Abstract
Bufavirus is a recently recognized member of the genus Protoparvovirus in the subfamily Parvovirinae. It has been reported that human bufavirus was detected predominantly in patients with diarrhoea in several countries. However, little is known about bufavirus or its close relatives in nonhuman mammals. In this study, we performed nested-PCR screening and identified bufavirus from 12 megabats of Pteropus spp. in Indonesia. Furthermore, we determined nearly the full genome sequence of a novel megabat-borne bufavirus, tentatively named megabat bufavirus 1. Phylogenetic analyses showed that megabat bufavirus 1 clustered with known protoparvoviruses, including human bufavirus but represented a distinct lineage of bufavirus. Our analyses also inferred phylogenetic relationships among animal-borne bufaviruses recently reported by other studies. Recombination analyses suggested that the most common recent ancestor of megabat bufavirus 1 might have arisen from multiple genetic recombination events. These results characterized megabat bufavirus 1 as the first protoparvovirus discovered from megabats and indicates the high genetic divergence of bufavirus.
Bufavirus (BuV) is a recently recognized parvovirus and was initially identified in the faeces of a child with diarrhoea in Burkina Faso in 20121. Then, multiple studies described BuV in the faeces of humans in Tunisia, Finland, the Netherlands, Bhutan, Thailand, China and Turkey1,2,3,4,5,6,7, indicating its worldwide distribution. BuV was detected predominantly in human patients with diarrhoea or gastroenteritis, while the pathogenicity of BuV remains to be elucidated because of the failure to recover infectious BuV from samples. All human-borne BuVs are phylogenetically distinct from other known members of the subfamily Parvovirinae and now belong to a new species, primate protoparvovirus 1 of the genus Protoparvovirus in the subfamily Parvovirinae8.
Previous viral metagenomic studies identified close relatives of human-borne BuV in animals. In an earlier study, we described distinct lineages of BuV in wild shrews and baboons in Zambia9. Other studies also reported different BuVs or related parvoviruses in laboratory rhesus monkeys in the USA10, wild rats in China11, domestic pigs in Hungary12 and in wild microbats in Hungary and China13,14. Although these previous studies have demonstrated the presence of animal-borne BuVs, to date there has been no phylogenetic analysis that has covered all published bufaviruses and related protoparvoviruses. Therefore, the phylogenetic relationships among these viruses are still poorly understood.
Bats are traditionally divided into two suborders: Megachiroptera (megabats) and Microchiroptera (microbats). Megabats and microbats mainly consist of frugivorous bats and insectivorous bats, respectively15. In addition to their biological differences, they are phylogenetically distinguishable16,17. Megabats are a potential reservoir of emerging zoonotic viruses, including Nipah virus, Hendra virus, SARS coronavirus and Ebola virus18,19. In addition, a metagenomic study showed that megabats harbour various mammalian viruses, some of which are genetically related to human viruses20. However, BuVs or related parvoviruses were detected in microbats but not in megabats. In the present study, we investigated BuVs in flying foxes (Pteropus spp.), which belong to the megabat family. Based on the obtained sequence data, we examined the phylogenetic relationships between the identified-BuVs and previously identified BuVs. One of the BuVs identified in this study was further characterized and we obtained a nearly full genome sequence.
Results
A total of 183 megabats consisting of four different species were included in this study (Table 1). Among these, Pteropus sp. and Dobsonia sp. were not identified at species level, but other genetically close species Pteropus hypomelanus and Dobsonia moluccensis, were included21. In an earlier study, BuV was detected from the spleen tissues and faeces of shrews9. Therefore, we used nested-PCR targeting for the NS1 gene of BuV using 183 spleens and 96 faeces samples (faecal samples were unavailable for a subset of megabats). As shown in Table 1, BuV was identified in three spleen and nine faeces specimens from Pteropus vampyrus, and one spleen specimen from Pteropus sp. Among these, one Pteropus vampyrus in Magelang was positive for BuV in both the spleen and faeces. With a limited number of samples, the detection rate of BuV in spleen (2%) was lower than that in faeces (9%).
Table 1. Sample information and nested-PCR screening results for bufaviruses.
| Bat species | Location collected | Year collected | No. of samples PCR-positive/No. of samples tested |
|
|---|---|---|---|---|
| Spleen | Faeces | |||
| Pteropus vampyrus | Panjalu district | 2010 | 0/26 | NT*** |
| Lima Puluh Kota district | 2011 | 2/20 | NT | |
| Surabaya district | 2012 | 0/3 | 2/3 | |
| Magelang district | 2012 | 1/20 | 7/19 | |
| Pteropus sp.* | Popayato district | 2011 | 0/4 | NT |
| Paguyaman district | 2011 | 1/23 | NT | |
| Paguyaman district | 2012 | 0/2 | 0/2 | |
| Paguyaman district | 2013 | 0/10 | 0/8 | |
| Soppeng district | 2014 | 0/7 | 0/7 | |
| Sidrap district | 2014 | 0/15 | 0/15 | |
| Acerodon celebensis | Paguyaman district | 2012 | 0/18 | 0/18 |
| Paguyaman district | 2013 | 0/18 | 0/7 | |
| Dobsonia sp.** | Paguyaman district | 2012 | 0/17 | 0/17 |
| Total | 4/183 | 9/96 | ||
*Genetically closely related to Pteropus hypomelanus. **Genetically closely related to Dobsonia moluccensis. ***Not tested because of unavailability of faeces.
Based on the nucleotide sequence identity, we tentatively grouped and named the identified BuVs into three strains: megabat bufavirus 1 (MgBuV1) originating from Pteropus vampyrus in Surabaya and Magelang (accession numbers LC085668 to LC085676), megabat bufavirus 2 (MgBuV2) from Pteropus vampyrus in Lima Puluh Kota (LC085665 and LC085666), and megabat bufavirus 3 (MgBuV3) from Pteropus sp. in Paguyaman (LC085667). The partial NS1 gene fragment of MgBuV1 showed 81–83% and 91–92% nucleotide identity to MgBuV2 and MgBuV3, respectively. The corresponding region of MgBuV2 was 80% identical to that of MgBuV3.
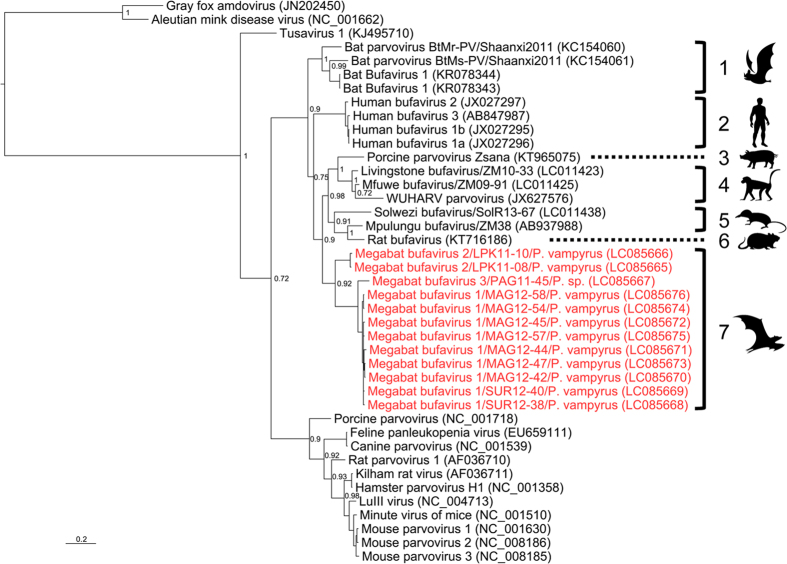
Currently, DDBJ/EMBL/GenBank databases contain complete or partial nucleotide sequences of BuV from humans, nonhuman primates, shrews, rats, pigs and microbats. A phylogenetic tree was generated on the basis of the identified sequences of NS1 gene in this study and a corresponding region of the available reference sequences of protoparvoviruses including BuVs (Fig. 1). Known BuVs formed subgroups by host were distinguishable from other protoparvoviruses (groups 1–6, Fig. 1). MgBuV1, MgBuV2 and MgBuV3 diverged from a common ancestor of known protoparvoviruses and were likely to form a distinct subgroup from other BuVs, including microbat-borne BuVs (group 7, Fig. 1).
Figure 1. Phylogenetic analysis based on the amplicon sequences and reference sequences of the NS1 gene from protoparvoviruses.
The analysed region corresponds to nucleotide positions 946 to 1379 in the human bufavirus 1a genome (GenBank accession number JX027296). The sequences identified in this study are coloured in red. The respective GenBank accession numbers of the viral sequences are shown in parentheses. Bayesian posterior probabilities are indicated at major nodes. The scale bar represents a distance of 0.2 substitutions per site. Images were provided by Ms. E. Hayashi with permission.
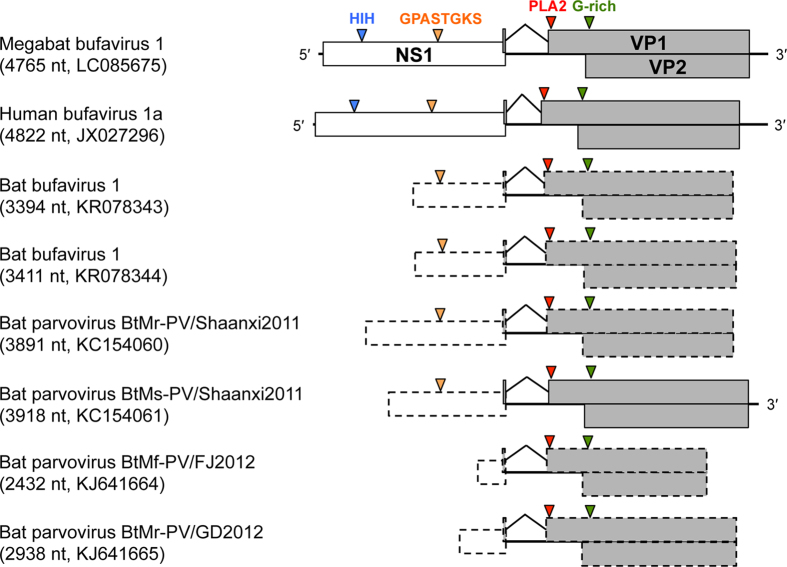
To reveal the whole genome sequence and genetically characterise further the identified BuVs, we subjected DNA samples isolated from spleens and faeces to next-generation sequencing. A small number of sequence reads from MgBuV1, but not MgBuV2 and MgBuV3, enabled to amplify the overlapping large fragments by PCR and determine nearly the whole genome sequence, including the complete protein-coding region. The obtained genome sequence of MgBuV1 (accession number LC085675) was 4765 nt in size and encoded three complete ORFs: the non-structural protein (NS1) and the viral capsid proteins (VP1 and VP2). These proteins showed 46–61% amino acid sequence identities to those of other BuVs of which complete genome sequences were available (Table 2). Parvovirus-conserved amino acid motifs were found in each viral protein of MgBuV1 (Fig. 2). NS1 contained a putative endonuclease metal coordination motif “HIH” and the helicase motif Walker A “GPASTGKS”. The VP1 of most parvovirus species possesses a phospholipase A2 (PLA2) motif that is required for viral entry22. Three potential sites responsible for the enzymatic activity of PLA2 were strictly conserved in the VP1 of MgBuV1: a calcium-binding loop site “YLGPG” and two catalytic sites “HDLEY” and “D”. The N-terminus of the VP1 and VP2 of MgBuV1 had a glycine-rich motif (G-rich) required for the cellular entry of parvovirus23.
Table 2. ORF length and pairwise amino acid identities in NS1, VP1 and VP2 between Megabat bufavirus 1 and the indicated bufaviruses.
| Accession no. | Genome size(nt) | NS1 |
VP1 |
VP2 |
||||
|---|---|---|---|---|---|---|---|---|
| Length(aa) | Identity(%) | Length(aa) | Identity(%) | Length(aa) | Identity(%) | |||
| Megabat bufavirus 1 | LC085675 | 4765 | 642 | 100 | 719 | 100 | 578 | 100 |
| Human bufavirus 1 | JQ918261 | 4912 | 671 | 46.0 | 707 | 59.3 | 569 | 57.5 |
| Human bufavirus 2 | JX027297 | 4562 | 673 | 52.6 | 707 | 61.4 | 569 | 60.0 |
| Human bufavirus 3 | AB847989 | 4766 | 673 | 53.5 | 710 | 61.4 | 572 | 59.7 |
| WUHARV parvovirus | JX627576 | 4909 | 665 | 59.8 | 737 | 60.8 | 569 | 60.4 |
| Mpulungu bufavirus | AB937988 | 4613 | 639 | 54.7 | 728 | 48.4 | 587 | 48.5 |
| Rat bufavirus | KT716186 | 4634 | 644 | 54.5 | 726 | 48.7 | 585 | 48.4 |
nt, nucleotide; aa, amino acid; ORF, Open Reading Frame.
Figure 2. Genome organization of megabat bufavirus 1 and microbat-borne bufaviruses reported previously.
The length of the determined nucleotide sequences and GenBank accession numbers of the viral sequences are shown in parentheses. Solid-lined or dashed-lined boxes indicate complete or incomplete sequenced ORFs, respectively. Blue, orange, red and green arrowheads on NS1 and VP1 indicate the position of conserved amino acid motifs of parvoviruses.
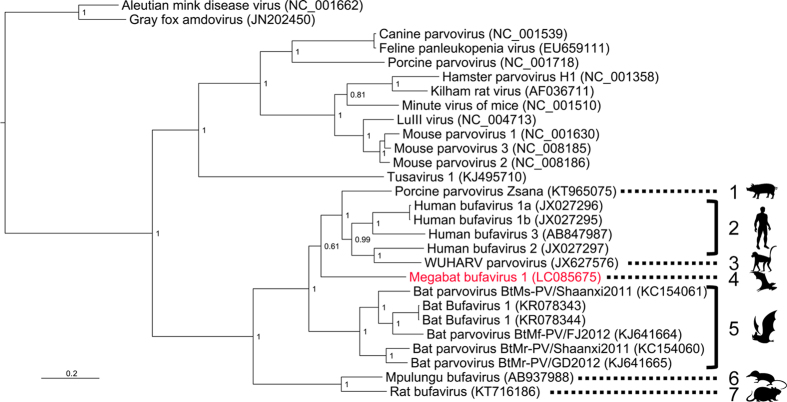
Recently, six partial genome sequences of human BuV-like parvovirus were identified from insectivorous microbats in Hungary and China13,14. These sequences covered a partial coding region of NS1 and almost complete to complete coding regions of VP1 and VP2 (Fig. 2). Therefore, using the coding region of VP2, we performed a phylogenetic analysis to infer the deeper phylogenetic relationships between these microbat-borne BuVs and megabat-borne MgBuV1 (Fig. 3). All microbat-borne BuVs formed a single cluster (group 5, Fig. 3). MgBuV1 branched from the common ancestor of human BuVs, non human primate-borne BuV (WUHARV parvovirus) and pig-borne BuV, and represented a distinct lineage of BuV (linage 4, Fig. 3). WUHARV parvovirus (linage 3, Fig. 3) fell into the cluster of human BuVs (group 2, Fig. 3), consistent with a previous study of past recombination events behind the origin of WUHARV parvovirus13.
Figure 3. Phylogenetic analysis based on the VP2 gene.
The sequence identified in this study is coloured in red. The respective GenBank accession numbers of the viral sequences are shown in parentheses. Bayesian posterior probabilities are indicated at each node. The scale bar represents a distance of 0.2 substitutions per site. Images were provided by Ms. E. Hayashi with permission.
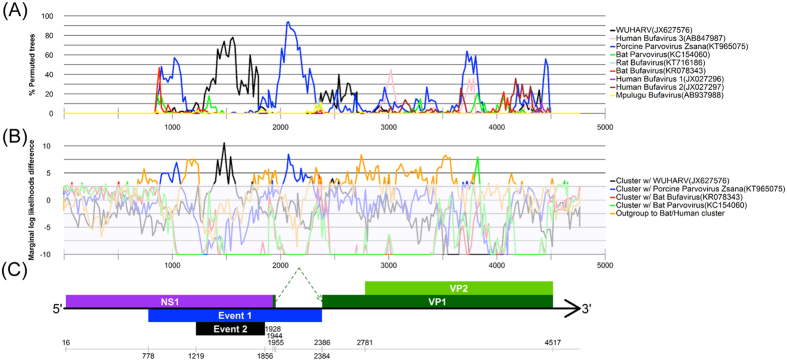
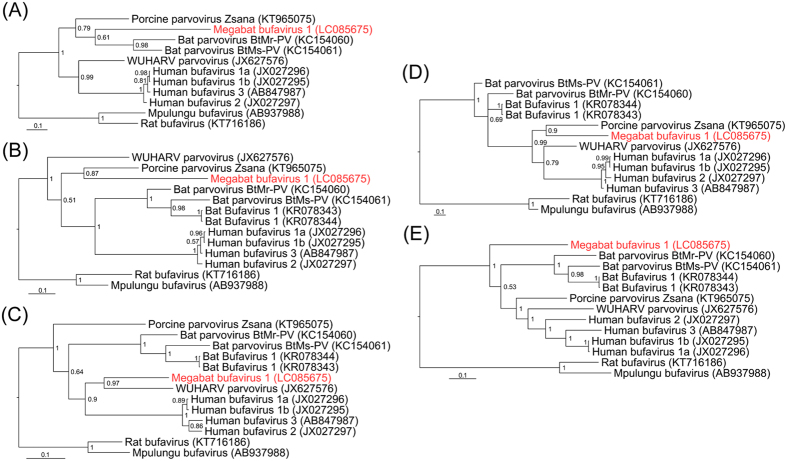
We searched for putative recombination events on the genome of MgBuV1 using two different approaches based on sliding window analyses, one using the Bootscan approach and the second using Bayesian inference (Fig. 4A,B). Both analyses suggested a mosaic genome structure in MgBuV1 and possible recombinations among ancestors of MgBuV1, WUHARV parvovirus and porcine parvovirus Zsana, around nucleotide positions 778 to 2384 of the MgBuV1 genome (Fig. 4C). These observations were supported by independent phylogenetic trees built from five regions bordered by the inferred recombination breakpoints (Fig. 5A–E). Among the BuVs and related protoparvoviruses, MgBuV1 branched from the ancestor of porcine parvovirus Zsana in the phylogenetic tree at position 778 to 1219 (Fig. 5B) and position 1856 to 2384 (Fig. 5D). However, MgBuV1 branched from its ancestor WUHARV parvovirus at position 1219 to 1856 in the phylogenetic tree (Fig. 5C). In contrast, MgBuV1 formed a distinct lineage in phylogenetic trees constructed from position 2384 to 4765 (Fig. 5E). Based on this evidence, we inferred the parsimonious origin of the mosaic genome in MgBuV1 from both recombination analyses (Fig. 4C).
Figure 4. Recombination analyses of megabat bufavirus 1.
Abscissae represent positions adjusted to reflect the position in the megabat bufavirus 1 genome. (A) Bootscan results supporting the clustering of megabat bufavirus 1 with other strains, which are color-coded as shown to the right. The ordinate shows the percentage of permuted trees supporting the clustering in each window. (B) Bayesian marginal likelihood support for the topological position of megabat bufavirus 1. The ordinate shows the log difference for the marginal likelihood of the model assuming the topological hypothesis and the model assuming the opposite. Lines below the threshold support (<3) are shown as a lighter colour. (C) Genome representation showing the supported recombination events and the position relative to the protein encoding regions. Event 1 and Event 2 show the supported recombination events between MgBuV1 and porcine parvovirus Zsana or between MgBuV1 and WUHARV parvovirus, respectively. Nucleotide positions 778, 1219, 1856 and 2384 show putative recombination breakpoints.
Figure 5. Separate phylogenetic analyses of five regions bordered by putative recombination breakpoints.
Phylogenetic trees were inferred from nucleotide positions 1–778 (A) 778–1219 (B) 1219–1856 (C) 1856–2384 (D) 2384–4765 (E) on the megabat bufavirus 1 genome. The respective GenBank accession numbers of the viral sequences are shown in parentheses. Bayesian posterior probabilities are indicated at each node. The scale bars represent a distance of 0.2 substitutions per site.
Discussion
Since the discovery of human BuV in 2012, its genetic divergence and worldwide distribution have been updated by a series of reports1,2,3,4,5,6,7. Although recent studies described BuVs in nonhuman mammals, little is known about the epidemiology of animal-borne BuVs.9,10,11,12,13,14. In this study, we report for the first time, a BuV identified in megabats. MgBuV1 is the fourth animal borne-BuV where the whole genome sequence of the protein-coding region has been determined. In addition, the detection of partial NS1 gene sequences different from MgBuV1 (called MgBuV2 and MgBuV3) indicates the presence of MgBuV1 variants. The discovery of megabat-borne BuVs facilitates a better understanding of the diversity and host-range of BuV.
MgBuV1 has a similar genome organization and is phylogenetically related to other mammalian-borne BuVs. According to the species demarcation criteria of the International Committee on Taxonomy of Viruses, parvoviruses in the same species should share >85% amino acid sequence identity in their whole NS18. As shown in Table 2, the NS1 of MgBuV1 showed <60% identity to those of other related parvoviruses. Taken together, these results suggest that MgBuV1 is a novel species of protoparvovirus harboured by megabats.
Recently, complete or partial genome sequences of BuVs related to human BuV were identified from nonhuman primates, shrews, rats, pigs and microbats9,10,11,12,13,14. However, the phylogenetic relationships among these viruses remains to be elucidated. Our phylogenetic analyses covered the available sequences of these BuVs and showed a large degree of genetic divergence of BuV in the genus Protoparvovirus. Both phylogenetic analyses based on NS1 and VP2 genes showed that a group of BuVs comprised host-specific lineages and megabat-borne BuVs were clearly distinguished from microbat-borne BuVs. While the phylogeny of BuV is likely to be associated with their host, further studies of BuVs in animals are needed to infer the virus-host coevolution.
Genome recombination is a compelling argument to explain the evolution of a variety of viruses24,25; furthermore, evidence for recombination events have been also observed in the genome of some protoparvoviruses13,26,27,28. In this study, we revealed putative recombined regions and phylogenetic incongruences across the MgBuV1 genome, suggesting genetic recombination between MgBuV1 and porcine parvovirus Zsana (event 1, Fig. 4C) and other recombination events between MgBuV1 and WUHARV parvovirus (event 2, Fig. 4C). These results imply that the most common recent ancestor of MgBuV1 might have arisen from multiple recombination events. Such events also imply the occurrence of dual infection with ancestors of MgBuV1, WUHARV parvovirus and porcine parvovirus Zsana in nature.
In conclusion, we demonstrated the presence of megabat-borne BuV, the first protoparvovirus discovered from megabats. Further genetic characterization revealed the phylogenetic distinctiveness of MgBuV1 and suggested a molecular evolution of MgBuV1 driven by multiple recombination events. These results provide new insights into the high genetic divergence of BuV and will help us to investigate further BuV in animals.
Methods
Ethics statement and sample collection
All experiments involving animals were performed in accordance with the ethical guidelines of the Animal Care and Use Committee of Veterinary Teaching Hospital, Bogor Agricultural University. The protocol was approved by the Animal Care and Use Committee of Veterinary Teaching Hospital, Bogor Agricultural University (permit number 05-2010 RSHP-IPB). Collection and exportation of samples from megabats were performed with the permission of the Directorate General of Livestock and Animal Health Services, Ministry of Agriculture, Republic of Indonesia. Faecal and spleen samples were collected from megabats at eight regions in Indonesia from 2010 to 2014. Some samples were used for different research projects, as reported previously21,29,30,31. Species were identified based on the morphological features and nucleotide sequence analysis of both mitochondrial 16S rRNA and cytochrome b as described previously21. Sample information is summarized in Table 1.
Nested-PCR screening
Faeces suspended in RNAlater reagent (Ambion, Life Technologies, Carlsbad, CA) were subjected to DNA extraction using a High Pure Viral Nucleic Acid kit (Roche Diagnostics GmbH, Mannheim, Germany) or QIAamp Viral RNA Mini Kit (Qiagen, Valencia CA). DNA from spleen tissues was prepared using DNAzol reagent (Molecular Research Center, Cincinnati, OH) or QIAamp DNA Mini kit (Qiagen). For nested PCR screen targeting for NS1 gene of BuV, we followed the method of Sasaki et al.9.
Complete genome sequencing
To determine the complete genome sequence of megabat BuV, a DNA library was prepared from the faecal-DNA samples using the Nextera XT DNA sample preparation kit (Illmina, San Diego, CA) and were sequenced on an illumina MiSeq instrument using the MiSeq Reagent Kit v3 600-cycle (Illumina) according to the manufacturer’s instructions. The sequence reads homologous to known BuVs were identified by read mapping using CLC Genomics Workbench (CLC bio, Aarhus, Denmark). The overlapping large fragments were amplified by conventional PCR with specific primer sets designed from each identified sequence read. The 3′ end of virus genome was amplified by the modified rapid amplification of cDNA ends as reported previously32. All the amplified fragments were assembled manually into a contiguous sequence.
Phylogenetic analyses
Sequences were aligned with MAFFT33 using the FFT-NS-I method. Phylogenetic inferences were performed on multiple sequence alignments with MrBayes 3.234 using a General Time Reversible model with a rate variation between sites modelled with a gamma distribution (+Γ) with six categories and allowing for a percentage of invariable sites (+I).
Recombination analyses
A recombination analysis was performed in Simplot 3.5.135 using the Bootscan algorithm with 200 nt window and 20 nt step under a Kimura (2-parameter) model36 and a Neighbor-Joining approach with 1000 repetitions.
Additionally, this study implemented a second analysis approach over the same sliding window that incorporated the power of Bayesian inference to support or reject hypotheses regarding the topology of the phylogenetic tree. For each window, a hypothesis of the clustering of two particular sequences was tested against the null hypothesis that the cluster was less likely than those sequences independently clustering with any other under the same model parameters. To maximize the inferential power of the approach, the marginal likelihoods of the models assuming opposite topological hypotheses were compared with the stepping stone37 sampling approach implemented in MrBayes 3.2. Then, the difference of the calculated marginal likelihoods for both models, i.e., the model assuming a clustering and the model without the clustering, represented the Bayes factor, which usually is interpreted as supporting the first model if the difference exceeds three and strongly supportive when it exceeds five38. All hypotheses were tested under the same model assumed for the phylogenetic analyses.
The dataset in both analyses was composed of sequences in the BuV cluster. Adopting a parsimonious approach, this study accepted only those events suggested by Bootscan that also had support from the Bayesian hypotheses testing.
Nucleotide sequences
All the nucleotide sequences determined in this study were deposited in the DDBJ/EMBL/GenBank databases under accession numbers LC085665−LC085676.
Additional Information
How to cite this article: Sasaki, M. et al. Divergent bufavirus harboured in megabats represents a new lineage of parvoviruses. Sci. Rep. 6, 24257; doi: 10.1038/srep24257 (2016).
Acknowledgments
We thank Ms. E. Hayashi for drawing the images used in the Figures 1 and 3. This study was supported by the Japan Initiative for Global Research Network of Infectious Diseases (J-GRID) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
Author Contributions M.S., T.K. and H.S. conceived the experiments, M.S., G.G., Y.W., K.I. and H.S. conducted the experiments and analysed the data. M.S., A.S., E.H., I.R., S.T., S.A., M.L., Z.A.K., M.S., S.K., I.N., T.K., Y.O. and H.S. contributed reagents/materials/analysis tools. M.S., G.G., A.S., E.H., K.I. and H.S. wrote the paper. All authors reviewed the manuscript.
References
- Phan T. G. et al. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J Virol 86, 11024–11030, 10.1128/JVI.01427-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiro T. et al. Novel human bufavirus genotype 3 in children with severe diarrhea, bhutan. Emerg Infect Dis 20, 1037–1039, 10.3201/eid2006.131430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S. L. et al. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg Infect Dis 20, 1218–1222, 10.3201/eid2007.140190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väisänen E. et al. Bufavirus in feces of patients with gastroenteritis, Finland. Emerg Infect Dis 20, 1078–1080, 10.3201/eid2006.131674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieochansin T., Vutithanachot V., Theamboonlers A. & Poovorawan Y. Bufavirus in fecal specimens of patients with and without diarrhea in Thailand. Arch Virol 160, 1781–1784, 10.1007/s00705-015-2441-z (2015). [DOI] [PubMed] [Google Scholar]
- Altay A. et al. Bufavirus genotype 3 in Turkish children with severe diarrhoea. Clin Microbiol Infect 21, 965.e961–964, 10.1016/j.cmi.2015.06.006 (2015). [DOI] [PubMed] [Google Scholar]
- Huang D. D. et al. Identification of Bufavirus-1 and Bufavirus-3 in Feces of Patients with Acute Diarrhea, China. Sci Rep 5, 13272, 10.1038/srep13272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore S. F. et al. The family Parvoviridae. Arch Virol 159, 1239–1247, 10.1007/s00705-013-1914-1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M. et al. Distinct Lineages of Bufavirus in Wild Shrews and Nonhuman Primates. Emerg Infect Dis 21, 1230–1233, 10.3201/eid2107.141969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley S. A. et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151, 253–266, 10.1016/j.cell.2012.09.024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. et al. Bufavirus Protoparvovirus in feces of wild rats in China. Virus Genes, 10.1007/s11262-015-1262-1 (2015). [DOI] [PubMed] [Google Scholar]
- Hargitai R. et al. Detection and genetic characterization of a novel parvovirus distantly related to human bufavirus in domestic pigs. Arch Virol, 10.1007/s00705-015-2732-4 (2016). [DOI] [PubMed] [Google Scholar]
- Kemenesi G. et al. Genetic diversity and recombination within bufaviruses: Detection of a novel strain in Hungarian bats. Infect Genet Evol 33, 288–292, 10.1016/j.meegid.2015.05.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J, 10.1038/ismej.2015.138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettigrew J. D. et al. Phylogenetic relations between microbats, megabats and primates (Mammalia: Chiroptera and Primates). Philos Trans R Soc Lond B Biol Sci 325, 489–559 (1989). [DOI] [PubMed] [Google Scholar]
- O’Leary M. A. et al. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339, 662–667, 10.1126/science.1229237 (2013). [DOI] [PubMed] [Google Scholar]
- Tsagkogeorga G., Parker J., Stupka E., Cotton J. A. & Rossiter S. J. Phylogenomic analyses elucidate the evolutionary relationships of bats. Curr Biol 23, 2262–2267, 10.1016/j.cub.2013.09.014 (2013). [DOI] [PubMed] [Google Scholar]
- Han H. J. et al. Bats as reservoirs of severe emerging infectious diseases. Virus Res 205, 1–6, 10.1016/j.virusres.2015.05.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. & Wang L. F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol 3, 84–91, 10.1016/j.coviro.2012.11.006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. S. et al. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology 441, 95–106, 10.1016/j.virol.2013.03.014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M. et al. Molecular detection of a novel paramyxovirus in fruit bats from Indonesia. Virol J 9, 240, 10.1186/1743-422X-9-240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr G. A., Zhang L. G. & Tattersall P. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci USA 102, 17148–17153, 10.1073/pnas.0508477102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos M. et al. A slender tract of glycine residues is required for translocation of the VP2 protein N-terminal domain through the parvovirus MVM capsid channel to initiate infection. Biochem J 455, 87–94, 10.1042/BJ20130503 (2013). [DOI] [PubMed] [Google Scholar]
- Simon-Loriere E. & Holmes E. C. Why do RNA viruses recombine? Nat Rev Microbiol 9, 617–626, 10.1038/nrmicro2614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P. et al. Recombination in Eukaryotic Single Stranded DNA Viruses. Viruses-Basel 3, 1699–1738, 10.3390/v3091699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton L. A., Hoelzer K., Parrish C. R. & Holmes E. C. Comparative analysis reveals frequent recombination in the parvoviruses. J Gen Virol 88, 3294–3301, 10.1099/vir.0.83255-0 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T. & Mochizuki M. Evidence for recombination between feline panleukopenia virus and canine parvovirus type 2. J Vet Med Sci 71, 403–408 (2009). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Evidence for natural recombination between mink enteritis virus and canine parvovirus. Virol J 9, 252, 10.1186/1743-422X-9-252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M. et al. Isolation and characterization of a novel alphaherpesvirus in fruit bats. J Virol 88, 9819–9829, 10.1128/JVI.01277-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anindita P. D. et al. Detection of coronavirus genomes in Moluccan naked-backed fruit bats in Indonesia. Arch Virol 160, 1113–1118, 10.1007/s00705-015-2342-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S. et al. Detection of novel polyomaviruses in fruit bats in Indonesia. Arch Virol 160, 1075–1082, 10.1007/s00705-015-2349-7 (2015). [DOI] [PubMed] [Google Scholar]
- Li Z., Yu M., Zhang H., Wang H. Y. & Wang L. F. Improved rapid amplification of cDNA ends (RACE) for mapping both the 5′ and 3′ terminal sequences of paramyxovirus genomes. J Virol Methods 130, 154–156, 10.1016/j.jviromet.2005.06.022 (2005). [DOI] [PubMed] [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780, 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61, 539–542, 10.1093/sysbio/sys029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K. S. et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73, 152–160 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16, 111–120 (1980). [DOI] [PubMed] [Google Scholar]
- Xie W., Lewis P. O., Fan Y., Kuo L. & Chen M. H. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol 60, 150–160, 10.1093/sysbio/syq085 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E. & Raftery A. E. Bayes factors. Journal of the american statistical association 90, 773–795 (1995). [Google Scholar]