNA amounts with 2195 mRNAs significantly higher compared to 249 mRNAs significantly lower (Fig. 4a). Up-regulated expressions were mainly related to biogenesis of cell wall and membrane and to signal transduction mechanisms while the down-regulated expressions were related to cell motility and carbohydrate transport and metabolism (Supplementary Table S3). These results are consistent with our phenotypic characterization and the literature (less motility, more biofilm formation and glycogen accumulation). The groups “Flagellum organization” and “Ciliary or flagellar motility” (containing flhDC gene, fli and flg operons) were enriched in the down-regulated mRNA levels correlating with reduced motility16,38. GO term “Single species biofilm formation” (including csg and ydeH genes) was enriched in the group of up-regulated mRNA levels but also yddV, yjjC and yliE mRNAs encoding GGDEF/EAL proteins involved in c-di-GMP metabolism15,33. All transcripts involved in glycogen synthesis (glgC, glgB, glgA and glgP/Y) were at least 3-fold up-regulated consistently with the high level of glycogen produced in MG1655(csrA51) in our experimental condition (Table 1). Moreover, numerous transcripts whose expressions are known to be controlled by CsrA including pck, cstA, hfq, relA and spoT have behavior consistent with the literature23,36,39,40.
Figure 4. Effect of the csrA51 and ΔcsrD mutations on mRNA levels.
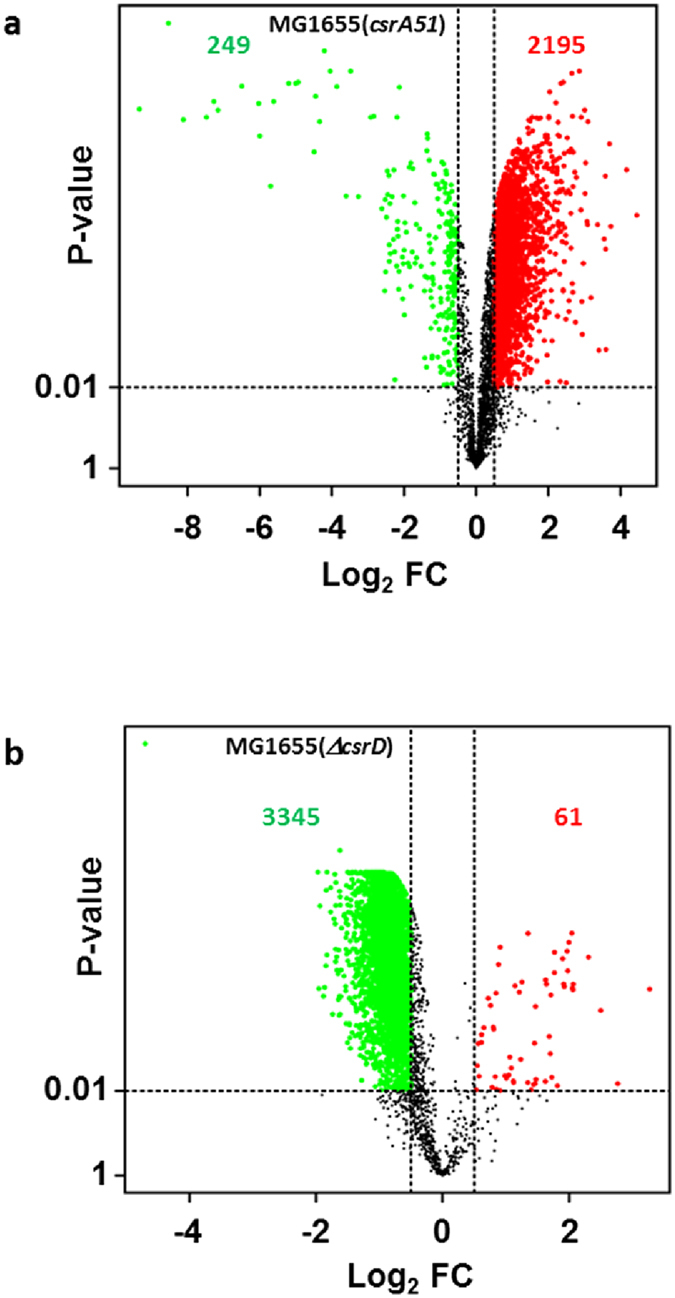
VolcanoPlot of the log2 fold change (Log2 FC) of mRNA amounts (n = 4254 mRNAs) (a) between the MG1655(csrA51) and MG1655 strains and (b) between the MG1655(ΔcsrD) and MG1655 strains. A P-value ≤ 0.01 (above the horizontal dashed line) and a log2 FC higher than 0.5 or lower than −0.5 (outside the vertical dashed lines) were required for fold change significance. Between the mutant strain and the MG1655 strain, the significantly up-regulated amounts were colored in red whereas the significantly down-regulated levels were colored in green.
A strong transcriptomic response was observed in the MG1655(ΔcsrD) strain compared to the MG1655 strain (Fig. 4b). A large majority of the genes were down-regulated (3345/4254) and only 61 were significantly up-regulated. In the MG1655(csrA51) strain, a connection between CsrA and CsrD expression is expected due to the negative regulation of csrD gene expression by CsrA15,17. Our results show that csrD mRNA quantity was strongly enhanced in the MG1655(csrA51) strain (4.6-fold increase). Considering this result, we have compared the transcriptomic responses in the MG1655(csrA51) and MG1655(ΔcsrD) strains. 77% (1878/2444) of differentially expressed mRNAs in MG1655(csrA51) were also differentially expressed in ΔcsrD and for most of them (1653/1878) the regulation was in opposite directions: most of the up-regulated mRNAs in MG1655(csrA51) were down-regulated in the MG1655(ΔcsrD) strain. Altogether these results show that mutations of CsrA and CsrD induce strong but opposite global regulation of mRNA levels and that the transcriptomic response observed in the MG1655(csrA51) strain could be at least partially mediated by CsrD.
CsrA and CsrD are involved in massive transcriptional regulation
To decipher the global roles of CsrA and CsrD at the transcriptional level, we integrated datasets of mRNA stability and mRNA quantity measured in the MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains. A difference in mRNA concentration between each mutant and the wild type strain can be the consequence of change in mRNA synthesis (transcription), degradation and/or mRNA dilution as cells divide not necessarily at the same rate. In our experiments, all the strains were cultivated at the same growth rate, so the effect of mRNA dilution on gene expression can be eliminated. Therefore, all changes in mRNA level in our study can only be explained by transcriptional and/or mRNA stability regulation.
In the MG1655(ΔcsrD) strain, no individual transcript stability was significantly modified whereas a negative transcriptomic response was observed for 3345 genes. Therefore, these changes in mRNA levels result mainly from down regulation of transcription. Similarly, in MG1655(csrA51), 542 mRNAs were regulated at the mRNA level (positively for 82%), but not in stability. This demonstrates that mutating csrA provokes a transcriptional response, which is generally positive. These results show that CsrD and CsrA regulate a large number of genes by acting only at the transcriptional level. Molecular validation of widespread transcriptional action of CsrA and CsrD was provided by transcriptional fusions using a lacZ reporter gene. We confirmed higher lacZ expression under the control of the glgB and ydeH promotors in the MG1655(csrA51) strain compared to the MG1655 strain, and the lower lacZ expression under the control of the frdA and csrC promotors in the MG1655(ΔcsrD) compared to the MG1655 strain (Supplementary Table S4).
To go deeper in the transcriptional regulation, we searched for changes in mRNA levels of genes related to the functional category “Transcription, DNA-dependent” in both MG1655(ΔcsrD) and MG1655(csrA51), compared to the MG1655 strain. Interestingly opposite regulation was observed in the mutant strains, as genes encoding protein involved in transcription were down-regulated in the MG1655(ΔcsrD) strain but up-regulated in MG1655(csrA51) (Table 2). These genes correspond to RNA polymerase (β and β’ subunits, rpoB and rpoC), sigma factors (rpoD, rpoE, rpoH, rpoN, rpoS and fecI), factors involved in the stringent response (relA, spoT and dksA) and global transcriptional regulators. More precisely, the transcriptional regulators were crp, cyaA and fruR/cra for regulating the carbon flow, fnr and arcA involved in regulation of the aerobic/anaerobic metabolisms and ihfA and ihfB encoding the IHF factor involved in chromosomal structure. Taken together with the up-regulated expression of csrD mRNA in the MG1655(csrA51) strain, these results support the conclusion that CsrD-mediated transcriptional regulation contributes to the transcriptional response observed in MG1655(csrA51).
Table 2. Effect of the ΔcsrD and csrA51 mutations on mRNA level of genes involved in the biological process “Transcription, DNA-dependent”.
| GeneId | GeneSym | MG1655(ΔcsrD) |
MG1655(csrA51) |
||
|---|---|---|---|---|---|
| FC | P-value | FC | P-value | ||
| b0080 | fruR/cra | 0.50 | 2.4E−07 | 1.40 | 1.6E−05 |
| b0145 | dksA | 0.53 | 7.4E−06 | 1.67 | 3.3E−05 |
| b0912 | ihfB | 0.60 | 8.2E−06 | 1.43 | 9.9E−05 |
| b1334 | fnr | 0.55 | 3.0E−05 | 1.31 | 4.1E−03 |
| b1712 | ihfA | 0.58 | 3.0E−05 | 1.30 | 2.8E−03 |
| b2573 | rpoE | 0.51 | 3.7E−05 | 1.36 | 4.3E−03 |
| b2741 | rpoS | 0.49 | 4.0E−05 | 1.28 | 1.9E−02 |
| b2784 | relA | 0.53 | 1.6E−05 | 2.05 | 1.0E−05 |
| b3067 | rpoD | 0.55 | 1.8E−07 | 1.26 | 3.5E−05 |
| b3202 | rpoN | 0.54 | 2.5E−07 | 2.06 | 1.3E−07 |
| b3357 | crp | 0.48 | 4.0E−07 | 1.39 | 4.1E−05 |
| b3461 | rpoH | 0.51 | 3.0E−05 | 2.28 | 1.2E−05 |
| b3650 | spoT | 0.58 | 1.2E−04 | 2.31 | 1.2E−05 |
| b3806 | cyaA | 0.53 | 2.2E−07 | 1.73 | 4.9E−07 |
| b3987 | rpoB | 0.69 | 5.8E−07 | 1.13 | 5.5E−04 |
| b3988 | rpoC | 0.53 | 3.7E−06 | 1.14 | 2.9E−02 |
| b4293 | fecI | 0.54 | 6.6E−05 | 2.13 | 2.3E−05 |
| b4401 | arcA | 0.50 | 3.3E−06 | 1.16 | 2.7E−02 |
GeneId corresponds to the gene Blattner number55, GeneSym to the gene name, FC to the fold change of mRNA amounts between the mutant strain and the MG1655 strain and P-value is the associated adjusted P-value.
CsrA is a global positive regulator of mRNA stability
To decipher the precise role of CsrA in large scale mRNA stability regulation, we considered variations in both mRNA stability and concentration between MG1655(csrA51) and MG1655. Stability and level variations in MG1655(csrA51) compared to MG1655 were schematically classified in three groups (Fig. 5a and Supplementary Table S5). Group I corresponded to a variation in mRNA amount not associated with a variation in stability. This group contained the above mentioned 542 mRNAs whose levels were under transcriptional control. In the two other groups, both the stability and quantity differed in MG1655(csrA51) compared to MG1655. In group II (n = 1217), stability and quantity varied in opposite direction (lower stability associated with a higher amount in MG1655(csrA51) compared to MG1655 or vice versa) whereas group III (n = 455) included mRNAs exhibiting stabilization with similar or a higher level and destabilization with a similar or lower level.
Figure 5.
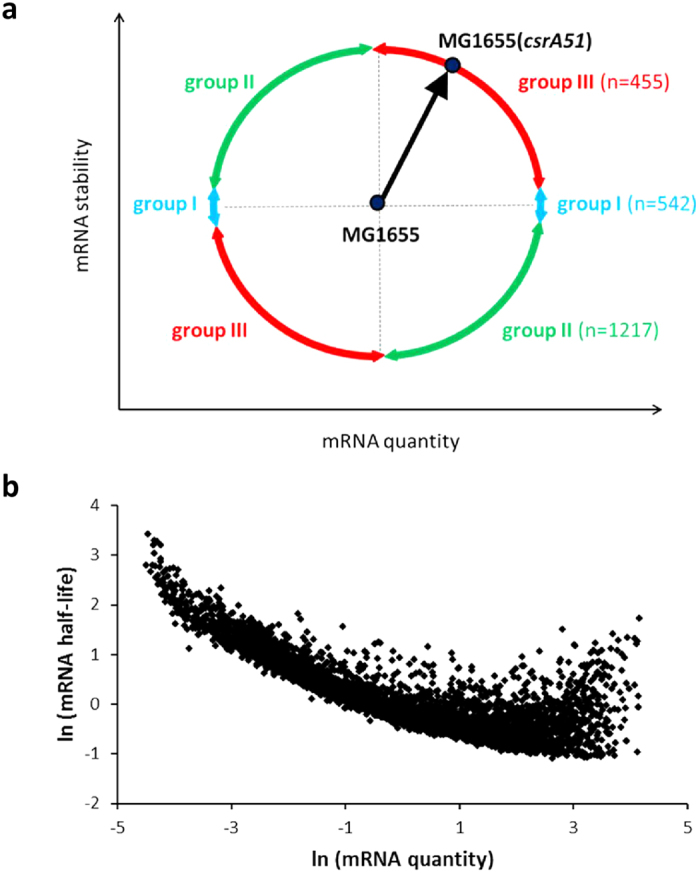
(a) Variations in mRNA stability and quantity between the strains MG1655(csrA51) and MG1655. mRNAs with variation in level but not in stability in MG1655(csrA51) compared to MG1655 are in group I, those with variations in stability and quantity in opposite directions are in group II, and those with variations in stability and quantity not in opposite directions are in group III. (b) Plots of transcript half-life (in min) as a function of transcript quantity (in arbitrary units per mg of dry cell weight) in the MG1655 strain (n = 3074 mRNAs) and MG1655(csrA51) (n = 4098 mRNAs). All the values were log-transformed and centered. The Pearson correlation coefficient is −0.81 with a P-value < 2.2E−16.
In a previous study, we have demonstrated in E. coli at the genome-wide scale that the quantity of an mRNA is the major determinant of its stability41. Plotting mRNA half-life as a function of its amount in MG1655(csrA51) and MG1655 showed a strong negative correlation (Fig. 5b). This indicates that for a given mRNA, a change in stability between MG1655(csrA51) and MG1655 can potentially be linked to a change in concentration. In other words, mRNA stabilization in MG1655(csrA51) may result from a lower mRNA amount whereas a destabilization may be due to a higher amount. This relationship has been validated at the molecular level (Nouaille et al., manuscript in preparation). Therefore, for the 1217 transcripts of group II with antagonistic regulation of stability and quantity in MG1655(csrA51), the regulation of mRNA stability could correspond to a direct effect of CsrA on mRNA stability or an indirect effect due to change in mRNA level depending on transcription regulation. The regulation of stability of these mRNAs by CsrA is thus complex due to overlapping direct and indirect effects. Most of the mRNAs (1215/1217) were destabilized with up-regulated level and functional classes such as “DNA replication” and “signal transduction” were enriched in this group (Supplementary Table S5). The presence of a consensus CsrA binding site RUACARGGAUGU27 was investigated in the −100/+100 nt region (+1 corresponding to the start codon) of the 1217 transcripts. 635 displayed at least one putative CsrA binding site.
For the 455 mRNAs of group III with stability and quantity varying in the same directions, variation in half-life cannot be explained by variation in amount according to the correlation shown in Fig. 5b. Consequently, it is likely that their stability is directly controlled by CsrA; these mRNAs were therefore predicted as putative direct post-transcriptional targets of CsrA. Many were novel CsrA targets, since cross-comparisons of our list with previous studies showed that only 10 and 93 were already in silico predicted28 or shown to co-purify with CsrA23, respectively (Supplementary Table S6). As previously, the presence of a consensus CsrA binding site was investigated in the 455 transcripts. We found that 180 mRNAs displayed unique putative CsrA binding site while 78 had at least 2 sites with hot spot localization in the 5′UTR region (Supplementary Table S6). Taken together, more than 65% (297/455) were predicted to experimentally bind CsrA23 and/or to have a putative CsrA binding site. These results suggest that most of the identified putative CsrA direct post-transcriptional targets are likely to interact physically with the CsrA protein although false positive experimental binding23 and degenerated CsrA binding motif cannot be totally excluded. We found that more than 88% (403/455) of the CsrA direct post-transcriptional targets were destabilized in the MG1655(csrA51) strain (Supplementary Table S6). Together with the results shown in Fig. 2b, our work suggests that CsrA has an important role in the stabilization of a large number of messengers.
Metabolic impact of the direct regulation of mRNA stability by CsrA
The predicted direct regulation by CsrA of the stability of 455 mRNAs is not expected to contribute in the same way to the control of metabolic adaptation. For 356 mRNAs, a change in stability between the two strains was not associated with a significant change in mRNA level, which is generally required to provoke protein concentration variations. For these mRNAs, stability regulation was counteracted by transcription regulation. These mRNAs were mainly related to translation and carbohydrate transport and metabolism (Supplementary Table S5). On the contrary, for 99 transcripts (Supplementary Table S7), changes in stability were associated with changes in mRNA amounts and might therefore contribute to metabolic differences between the two strains.
More precisely, 64 mRNAs were destabilized with down-regulated level whereas 35 were stabilized with up-regulated level between the MG1655(csrA51) and MG1655 strains (Supplementary Table S7). Among the destabilized and down-regulated mRNAs in the MG1655(csrA51) strain, we found tpiA in agreement with a previous result showing that the triose-phosphate isomerase activity was positively regulated by CsrA activity42. We also identified the zwf transcript, previously reported to co-purify with CsrA23, but its stability and amount have never previously been shown to be regulated by the Csr system. Our results clearly showed down-regulated mRNA stability and quantity of genes encoding carbohydrate transport (araF, malE, alsB, idnT, melB, glpF, srlAEB and agaVW) and certain metabolic pathways, e.g. idonate/gluconate metabolic process (idnDOT) and galacturonate catabolic process (uxaCA) in MG1655(csrA51) (Supplementary Table S5). mRNA stability and level of a large number of metabolic enzymes (aldA, ldhA, udp, yfaU, hdhA, glpK, dmlA, gloB, aes, fdhF, speB, tpx, thiH, ydiB, ilvC, pptA, ebgC and frlB) were found to be also negatively regulated in MG1655(csrA51).
In the stabilized and up-regulated mRNAs in the MG1655(csrA51) strain, several genes belonged to the glycogenesis and gluconeogenesis pathways: maeA, maeB, pck, gapA, pgm and glgB (Supplementary Table S5). The expression and/or activity of some of these genes/proteins were known to be negatively controlled by CsrA36,42. Our results provide a better understanding of the CsrA effect on these targets, the regulator acting at the level of both mRNA stability and quantity. Expression (stability and level) of several genes of the Krebs cycle and the amino acid metabolism were found to be upregulated in the MG1655(csrA51) strain: acnB, fumA, sdhC, leuB, leuC, glnA and asd. We observed that the messengers of the mannose PTS permease (manXYZ), of the pyridine nucleotide transhydrogenase (pntAB) and of the fatty acid synthesis (fabD and fabF) were stabilized and exhibited higher level in the MG1655(csrA51) strain. The csrA51 mutation also stabilizes and increases the amount of mRNAs related to transcription (rpoD encoding the vegetative sigma factor σ70 of RNA polymerase) and to translation (ribosomal rplP, rplN and rpsS transcripts). This and the accumulation of glycogen (Table 1) could partially explain the slightly higher biomass yield in the MG1655(csrA51) strain compared to the MG1655 (Table 1).
For the 99 mRNAs whose level and stability were regulated in the MG1655(csrA51) strain, we examined the contribution of stability regulation to the control of the mRNA amount. Regulatory coefficients were calculated between MG1655(csrA51) and the MG1655 strain. This quantifies the respective effects of transcription regulation and stability regulation on mRNA amount3. For a large majority of these mRNAs (85/99), the regulation of stability predominantly controlled mRNA level. This indicates that it was a change in stability that provoked differential mRNA amount in the MG1655(csrA51) strain and this is true even if transcription regulation is present; we called this type of regulation degradational control of the mRNA quantity (Supplementary Table S7). Altogether these results provide the first demonstration of how direct regulation (positive/negative) of mRNA stability by CsrA regulates a hundred mRNA amounts independently of transcription regulation and therefore contributes to control the global metabolism.
Discussion
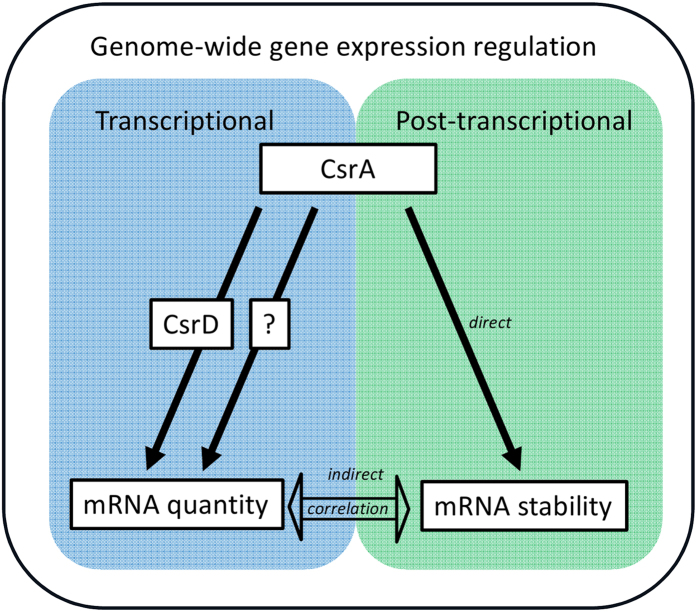
In this study, we applied for the first time an integrative biology approach to decipher the transcriptional and post-transcriptional roles of CsrA and CsrD. Although the Csr system is generally believed to be involved in a post-transcriptional response, the protein CsrD was demonstrated here to be involved in a massive positive transcriptional regulation. In contrast, mutating the csrA gene induces both transcriptional and post-transcriptional responses. Transcription of more than 500 genes and stability of more than 1600 mRNAs differ between the MG1655(csrA51) and MG1655 strains. Opposite transcriptional responses were obtained in the MG1655(csrA51) and MG1655(ΔcsrD) strains. Since we confirmed the strong up-regulation of csrD expression in MG1655(csrA51), it is likely that the transcriptional response observed in MG1655(csrA51) was at least partially mediated by CsrD. The importance of the negative feedback created by CsrA regulation of CsrD expression was previously highlighted in the dynamic response of the Csr system43. The time of turning on and off expression of CsrA target proteins was dependent on the CsrD expression level. Our study shows that the regulatory control of the Csr system involves genome-wide transcriptional and post-transcriptional regulation of gene expression connected by CsrA and CsrD (Fig. 6).
Figure 6. Scheme of the connections described in this study between the transcriptional and post-transcriptional regulatory networks, CsrA and CsrD, which are involved in the genome-wide regulation of gene expression.
Filled arrows represent a connection type “act on” while the empty arrow represents a correlation.
Several mechanisms of CsrD action at the transcriptional level can be proposed. This may occur indirectly via transcriptional factors whose expressions are CsrD-dependent. Many transcriptional regulators are down-regulated in the absence of CsrD. The actions of CsrD on transcriptional regulators could involve regulation of enzymes of the c-di-GMP signaling pathway, as shown for CsgD26. Another possibility is that CsrD regulates the turnover of sRNAs other than CsrB and CsrC17, which in turn control transcriptional regulator expression at the post-transcriptional level. Finally, we cannot exclude a direct transcriptional effect of CsrD on targeted genes through a still unknown mechanism.
A strong negative correlation between mRNA half-life and its amount has been observed here for the whole genome, indicating that as a general trend mRNA stabilization can be due to a decrease in mRNA amount and vice versa. The regulation of mRNA stability by CsrA was complex for the 1217 transcripts with opposite variations of stability and quantity in MG1655(csrA51). For these genes, direct effect of CsrA on stability and the potential indirect effect of CsrA on stability due to change in mRNA level cannot not be discriminated (Fig. 6). Nevertheless for more than 400 mRNAs with stability varying in the same direction as mRNA quantity or not associated with mRNA quantity variation, we predicted that CsrA regulates directly their stability. For the majority of these mRNAs, control may be attributed to a physical interaction between CsrA and the mRNA molecule as indicated by the prediction of a putative binding site and/or high throughput binding analysis. For one third of these mRNAs, there was no experimental proof of CsrA binding23 and we did not identify any consensus CsrA binding site. Either the sequences of the CsrA binding site were highly degenerated compared to the consensus in these mRNAs or CsrA may act on their stability through other post-transcriptional regulators (such as the RNA-binding protein Hfq) whose expression is CsrA-dependent39. About 88% of the half-lives directly regulated by CsrA were shorter in the MG1655(csrA51) strain than in the MG1655 strain. This result is surprising because, to date, most of the known targets of CsrA are repressed and/or destabilized, and at the molecular level, examples of repression are more numerous than positive regulation11,13,44,45,46. Thus, contrary to earlier studies on selected targets, our genome-wide analysis shows that CsrA has a significant role as a positive regulator of mRNA stability. The protective effect of CsrA binding on mRNA by sequestering RNase E cleavage sites, only described until now in the case of flhDC45, might be common.
In terms of gene expression regulation, for one hundred mRNAs, the direct regulation of mRNA stability by CsrA led to a significant variation in mRNA concentration. We confirmed previously identified metabolic targets of CsrA such as glgB, pgm, tpiA and pck mRNAs36,42. In addition, many new direct mRNA targets of CsrA were discovered with a potential metabolic impact via mRNA level changes. Functional analysis of these transcripts underlined that the Csr system is strongly involved in the control of E. coli metabolism by regulating expression of enzymes involved in the central carbon metabolism and carbohydrate transport but also in other specific metabolic pathways (syntheses of amino acids and fatty acids). In the MG1655(csrA51) strain, mRNA stability regulation favors gluconeogenesis especially anaplerotic reactions (e.g. malic enzyme, PEP carboxykinase), whereas the transport of glycolytic substrates (such as sorbitol, melibiose, gluconate, allose, maltose and arabinose) is down-regulated. Therefore, compared to MG1655, the MG1655(csrA51) strain is more likely to efficiently metabolize gluconeogenic substrates. Consequently, our results provide the first evidence at the genome-wide scale of the role of CsrA-dependent regulation of transcript stability in metabolic adaptation.
Our study provides new insights into the Csr regulatory network. We determined the extent, the nature and the metabolic effect of the CsrD and CsrA regulations. Our results highlight the superimposition at the genome-wide scale of the transcriptional and post-transcriptional controls that likely contribute to the robustness and dynamics of gene expression reprogramming during bacterial adaptation.
Methods
Bacterial strains and plasmids
E. coli K-12 MG1655 strain (λ− F− rph−1) was the genetic background for mutation of csrA and csrD genes. Mutagenesis was carried out using λ red recombination47 and primer pair OKT-24/OKT-25. As the csrA gene is essential29,30, the ORF was mutated using the csrA mutant obtained by Romeo and coworkers as a model36. The MG1655(csrA51) strain expresses a truncated variant of CsrA in which 11 amino acids at the C-terminal end of the protein have been deleted. The MG1655(ΔcsrD) strain has a total deletion of the csrD gene, which was obtained from the KEIO collection29. The mutations were transferred into E. coli K-12 MG1655 wild type background by P1 phage-mediated transduction before removing the antibiotic-resistance cassette by FLP recombination. MG1655(csrA51) strain was complemented by a low copy number plasmid containing the wild type csrA gene. Briefly, csrA was amplified using primer pair OCT-29/OCT-30 and cloned in pSAB1148 at EcoRI site. To perform gene expression assays, csrC-lacZ, glgB-lacZ, frdA-lacZ and ydeH-lacZ transcriptional fusions were constructed. The transcriptional fusions were cloned in pJYB79 (CmR), a plasmid that was constructed by inserting the omegon kan from pHP45-kan into pACYC184 using the restriction site BamHI and subsequently by cloning at PstI site a PCR amplified fragment containing plac-lacZ from E. coli. First, a 5′UTR sequence containing a ribosome binding site and a start codon (GAATTCCCGGGGATCCTAAGTAAGTAAGGAGAAAAAAATGGCTGATCCC) was fused to lacZ 10th codon. Then, the vector was amplified by PCR with primers MBO-200/-199 and assembled with the promoter regions using In-Fusion® HD cloning kit (Clontech). The promoters regions upstream of the transcription starts of csrC (249 nt), glgB (200 nt), frdA (205 nt) and ydeH (421 nt) were amplified on MG1655 genomic DNA with oligonucleotide pairs MBO-137/-138, MBO-203/-204, MBO-207/-208 and MBO-197/-198, respectively. For glgB, frdA and ydeH, the promoter regions that were chosen using information available on the EcoCyc E. coli Database listing previously characterized transcriptional regulators. For csrC, the promoter region is as used in Suzuki et al.17. The strains and plasmids were validated by sequencing. The oligonucleotides are listed in Table S8.
Culture conditions
For RNA extraction and microarray analysis, cells were grown in continuous culture at 37 °C in M9 minimal medium supplemented with glucose as described3. The MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains were cultured at the same growth rate, μ = 0.10 h−1, which corresponds to doubling time of 6.9 h. Each culture was repeated three times to provide independent biological replicates. Biomass was estimated from absorbance at 600 nm (Libra S4, Biochrom): 1 unit of absorbance corresponding to 0.42 g of dry cell weight l−1 for the MG1655 strain, 0.27 g of dry cell weight l−1 for the MG1655(csrA51) strain and 0.44 g of dry cell weight l−1 for the MG1655(ΔcsrD) strain. To determine the maximal growth rate of each strain, the strains were grown in batch culture (identical medium, oxygen concentration, pH and temperature) and the rates were determined in exponential growth phase.
Sampling and RNA extraction
Sampling and RNA extraction were conducted as described3 with additional cell samplings after rifampicin addition at 9, 15, 20 and 30 min.
Northern Blot
For each sample, 10 μg of total RNA was denatured for 5 min at 95 °C in RNA loading buffer (95% [v/v] formamide, 0.1% [w/v] xylene cyanole, 0.1% [w/v] bromphenol blue, 10 mM EDTA), separated on a 7 M urea/6% polyacrylamide gel at 250 V and transferred to Hybond-XL membranes (GE Healthcare) by electroblotting (1 h, 50V, 4 °C) in 1X TBE. After UV crosslinking, the membranes were hybridized overnight in Roti®-Hybri-Quick (Roth) at 65 °C with [32P] body-labeled riboprobes specific of CsrB (primers OCT-50/OCT-51) or CsrC (primers OCT-52/OCT-53) or at 42 °C with a 5S specific [32P] end-labeled oligonucleotide (MBO-59). Hybridization with CsrB or CsrC riboprobe is followed by 15 min washes in 2X, 1X and 0.1X SSC/0.1% SDS solutions at 65 °C. Hybridization with 5S oligonucleotide is followed by 15 min washes in 5X, 1X and 0.1X/0.1% SDS solutions at 42 °C. Hybridization signals were quantified on a PhosphorImager (Typhoon Trio – GE Healthcare) with MultiGauge software (Fujifilm).
Microarray procedures
A double-stranded cDNA synthesis kit (InvitroGen) was used to produce cDNA from 2 μg of total RNA. cDNA (1 μg) was labeled using the one color DNA labeling kit (Nimblegen – Roche) and labeled cDNA (2 μg) was hybridized onto E. coli K-12 gene expression arrays (Nimblegen – Roche) for 17 h at 42 °C according to the manufacturers’ instructions. Arrays were washed and then scanned with a MS200 Microarray Scanner (Nimblegen – Roche). The images were analyzed with DEVA 1.2.1 software. Only raw data were used for further analyses. All array procedures were performed by the GeT-Biopuces platform (http://get.genotoul.fr).
mRNA level and half-life determination
mRNA quantity determination by transcriptomic analysis was conducted as described3. Intensity values were multiplied by the total RNA extraction yield (in μg total RNA per mg of dry cell weight) to provide mRNA amount value in arbitrary units per mg of dry cell weight. RNA extraction yields were 13.1 ± 2.2, 8.7 ± 0.9 and 8.9 ± 1.5 μg RNA per mg of dry cell weight for the MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains, respectively. Differences in mRNA level were evaluated as described in3. The P-values were adjusted for multiple testing by the ‘Benjamini and Hochberg’ (BH) false discovery rate method49. Differences in mRNA quantity were considered as significant for adjusted P-values lower than 1% and log2 fold change (Log2 FC) higher than 0.5 or lower than −0.5. mRNA half-life determination was performed as previously described3. The statistical significance of differences in half-life was evaluated using the probability value of interaction between time and the type of strain (MG1655, MG1655(csrA51) or MG1655(ΔcsrD)) in a global model of linear regression. A statistical threshold of 10% was used for adjusted P-values by the “BH” false discovery rate method49.
Determination of mRNA level regulatory coefficients
For the selected mRNAs, the coefficient ρD, corresponding to the relative contribution of the mRNA stability regulation in the control of mRNA level was calculated as previously described3 using data of mRNA stability and quantity obtained in the MG1655(csrA51) and MG1655 strains. Regulatory classes were defined according to the ρD value: 0 < ρD < 0.6, mRNA quantity mainly controlled by transcription or controlled by both transcription and mRNA stability; 0.6 < ρD, mRNA quantity mainly controlled by mRNA stability.
Enrichment methods
R free statistical software (www.r-project.org) was used for enrichment methods. Functional categories enriched in transcript subgroups were determined by a hypergeometric test on data using the Biological Process of Gene Ontology annotation database (GO project; http://www.geneontology.org/). Only GO terms with associated P-value ≤ 0.05 were considered as significant.
In silico research of CsrA binding sites
To determine if an mRNA contained a potential CsrA binding site, the software PatScan was used50. The SELEX-derived consensus RUACARGGAUGU was searched in a −100/+100 nt window (+1 corresponding to the start codon) allowing a maximum of 5 mismatches. Subsequently, only sequences that included a conserved GGA motif were considered as a potential CsrA binding site.
Analytical methods
Glucose and acetate concentrations were measured by HPLC coupled to a refractometer and with UV detection. The device was equipped with a Bio-Rad HPX87H column maintained at a temperature of 48 °C and 5 mM H2SO4 was used as the eluent, at a flow rate of 0.5 ml min−1. Glycogen staining by iodine vapor, biofilm formation by cristal violet staining and motility assays on 0.3% agar plate were carried out as previously described51. The intracellular glycogen quantification was done as previously described52. Briefly, the cells were lysed to extract the glycogen which was then hydrolyzed by amyloglucosidase into glucose subunits. The glucose subunits were then quantified using glucose oxidase coupled to the colorimetric reagent o-dianisidine dihydrochloride. Three independent experiments were performed for each assay.
The csrC-lacZ, glgB-lacZ, frdA-lacZ and ydeH-lacZ gene expression assays were performed using cells grown in batch culture at 37 °C in M9 minimal medium supplemented with fructose (2.7 g.L−1), casamino acids (0.2%) and chloramphenicol (25 μg.ml−1). Under these conditions, the MG1655, MG1655(csrA51) and MG1655(ΔcsrD) strains have similar growth rates (0.39 h−1, 0.39 h−1 and 0.31 h−1 respectively). These conditions aimed at mimicking growth in continuous cultures. In exponential growth, the cells were centrifuged, suspended in Tris (6 mM)/Tricarballylate (1.8 mM) buffer pH 7.2 and lysed mechanically with glass beads. β-galactosidase activity was assayed using 2.5 mM orthonitrophenyl-β-galactoside53. Total protein was measured by the Bradford method with bovine serum albumin as the protein standard54. Three independent experiments were performed for each assay.
Additional Information
How to cite this article: Esquerré, T. et al. The Csr system regulates genome-wide mRNA stability and transcription and thus gene expression in Escherichia coli. Sci. Rep. 6, 25057; doi: 10.1038/srep25057 (2016).
Supplementary Material
Acknowledgments
This work was funded by the Université de Toulouse, the Région Midi-Pyrénées and the Agence Nationale de la Recherche (ANR-13-BSV6-0005-1). The authors thank S. Mondiel and I. Canal for technical help, C. Gaspin for motif search with PatScan and J.-Y. Bouet for the pJYB79 plasmid.
Footnotes
Author Contributions T.E., C.T. and M.B. experimental work and data acquisition; T.E., M.B., A.J.C., L.G. and M.C.B. designed the research, analyzed the data and wrote the paper.
References
- Dahan O., Gingold H. & Pilpel Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends in genetics: TIG 27, 316–322, doi: 10.1016/j.tig.2011.05.008 (2011). [DOI] [PubMed] [Google Scholar]
- Schwanhausser B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342, doi: 10.1038/nature10098 (2011). [DOI] [PubMed] [Google Scholar]
- Esquerré T. et al. Dual role of transcription and transcript stability in the regulation of gene expression in Escherichia coli cells cultured on glucose at different growth rates. Nucleic acids research 42, 2460–2472, doi: 10.1093/nar/gkt1150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad T. R. et al. Global analysis of mRNA stability in Mycobacterium tuberculosis. Nucleic acids research 41, 509–517, doi: 10.1093/nar/gks1019 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Antonio A. Escherichia coli transcriptional regulatory network. Network Biology 1, 21–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoumieux S. et al. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Molecular systems biology 9, 634, doi: 10.1038/msb.2012.70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. & Luisi B. F. Hfq and its constellation of RNA. Nature reviews. Microbiology 9, 578–589, doi: 10.1038/nrmicro2615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Salvail H., Desnoyers G. & Arguin M. Small RNAs controlling iron metabolism. Current opinion in microbiology 10, 140–145, doi: 10.1016/j.mib.2007.03.013 (2007). [DOI] [PubMed] [Google Scholar]
- Papenfort K., Sun Y., Miyakoshi M., Vanderpool C. K. & Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153, 426–437, doi: 10.1016/j.cell.2013.03.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans J. & Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cellular and molecular life sciences : CMLS 67, 2897–2908, doi: 10.1007/s00018-010-0381-z (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T., Vakulskas C. A. & Babitzke P. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environmental microbiology 15, 313–324, doi: 10.1111/j.1462-2920.2012.02794.x (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Y., Yang H. & Romeo T. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. Journal of bacteriology 177, 2663–2672 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. S., Morozov I., Suzuki K., Romeo T. & Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Molecular microbiology 44, 1599–1610 (2002). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Molecular microbiology 56, 1648–1663, doi: 10.1111/j.1365-2958.2005.04648.x (2005). [DOI] [PubMed] [Google Scholar]
- Jonas K. et al. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Molecular microbiology 70, 236–257, doi: 10.1111/j.1365-2958.2008.06411.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B. L. et al. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Molecular microbiology 40, 245–256 (2001). [DOI] [PubMed] [Google Scholar]
- Suzuki K., Babitzke P., Kushner S. R. & Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes & development 20, 2605–2617, doi: 10.1101/gad.1461606 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho M. I. et al. Effects of the Global Regulator CsrA on the BarA/UvrY Two-Component Signaling System. Journal of bacteriology 197, 983–991, doi: 10.1128/JB.02325-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Y. et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. The Journal of biological chemistry 272, 17502–17510 (1997). [DOI] [PubMed] [Google Scholar]
- Weilbacher T. et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Molecular microbiology 48, 657–670 (2003). [DOI] [PubMed] [Google Scholar]
- Babitzke P. & Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Current opinion in microbiology 10, 156–163, doi: 10.1016/j.mib.2007.03.007 (2007). [DOI] [PubMed] [Google Scholar]
- Jorgensen M. G., Thomason M. K., Havelund J., Valentin-Hansen P. & Storz G. Dual function of the McaS small RNA in controlling biofilm formation. Genes & development 27, 1132–1145, doi: 10.1101/gad.214734.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. N. et al. Circuitry linking the Csr and stringent response global regulatory systems. Molecular microbiology 80, 1561–1580, doi: 10.1111/j.1365-2958.2011.07663.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. et al. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. Journal of bacteriology 184, 5130–5140 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K. & Melefors O. The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS microbiology letters 297, 80–86, doi: 10.1111/j.1574-6968.2009.01661.x (2009). [DOI] [PubMed] [Google Scholar]
- Sommerfeldt N. et al. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155, 1318–1331, doi: 10.1099/mic.0.024257-0 (2009). [DOI] [PubMed] [Google Scholar]
- Dubey A. K., Baker C. S., Romeo T. & Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. Rna 11, 1579–1587, doi: 10.1261/rna.2990205 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni P. R. et al. A sequence-based approach for prediction of CsrA/RsmA targets in bacteria with experimental validation in Pseudomonas aeruginosa. Nucleic acids research 42, 6811–6825, doi: 10.1093/nar/gku309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology 2, 2006 0008, doi: 10.1038/msb4100050 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans J. & Van Melderen L. Conditional essentiality of the csrA gene in Escherichia coli. Journal of bacteriology 191, 1722–1724, doi: 10.1128/JB.01573-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu M. Y. & Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. Journal of bacteriology 178, 1012–1017 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Shin S., LaPorte D., Wolfe A. J. & Romeo T. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. Journal of bacteriology 182, 1632–1640 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. W. et al. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. Journal of bacteriology 184, 290–301 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannuri A. et al. Translational repression of NhaR, a novel pathway for multi-tier regulation of biofilm circuitry by CsrA. Journal of bacteriology 194, 79–89, doi: 10.1128/JB.06209-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Y. & Romeo T. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. Journal of bacteriology 179, 4639–4642 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T., Gong M., Liu M. Y. & Brun-Zinkernagel A. M. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. Journal of bacteriology 175, 4744–4755 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y. et al. Regulation of CsrB/C sRNA decay by EIIA of the phosphoenolpyruvate: carbohydrate phosphotransferase system. Molecular microbiology, doi: 10.1111/mmi.13259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon S. D. et al. Global regulation by CsrA in Salmonella typhimurium. Molecular microbiology 48, 1633–1645 (2003). [DOI] [PubMed] [Google Scholar]
- Baker C. S. et al. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. Journal of bacteriology 189, 5472–5481, doi: 10.1128/JB.00529-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey A. K. et al. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. Journal of bacteriology 185, 4450–4460 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquerre T. et al. Genome-wide investigation of mRNA lifetime determinants in Escherichia coli cells cultured at different growth rates. BMC Genomics 16, 275, doi: 10.1186/s12864-015-1482-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis N. A., Yang H. & Romeo T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. The Journal of biological chemistry 270, 29096–29104 (1995). [DOI] [PubMed] [Google Scholar]
- Adamson D. N. & Lim H. N. Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation. Proceedings of the National Academy of Sciences of the United States of America 110, 13120–13125, doi: 10.1073/pnas.1308476110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H. et al. CsrA represses translation of sdiA, which encodes the N-acylhomoserine-L-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. Journal of bacteriology 193, 6162–6170, doi: 10.1128/JB.05975-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin A. V. et al. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Molecular microbiology 87, 851–866, doi: 10.1111/mmi.12136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson-Fortin L. M., Vakulskas C. A., Yakhnin H., Babitzke P. & Romeo T. Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. Journal of molecular biology 425, 3662–3677, doi: 10.1016/j.jmb.2012.12.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A. & Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America 97, 6640–6645, doi: 10.1073/pnas.120163297 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Bara S. & Carpousis A. J. Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the gammaproteobacteria. Journal of bacteriology 192, 5413–5423, doi: 10.1128/JB.00592-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300, doi: 10.1128/JB.187.24.8237-8246.2005 (1995). [DOI] [Google Scholar]
- Dsouza M., Larsen N. & Overbeek R. Searching for patterns in genomic data. Trends in genetics: TIG 13, 497–498 (1997). [DOI] [PubMed] [Google Scholar]
- Revelles O. et al. The carbon storage regulator (Csr) system exerts a nutrient-specific control over central metabolism in Escherichia coli strain Nissle 1917. Plos one 8, e66386, doi: 10.1371/journal.pone.0066386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrou J. L. & Francois J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Analytical biochemistry 248, 186–188, doi: 10.1006/abio.1997.2138 (1997). [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Biology. (Cold Spring Harbor Laboratory, 1972). [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Blattner F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.