Abstract
Background
Wild ginseng, Panax ginseng Meyer, is an endangered species of medicinal plants. In the present study, we analyzed variations within the ribosomal DNA (rDNA) cluster to gain insight into the genetic diversity of the Oriental ginseng, P. ginseng, at artificial plant cultivation.
Methods
The roots of wild P. ginseng plants were sampled from a nonprotected natural population of the Russian Far East. The slides were prepared from leaf tissues using the squash technique for cytogenetic analysis. The 18S rDNA sequences were cloned and sequenced. The distribution of nucleotide diversity, recombination events, and interspecific phylogenies for the total 18S rDNA sequence data set was also examined.
Results
In mesophyll cells, mononucleolar nuclei were estimated to be dominant (75.7%), while the remaining nuclei contained two to four nucleoli. Among the analyzed 18S rDNA clones, 20% were identical to the 18S rDNA sequence of P. ginseng from Japan, and other clones differed in one to six substitutions. The nucleotide polymorphism was more expressed at the positions 440–640 bp, and distributed in variable regions, expansion segments, and conservative elements of core structure. The phylogenetic analysis confirmed conspecificity of ginseng plants cultivated in different regions, with two fixed mutations between P. ginseng and other species.
Conclusion
This study identified the evidences of the intragenomic nucleotide polymorphism in the 18S rDNA sequences of P. ginseng. These data suggest that, in cultivated plants, the observed genome instability may influence the synthesis of biologically active compounds, which are widely used in traditional medicine.
Keywords: concerted evolution, nucleolus, Panax ginseng, plantation, 18S rDNA
1. Introduction
Wild ginseng, Panax ginseng Meyer, is an endangered species of medicinal plants belonging to the relict family Araliaceae, which has been used as a source of biologically active substances for thousands of years. The domestication and cultivation in vitro of P. ginseng aim to make up for natural resources of the valuable medicinal remedy for various diseases and a diet supplement for the elderly. However, under conditions of artificial reproduction, the medical properties of ginseng are believed to have become weaker, and the amount of the main biologically active compound—ginsenosides—decreases [1]. The nature of this phenomenon remains insufficiently studied.
In most eukaryotes, the nuclear ribosomal DNA (rDNA) occurs in tandem arrays at one or several loci. Each repeat unit contains the 18S, 5.8S, and 28S rDNA subunits; the internal transcribed spacers; and the intergenic spacer, which separates transcribed units. The molecular evolution of the ribosomal RNA (rRNA) genes fundamentally differs from the evolution of single-copy genes. The individual rDNA repeats do not evolve independently of each other; they homogenize horizontally. Therefore, only small differences are usually observed between the rDNA sequences within the tandem unit in any species, while they accumulate between species. The ability of all units to change their rDNA sequences in a highly organized manner was described as concerted evolution [2], [3], [4], [5], [6].
The concerted evolution is important to maintain a significant number of the active rDNA units, and homogenization aims to reduce mutational load, and it is supported by selection [7]. However, a lot of studies show that, in many taxa, the homogenization rate may be too low to prevent significant levels of intraspecific polymorphism of the rDNA repeats. This variation is often limited to noncoding regions of the rDNA, such as internal transcribed spacers and intergenic spacer [8], [9], [10], [11], [12]. At the same time, there are an increasing number of studies, which revealed the presence of divergent rDNA paralogs and pseudogenes whose coding regions are free from functional obligation in different taxa [12], [13], [14], [15], [16], [17]. Polyploidy, interspecific hybridization, loci number, and localization of nuclear-organization regions (NORs) on nonhomologous chromosomes, large genome size, and higher mutation rates in comparison with the rate of sequence homogenization are usually discussed as the possible reasons for the incomplete development of concerted-evolution mechanisms in the rDNA repeats [12], [15], [16], [18], [19], [20], [21], [22], [23], [24], [25].
The nucleolus assembles around the clusters of ribosomal gene repeats, and is easily visible in alive or fixed cells by phase-contrast or differential-interference-contrast optics. Currently, the nucleolus is recognized as a multifunctional nuclear subcompartment of the eukaryotic cell, which is involved in many important biological processes, including cellular stress responses and aging, checkpoint control, messenger RNA export and modification, and also protein degradation and sequestration [26], [27], [28], [29], [30], [31], [32]. Additionally, the nucleolus is the organizing center for different chromosome domains, and therefore, may be associated with the genetic and epigenetic regulation of eukaryotic genome [26], [31].
In the present study, we analyzed variations within the rDNA cluster to gain insight into the genetic diversity of Oriental ginseng, P. ginseng, at artificial plant cultivation. This genetic information may be important for our understanding of the mechanisms involved in the loss of ability to actively synthesize ginsenosides by cultivated plants.
2. Materials and methods
2.1. Plant source
The roots of wild P. ginseng Meyer plants were sampled from a nonprotected natural population in Sikhote-Alin mountain range, Russian Far East. The collected roots were transferred to an open experimental nursery, and grown under conditions that were similar to the ginseng natural habitat (Spassky District of the Primorsky Region, southern Russian Far East). For investigations, the leaves of 5-yr-old ginseng plants were collected at the beginning of the vegetation period.
2.2. Cytogenetic analysis
The cytogenetic technique was performed as described previously [33]. The leaf tissues (volume: 0.5–1 mL) were pretreated in 0.2% colchicine solution for 2–3 h at room temperature (∼22°C), fixed in 3:1 ethanol:acetic acid mixture, and stained with aceto-hematoxylin. The slides were prepared using the squash technique. A 50% solution of silver nitrate was used to stain the nucleoli [34]. The number of nucleoli was counted in 500–1,000 cells.
2.3. Isolation of total DNA and polymerase-chain-reaction amplification
The isolation of total DNA was performed as described previously [35]. Amplification reactions were performed in 20 μL volumes containing 100 ng template DNA, 10mM Tris-HCl (pH 8.5), 50mM KCl, 2.5mM MgCl2, 0.01% gelatin (w/v), 0.1mM Triton X-100, 0.25mM each deoxynucleotide triphosphate, and 10pM each primer. We also used a mix (1:8) of Pfu and Taq DNA polymerases (Fermentas, Vilnius, Lithuania) to minimize amplification errors. The polymerase-chain-reaction (PCR) primer sequences and temperature conditions were those as in [36].
2.4. Cloning and sequencing of the 18S rDNA sequences
The PCR products of two independent PCR reactions were separately cloned into the pTZ57R/T plasmid using the InsTAclone PCR product kit (Fermentas). The PCR products were used for cloning procedures without any modification. Clones were generated according to the protocol recommended by the manufacturer and supplied with the cloning kit. The clones were amplified using M13 (−20) (Syntol, Moscow, Russia) primers for colony screening and sequencing. The sequence of each clone was determined by an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) at the Instrumental Centre of Biotechnology and Gene Engineering of the Institute of Biology and Soil Science, Far Eastern Branch of the Russian Academy of Sciences using additional nested primers [37]. The nucleotide-sequence data of 18S rRNA gene were recorded in GenBank nucleotide-sequence databases with the following accession numbers: KC593794–KC593823.
2.5. Statistical analysis and phylogenetic reconstructions
The Basic Local Alignment Search Tool program was used for the sequence analysis, and the sequence assembly and multiple alignments were performed using the MEGA 5 software package [38]. The distribution of nucleotide diversity (π) along the gene sequence (a “sliding-window” approach; window length of 10 sites and step size of 10 sites) was analyzed using DnaSP version 5 [39]. The recombination events (and breakpoints) were analyzed using various recombination-detection methods (RDP, GENECONV, MaxChi, Chimera, SiScan, and Seq) implemented in the RDP software [40].
The interspecific phylogenies for the total data set were reconstructed with the neighbor-joining (NJ) methods in the PAUP 4.0 b10 program (Sinauer Associates Inc., Sunderland, MA, USA). The model of evolution was selected by hierarchical likelihood ratio tests in Modeltest 3.07 [41]. NJ and maximum-likelihood trees were constructed using 1,000 and 100 bootstrap replicates, respectively.
3. Results
3.1. Cytogenetic analysis
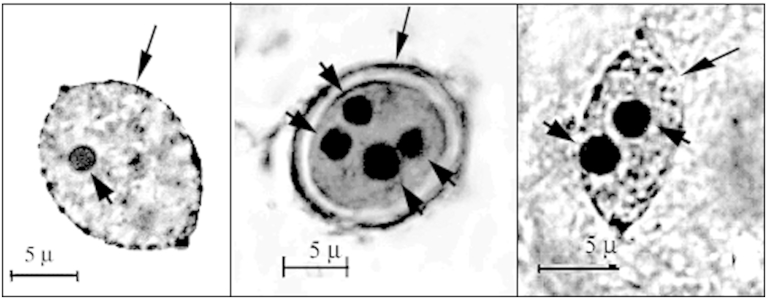
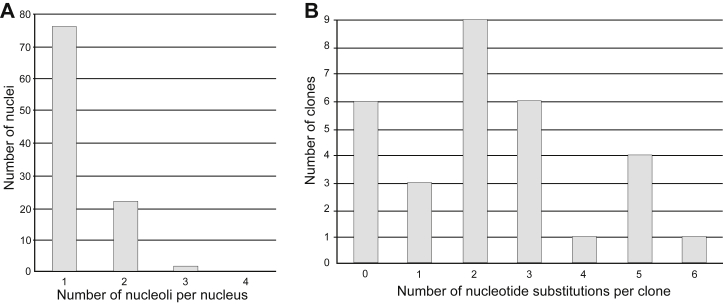
The cytogenetic analysis has shown that, in the cultivated P. ginseng plant, the number of nucleoli in the interphase nuclei of the leaf mesophyll cells varies from one to four (Fig. 1). Totally, the percentages of nuclei with a single nucleolus and those with two, three, and four nucleoli were estimated to be 75.7% and 24.3%, respectively (Table 1; Fig. 2A). The nuclei/nucleoli ratio of the area ranged from 28.4 in one nucleolus cells to 15.1 in cells with four nucleoli (Table 1). The nucleoli of single cells were estimated to be of the same or different sizes, which form descending series.
Fig. 1.
Nuclei of mesophyll cells in the Panax ginseng leaves with different number of nucleoli. The nucleus with four nucleoli was obtained after leaf maceration. Long and short arrows indicate nuclei and nucleoli, respectively.
Table 1.
Nuclei/nucleoli characteristics of Panax ginseng leaf mesophyll cells
| Parameters | No. of nucleoli in interphase nuclei |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| % of cells (n = 500, occurrence, 100%) | 75.7 ± 10 | 22 ± 4 | 1.8 ± 0.3 | 0.5 ± 0.02 |
| Nucleus area (μm2) | 142 ± 7 | 132 ± 8 | 130 ± 9 | 124 ± 6 |
| Nucleoli area (μm2) | 5 ± 0.2 | 4.1 ± 0.2 | 3.5 ± 0.3 | 3.1 ± 0.2 |
| 2.5 ± 0.1 | 2.3 ± 0.1 | 2.5 ± 0.1 | ||
| 1.8 ± 0.08 | ||||
| 0.8 ± 0.06 | ||||
| Nuclei/nucleoli ratio of area | 28.4 ± 1.1 | 20 ± 0.8 | 16.6 ± 0.7 | 15.1 ± 0.7 |
Data are presented as mean ± standard deviation
Fig. 2.
Distribution of nucleoli in ginseng cells and nucleotide substitutions in the 18S rDNA clones. (A) The numbers of nucleoli per nucleus and (B) nucleotide substitutions per the 18S ribosomal DNA clone for cultivated ginseng plants.
3.2. Allelic and nucleotide polymorphisms of 18S rDNA sequences in cultivated P. ginseng
A total of 30 Escherichia coli colonies with the 18S rDNA insertion were selected for our analysis. The full-length sequences for the 18S rRNA gene were found to be 1,809 or 1,808 bp in length due to a single deletion at the position 1,073/1,804 bp. Six of these clones (20%) showed a complete identity with the 18S rDNA sequence of the cultivated P. ginseng from Japan (GenBank accession number D83275) [36]. Among 24 (80%) other clones, a deviation from the GenBank sequence was estimated at 67 different positions (Fig. 3), with most clones (50%) exhibiting differences in two to three nucleotide substitutions. Clones with one and four to six nucleotide substitutions accounted for 10% and 20% of the sequences, respectively (see Fig. 2B). This observed heterogeneity is mainly due to singleton nucleotide substitutions, which resulted in 25 alleles for the 18S rRNA gene. The average number of substitutions per gene sequence was estimated to be 2.79 (i.e., 0.15%).
Fig. 3.
The variable sites for full sequences of the cloned Panax ginseng 18S ribosomal DNA. The nucleotide positions are indicated on the top of alignment.
The majority of polymorphisms corresponded to transitions (88.1%), and C→T transitions were the rarest (9.52%), while other types of transitions occurred with similar frequencies (23.81–28.57%). The transversions were represented by only four types of nucleotide substitutions (Table 2), and the transition/transversion ratio was 7.4.
Table 2.
Nucleotide substitutions in the 18S rDNA clones of cultivated Panax ginseng plant
| Type of nucleotide substitutions | Transitions |
Transversions |
||||||
|---|---|---|---|---|---|---|---|---|
| A → G | T → C | G → A | C → T | T → G | G → T | C → G | C → A | |
| No. of nucleotide substitutions, n (%) | 10 (23.81) | 12 (28.57) | 11 (26.19) | 4 (9.52) | 1 (2.38) | 2 (4.76) | 1 (2.38) | 1 (2.38) |
n, number of clones; rDNA, ribosomal DNA
All six recombination-detection methods implemented in the RDP software failed to detect the events (p < 0.001 for each method) in the 18S rDNA sequences of P. ginseng.
3.3. Distribution of nucleotide diversity
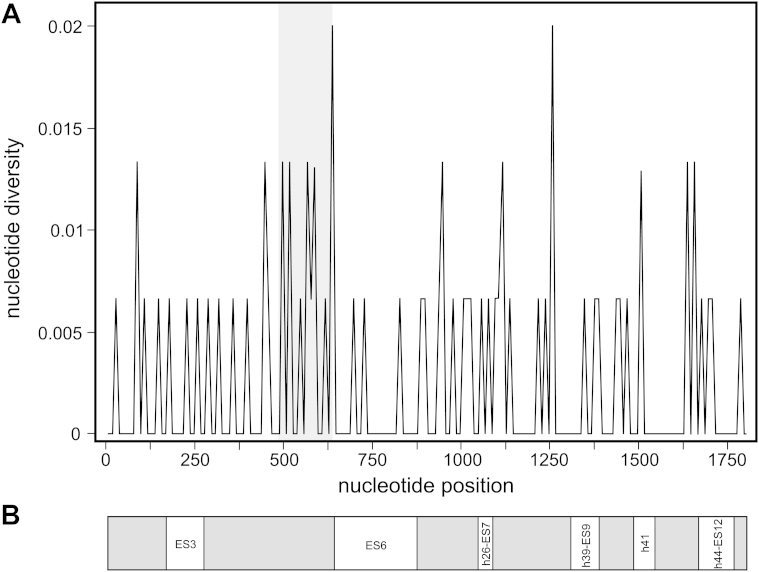
The distribution pattern of nucleotide diversity along the 18S rDNA sequences (Fig. 4) revealed the nucleotide polymorphism both in variable and expansion segments (ESs), as well as in the conservative core structure. The first third of the gene sequence revealed a higher frequency of nucleotide substitutions, especially at the positions 440–640 bp. The main differences between P. ginseng and other species of the genus Panax appeared to be localized in this region and in the neighboring part of the ES6 at the positions 712–729 bp. Additionally, two fixed mutations at the positions 497 bp and 501 bp distinguished both the P. ginseng 18S rDNA clones and the GenBank sequence from other species of the genus Panax (see Fig. 4).
Fig. 4.
Distribution of nucleoli in ginseng cells and nucleotide substitutions in the 18S rDNA clones. (A) The distribution pattern of nucleotide diversity along the 18S ribosomal RNA gene sequences of the cultivated plant Panax ginseng as computed using the sliding-window option (B) with localization of the secondary structure's elements. The region with most nucleotide variability among clones is in gray color, and those of Panax species are in brackets.
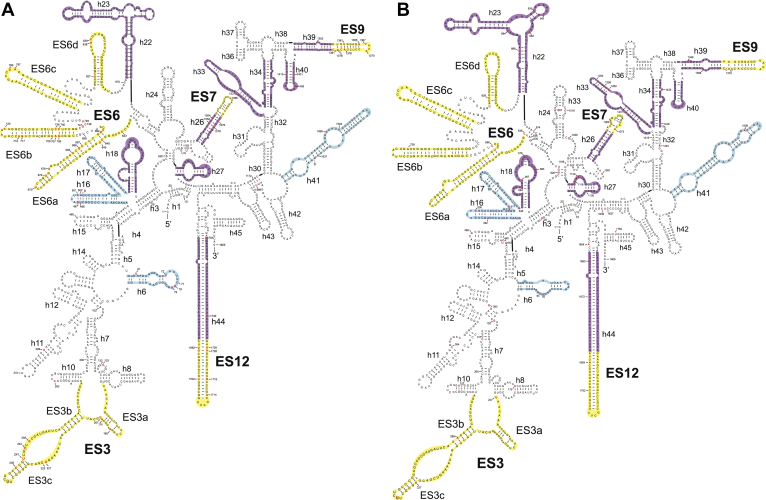
The general folding of the 18S rRNA is highly conservative in a wide number of organisms. For eukaryotes, the 18S rRNA secondary structure for wheat [42] is the most modern, and therefore, we used it for our data as a reference. The nucleotide substitutions of the P. ginseng 18S rRNA were localized in different variable regions, ESs, and also in the main functional domains, including the highly important helices 18, 27, and 34 (Fig. 5). In total, we analyzed 14 conservative and variable helices, ES3, ES6, ES9, and ES12 regions to examine whether these mutations affect the secondary structure or not. The obtained folding patterns demonstrated some differences between the 18S rRNA structure of Triticum aestivum and those of P. ginseng, which are identical to the GenBank sequence in ES6a, ES9, and variable (h41) and conservative (h33) helices (Fig. 5A). For the individual 18S rRNA molecules of P. ginseng, the obtained folding patterns revealed additional differences in variable h6 and conservative h22 (Fig. 5B).
Fig. 5.
The putative scheme of the 18S ribosomal RNA (rRNA) secondary structure for Panax ginseng based on the data for the wheat 18S rRNA molecule. The nucleotide substitutions are highlighted in red letters, deletions are indicated as red “D,” and the insertion is in red letter in a circle. Yellow, blue, and lilac regions indicate de novo modeled elements of the expansion segments, variable and conservative helices, respectively. (A) and (B) Comparison of the 18S rRNA sequences of P. ginseng, which are identical to the GenBank sequence (accession number D83275) with that of Triticum aestivum and 18S rRNA sequences of P. ginseng among themselves, respectively.
3.4. Phylogenetic reconstructions
The model test detected that the Tamura–Nei substitution model of evolution with gamma-distribution shape parameter 1.5251 provides the best fit for the 18S rDNA data. Therefore, the Tamura–Nei genetic distance for the estimation of intraspecific phylogenetic relationships was used in our study.
The genetic distances among the cloned 18S rRNA gene sequences ranged from 0.0006 to 0.0063, with an average of 0.0026 for the total data set. The average value of genetic distances among 13 Panax taxa (GenBank sequences) was estimated to be 0.0015. The genetic distances between the rDNA sequences of cultivated and wild P. ginseng plants and those between P. ginseng and other Panax species were estimated to be 0.0016 and 0.0018, respectively (Table 3).
Table 3.
Genetic distances between 18S rDNA sequences of species from the genus Panax
| Comparison groups | D |
|---|---|
| Clones | 0.0026 (0.0006–0.0063) |
| Clones vs. Panax ginseng | 0.0016 (0.0006–0.0034) |
| Clones vs. Panax species | 0.0030 (0.0006–0.0068) |
| P. ginseng vs. Panax species | 0.0018 (0.0011–0.0034) |
| Panax species | 0.0015 (0.0006–0.0051) |
The range of D values in pairwise comparisons of individual sequences is indicated in brackets
D, genetic distances; rDNA, ribosomal DNA
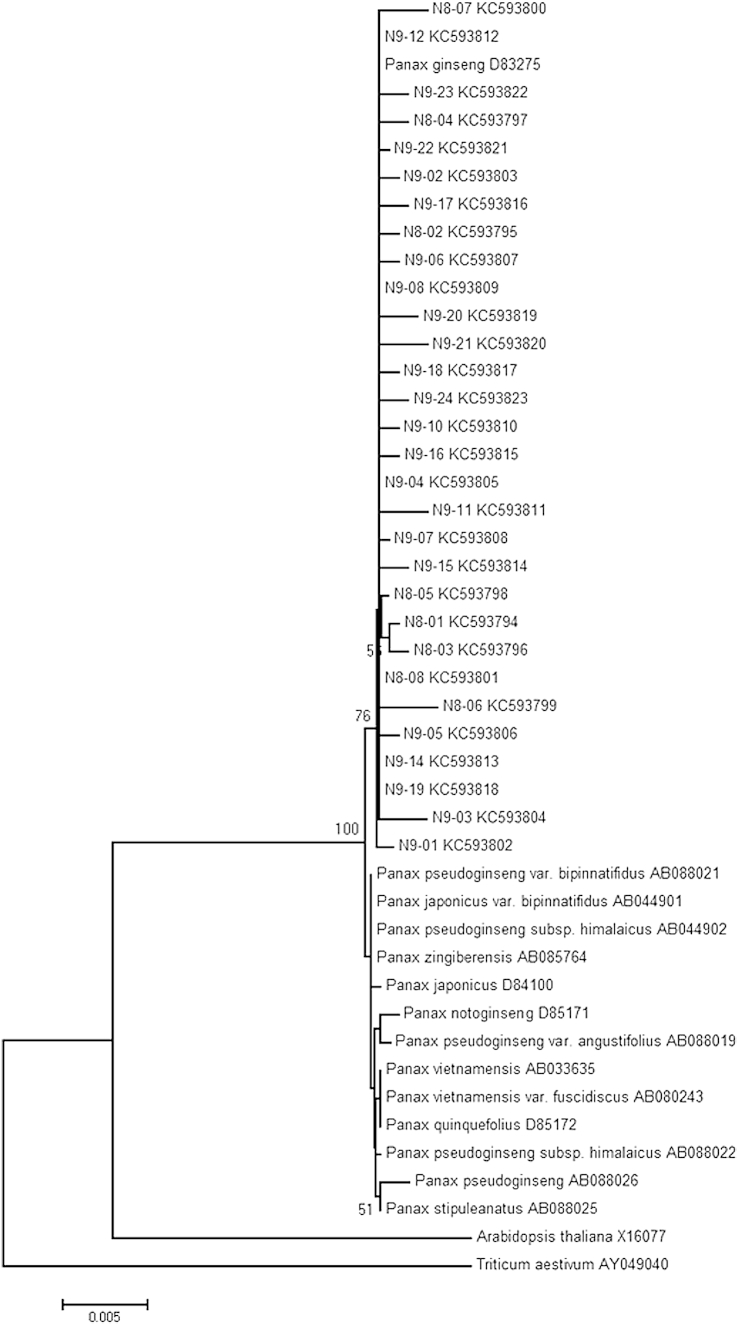
The topology of the NJ tree (Fig. 6) indicated the statistically supported (79%) clustering of the clones with the GenBank sequence of P. ginseng from the plantation in Japan, as well as a low interspecific differentiation and monophyly (100%) of the Panax species.
Fig. 6.
The neighbor-joining tree reconstructed on the basis of the Tamura–Nei substitution model of evolution with gamma distribution shape parameter 1.52 for cloned and direct sequenced 18S ribosomal DNA sequences of Panax ginseng and other congeneric species. The bootstrap values higher than 50 are indicated at the branching nodes. 1–24, individual clones; 8 and 9, two independent polymerase chain reactions; N, native plant.
4. Discussion
Polyploidy and hybridization are the most often discussed reasons for nucleotide variability in the rDNA sequences [16], [18], [21], [25], [43]. Interspecific hybridization is known to be a major force of plant evolution; however, there are no available evidences of such events for the species from the genus Panax. In our study, the phylogenetic analysis strongly supported the conspecificity of Oriental-ginseng plants cultivated in Russia and Japan, and their differentiation from other species of the genus Panax, suggesting that the intragenomic polymorphisms of P. ginseng reflect the species-specific nucleotide substitutions. Taking into account a large species range of P. ginseng, the intragenomic polymorphism of this species may be due (at least partially) to interbreeding among conspecific and genetically different populations in their recent history. However, this assumption contradicts the low level of genetic variability in natural populations of this relict plant based on allozyme and random amplified polymorphic DNA markers [44], [45], [46]. The phylogenetic reconstructions also supported monophyly and the close relations of 13 species and intraspecific forms from the genus Panax collected in different countries of Asia and North America, and these results are in concordance with previously reported data [47].
On the contrary, according to the estimated chromosome number (2n = 44 or 2n = 48), Oriental ginseng, P. ginseng, can be a natural tetraploid with two polyploidization events, which occurred during the evolution of this plant because the basic chromosome number in the family Araliaceae was 12 [48], [49]. At the same time, the species with aneuploid chromosome numbers (n = 9) were found in a closely related family Apiaceae, and therefore, it was repeatedly hypothesized that the basic number of chromosomes in the order of Apiales may be equal to six [50], [51]. Thus, P. ginseng may be a natural polyploid (tetra- or octoploid), and hence, intact plants may show genomic instability.
It has long been known that the genomic instability of polyploid cells is more expressed in cultivated plants, including P. ginseng, manifesting itself in mixoploidy and chimeras [52], [53]. In the cultivated plants and longtime cultivated cell lines, mixoploidy was more pronounced, and the number of polyploid cells was higher than that for wild plants [54]. Additionally, the changes in flavonoid and saponin levels during the cell cultivation were suggested to be the result of the changes in ploidy. In Atragene speciosa, the polyploidy promoted a substantial increase in the saponin quantity [55], although in P. ginseng the polyploidy was not accompanied by the accumulation of triterpene glycosides [56].
In this study, we detected an increased number of nucleoli in almost a quarter of the cells in phototrophic leaf tissue, which is 2.5 times higher in comparison with the root meristem of cultivated Oriental-ginseng seedling [33]. The number of nucleoli in the nucleus can change due to different factors. In the first hybrid generation of an onion seed, the number of cells with two nucleoli was two times higher in comparison with the parental forms [57]. In the artificial plantations in contaminated areas, the proportion of cells with a high number of nucleoli (4–7) increased up to 80% in the root meristem of a seedling of Pinus pallasiana [58]. The number of nucleoli in the long-time cultivated cell lines of Oriental ginseng ranges from one to nine, with the modal classes of 3.4 and 5 nucleoli [33]. We hypothesized that an increased number of nucleoli in the leaves of P. ginseng observed in the present study can be initiated by the changes in adaptive strategy during plantation (i.e., from the stress tolerant in wild plants to the competitive and ruderal strategies of survival in cultivated plants). These alterations were shown to be associated with an increase in the acceptor load on the leaves, which are known to be a donor organ in ginseng [59]. Interestingly, the morphology of ginseng roots is mainly determined by the habitat, and therefore, the roots of the wild and cultivated plants are well distinguished. It has been recently reported that this habitat-induced reciprocal transformation is accompanied by an alteration in DNA methylation [60]. Additionally, we can suggest that, in three to four nucleoli cells, the biosynthesis is more intensive than in one to two nucleoli cells, because the observed decrease in the nuclei/nucleoli ratio of the area implies the increase in the activity of protein synthesis in the cell [57]. Thus, a total level of biosynthesis (including rDNA expression) in the cultivated ginseng exhibiting a greater number of three to four nucleoli cells might be higher than that of a native plant.
The variation in the nucleolus number and the sequence polymorphism of the 18S rDNA are believed to be associated with the genome instability due to polyploidy. Both polyploidy and the number/location of nucleoli can be the reasons for the 18S rDNA polymorphism (see Introduction). Our exploration on whether the nucleotide polymorphism of 18S rDNA is associated with variation in the number of nucleoli in the cell nuclei revealed no correlation between the patterns of molecular and cytogenetic variability, suggesting that these characteristics are able to vary independently to some degree. The tissue specificity of nucleotide variability within the rDNA clusters may be regarded as a possible reason for this discrepancy, although (unlike tissue-specific gene expression) no evidences of such variability have been reported until present. Despite the fact that we cannot identify the inactivated NOR loci by means of silver staining, we can assume their presence on the basis of the polyploid nature of ginseng, and consider it as a true reason for the observed discrepancy. In polyploid plants, a large number of the rDNA repeats are packed into transcriptionally inactive chromatin, the formation and patterns of the rRNA gene expression of which are believed to be under strong genetic and epigenetic (cytosine methylation, histone acetylation, and silencing initiated by small interfering RNAs) control [26], [31]. It was also predicted that reduced or eliminated homogenization, as well as the rDNA sequence divergence, are the consequences of the epigenetic silencing of the rDNA loci [13]. Thus, polyploidy and epigenetic factors associated with ecological adaptation can be considered as the real reasons for the molecular and cytogenetic variations of the ginseng ribosomal clusters, which are implemented through different cellular and genomic mechanisms.
Usually, the rDNA sequences are rapidly homogenized within the genome; however, a number of exceptions from this rule are known at present (see above), and no universal explanation was proposed for this phenomenon. In plants, among which polyploidy is widespread, the intraspecific variation of the rDNA sequences is often associated with the location of NORs on nonhomologous chromosomes [12], [18]. When organisms are polyploid, the concerted evolution may not be effective because they are likely to have the rDNA loci on nonhomologous chromosomes that can prevent interloci recombination, and thereby influence the divergence of the rDNA sequences [12], [18], [25]. Unfortunately, without information about the location of P. ginseng NORs (based on the silver staining and fluorescence in situ hybridization technique), it is difficult to link the lack of the rDNA sequence homogenization with their location on chromosomes. However, the rDNA polymorphism revealed in the cultivated ginseng can indicate that the mutation rate exceeds the rate of concerted evolution, which is too low to avoid intragenomic polymorphism in the rDNA loci of this species. Besides, our results clearly indicated the presence of nucleotide polymorphism among the 18S rDNA sequences in P. ginseng cells with any number of nucleoli; consequently, the 18S rDNA of the native plant may also be assumed to be possibly polymorphic. Additional wide-scale investigations are needed to evaluate in how many taxa the rDNA does not evolve in the concerted-evolution manner, and to clarify why the concerted-evolution mechanisms do not effectively work in some cases, including P. ginseng. In any case, the multiplicity of allele variants for the 18S rRNA gene can be regarded as an evolutionary advantage that, in theory, gives an opportunity to develop genes with new or improved properties, increasing the viability of the species as a whole.
In this study, the intragenomic sequence variation was substantial if based on the number of unique alleles, but low enough when we took into account the number of polymorphic sites per gene sequence. For example, in diatom species, the observed intragenomic nucleotide polymorphism for the 18S rDNA is higher: 0.57–1.81% [61]. In the Far Eastern sturgeons, the evolutionary history of which was closely associated with polyploidy and interspecific-hybridization events, the nucleotide polymorphism for the 18S rDNA is significantly higher: 3.1% in species and 4.3% in their hybrids [62], [63].
At least three methodological aspects are known to influence the results of nucleotide polymorphism in the cloned DNA sequences: (1) the polymerase-mediated errors during PCR. We can rule out that nucleotide polymorphisms observed in our study are due to PCR errors because we used the Pfu polymerase for which incorporation of a wrong nucleotide is low enough, ∼1.3 × 10−6 errors per nucleotide [64]. (2) The replication error during cloning can also be a reason of the false-positive polymorphisms. For E. coli, this value was estimated to be 1.1 × 10−9 errors per nucleotide [65] that is quite low to be critical for our results. (3) The PCR-mediated recombination is a common event in the amplification of sequences from the multigene families, such as the rDNA, that may be a reason for an existence of new allelic variants [66]. However, no recombination events were detected when we used different statistical approaches. Therefore, the number of unique alleles, which were identified in our study, should be close to the real value.
Our results showed the existence of the intragenomic sequence variation among the 18S rDNA clones both within nonconservative regions, including ES, and the conservative core structure. In theory, the nucleotide substitutions located in the functionally significant regions of the 18S rRNA molecule, such as helices 18, 34, and 27, which are responsible for the decoding and translation accuracy, respectively, may lead to errors during protein synthesis or the loss of translational activity. The question about the functional role of ES in eukaryotes remains open. In Tetrahymena thermophila, for example, the 119 bp insertion in ES near the 3′-end of the 26S rRNA is tolerant, while the deletion or substitutions in ES 27L can highly decrease the cell growth or induce the accumulation of 27S pre-RNA, and even can be lethal [42]. In addition, the rDNA could serve as a “sensor” of DNA damage and a “shock absorber,” which prevents genomic damage [29]. The rDNA damage can lead to the formation of an unusual secondary structure, which may cause the inhibition of DNA replication and the emergence of “hot-spot” recombination inducing the translocation and change in the number of repeats. The extra copies that are not involved in transcription are necessary to facilitate damage repair [30]. Altogether, these evidences permit us to suggest that the observed nucleotide variability of the 18S rDNA sequences in P. ginseng leaves may affect the synthesis of highly specific, biologically active substances during the plant cultivation.
Thus, we believe it was the first attempt to describe the intragenomic variability of the 18S rDNA sequences, including the generation of secondary structures for the functionally important and variable domains of the 18S rRNA molecule in the cultivated ginseng, and to exclude the variation in the active nucleolus number and interspecific-hybridization events as actual reasons for the 18S rDNA polymorphism in this plant. The revealed cytogenetic and molecular variabilities are regarded to be associated with polyploidy, epigenetic factors, and alterations in the plant's adaptive strategy that may lead to the decrease in the synthesis of valuable biologically active compounds during P. ginseng plantation. Additional studies are needed to clarify the mechanisms of concerted evolution in ginseng.
Conflicts of interest
The authors have no potential conflicts of interest.
Acknowledgments
This research was partially supported by a grant from the Far Eastern Branch of the Russian Academy of Sciences (project number 12-I-П6-03).
References
- 1.Zhuravlev Y.N., Kolyada A.S. Dalnauka; Vladivostok: 1996. Araliaceae: ginseng and others. [in Russian] [Google Scholar]
- 2.Arnheim N. Concerted evolution in multigene families. In: Nei M., Koehn R., editors. Evolution of genes and proteins. Sinauer; Sunderland: 1983. pp. 38–61. [Google Scholar]
- 3.Brown D.D., Wensink P.C., Jordan E. A comparison of the ribosomal DNA’s of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972;63:57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- 4.Dover G.A. Concerted evolution, molecular drive and natural selection. Curr Biol. 1994;4:1165–1166. doi: 10.1016/s0960-9822(00)00265-7. [DOI] [PubMed] [Google Scholar]
- 5.Nei M., Rooney A.P. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohta T. Evolution of gene families. Gene. 2000;259:45–52. doi: 10.1016/s0378-1119(00)00428-5. [DOI] [PubMed] [Google Scholar]
- 7.Ohta T. The mutational load of a multigene family with uniform members. Genet Res. 1989;53:141–145. doi: 10.1017/s0016672300028020. [DOI] [PubMed] [Google Scholar]
- 8.Fenton B., Malloch G., Germa F. A study of variation in rDNA ITS regions shows that two haplotypes coexist within a single aphid genome. Genome. 1998;41:337–345. [PubMed] [Google Scholar]
- 9.Harris D.J., Crandall K.A. Intra-genomic variation within ITS1 and ITS2 of freshwater crayfishes (Decapoda: Cambaridae): implications for phylogenetic and microsatellite studies. Mol Biol Evol. 2000;17:284–291. doi: 10.1093/oxfordjournals.molbev.a026308. [DOI] [PubMed] [Google Scholar]
- 10.Parkin E.J., Butlin R.K. Within- and between-individual sequence variation among ITS1 copies in the meadow grasshopper Chorthippus parallelus indicates frequent intrachromosomal gene conversion. Mol Biol Evol. 2004;27:1595–1601. doi: 10.1093/molbev/msh163. [DOI] [PubMed] [Google Scholar]
- 11.Xu H., Wang Z., Ding X., Zhou K., Xu L. Differentiation of Dendrobium species used as “Huangcao Shihu” by rDNA ITS sequence analysis. Planta Med. 2005;71:1–3. doi: 10.1055/s-2005-916228. [DOI] [PubMed] [Google Scholar]
- 12.Matyášek R., Renny-Byfield S., Fulnecek J., Macas J., Grandbastien M.-A., Nichols R., Leitch A., Kovarík A. Next generation sequencing analysis reveals a relationship between rDNA unit diversity and locus number in Nicotiana diploids. BMC Genomics. 2012;13:722. doi: 10.1186/1471-2164-13-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovarik A., Dadejova M., Lim Y.K., Chase M.W., Clarkson J.J., Knapp S., Leitch A.R. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Ann Bot. 2008;101:815–823. doi: 10.1093/aob/mcn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller I., Chintauan-Marquier I.C., Veltsos P., Nichols R.A. Ribosomal DNA in the grasshopper Podisma pedestris: escape from concerted evolution. Genetics. 2006;174:863–874. doi: 10.1534/genetics.106.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Márquez L.M., Miller D.J., Mackenzie J.B., Van Oppen M.J.M. Pseudogenes contribute to extreme diversity of nuclear ribosomal DNA in the hard coral Acropora. Mol Biol Evol. 2003;20:1077–1086. doi: 10.1093/molbev/msg122. [DOI] [PubMed] [Google Scholar]
- 16.Muir G., Fleming C.C., Schlötterer C. Three divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Mol Biol Evol. 2001;18:112–119. doi: 10.1093/oxfordjournals.molbev.a003785. [DOI] [PubMed] [Google Scholar]
- 17.Ruggiero M.V., Procaccini G. The rDNA ITS region in the lessepsian marine angiosperm Halophila stipulacea (Forssk.) Aschers. (Hydrocharitaceae): intragenomic variability and putative pseudogenic sequences. J Mol Evol. 2004;58:115–121. doi: 10.1007/s00239-003-2536-0. [DOI] [PubMed] [Google Scholar]
- 18.Campbell C.S., Wojciechowski M.F., Baldwin B.G., Alice L.A., Donoghue M.J. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae) Mol Biol Evol. 1997;14:81–90. doi: 10.1093/oxfordjournals.molbev.a025705. [DOI] [PubMed] [Google Scholar]
- 19.Crease T.J., Lynch M. Ribosomal DNA variation in Daphnia pulex. Genetics. 1991;141:1327–1337. doi: 10.1093/genetics/141.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieger J., Fuerst P.A. Characterization of nuclear 18S rRNA gene sequence diversity and expression in an individual lake sturgeon (Acipenser fulvescens) J Appl Ichthyol. 2004;20:433–439. [Google Scholar]
- 21.Liu Z.L., Zhang D., Wang Z.Q., Ma X.F., Wang X.R. Intragenomic and interspecific 5S rDNA sequence variation in five Asian pines. Am J Bot. 2003;90:17–24. doi: 10.3732/ajb.90.1.17. [DOI] [PubMed] [Google Scholar]
- 22.Pillet L., Fontaine D., Pawlowski J. Intra-genomic ribosomal RNA polymorphism and morphological variation in Elphidium macellum suggests inter-specific hybridization in foraminifera. PLoS ONE. 2012;7:e32373. doi: 10.1371/journal.pone.0032373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robles F., de la Herrán R., Ludwig A., Ruiz-Rejón C., Ruiz-Rejón M., Garrido-Ramos M. Genomic organization and evolution of the 5S ribosomal DNA in the ancient fish sturgeon. Genome. 2005;48:18–28. doi: 10.1139/g04-077. [DOI] [PubMed] [Google Scholar]
- 24.Wei N.-W.V., Wallace C.C., Dai C.-F., Pillay K.R.M., Chen C.A. Analyses of the ribosomal internal transcribed spacers (ITS) and the 5.8S gene indicate that extremely high rDNA heterogeneity is a unique feature in the Scleractinian coral genus Acropora (Scleractinia; Acroporidae) Zool Stud. 2006;45:404–418. [Google Scholar]
- 25.Wendel J.F. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 26.Bártová E., Harničarová Horáková A., Uhlířová R., Raška I., Galiová G., Orlova D., Kozubek S. Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments. J Histochem Cytochem. 2010;58:391–403. doi: 10.1369/jhc.2009.955435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catalano A., O’Day D.H. Rad53 homologue forkhead-associated kinase A (FhkA) and Ca2+-binding protein 4a (CBP4a) are nucleolar proteins that differentially redistribute during mitosis in Dictyostelium. Cell Div. 2013;8:4. doi: 10.1186/1747-1028-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T., Heck D.J., Nomura M., Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi T. A new role of the rDNA and nucleolus in the nucleus rDNA instability maintains genome integrity. Bioessays. 2008;30:267–272. doi: 10.1002/bies.20723. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol Life Sci. 2011;68:1395–1403. doi: 10.1007/s00018-010-0613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olausson K.H., Nistér M., Londsröm M.S. p53-Dependent and -independent nucleolar stress responses. Cell. 2012;1:774–798. doi: 10.3390/cells1040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Show P., Brown J. Nucleoli: composition, function, and dynamics. Plant Physiol. 2012;158:44–51. doi: 10.1104/pp.111.188052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khrolenko Y.A., Burundukova O.L., Lauve L.S., Muzarok T.I., Makhan’kov V.V., Zhuravlev Y.N. Characterization of the variability of nucleoli in the cells of Panax ginseng Meyer in vivo and in vitro. J Ginseng Res. 2012;36:322–326. doi: 10.5142/jgr.2012.36.3.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muratova E.N. Nucleolus staining methods for karyotype analysis of conifers. Bot J. 1995;80:82–86. [Google Scholar]
- 35.Kiselev K.V., Bulgakov V.P. Stability of the rolC gene and its expression in 15-year-old cell cultures of Panax ginseng. Appl Biochem Microbiol. 2009;45:252–258. [PubMed] [Google Scholar]
- 36.Fushimi H., Komatsu K., Isobe M., Namba T. 18S ribosomal RNA gene sequences of three Panax species and the corresponding ginseng drugs. Biol Pharm Bull. 1996;19:1530–1532. doi: 10.1248/bpb.19.1530. [DOI] [PubMed] [Google Scholar]
- 37.Krieger J., Hett A.K., Fuerst P.A., Birstein V.J., Ludwig A. Unusual intraindividual variation of the nuclear 18S rRNA gene is widespread within the Acipenseridae. J Hered. 2006;97:218–225. doi: 10.1093/jhered/esj035. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 40.Martin D.P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 42.Armache J.P., Jarasch A., Anger A.M., Villa E., Becker T., Bhushan S., Jossinet F., Habeck M., Dindar G., Franckenberg S. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krieger J., Fuerst P.A. Evidence of multiple alleles of the nuclear 18S ribosomal RNA gene in sturgeon. Appl Ichthyol. 2002;18:290–297. [Google Scholar]
- 44.Koren O.G., Potenko V.V., Zhuravlev Y.N. Inheritance and variation of allozymes in Panax ginseng C.A. Meyer (Araliaceae) Int J Plant Sci. 2003;164:189–195. [Google Scholar]
- 45.Kozyrenko M.M., Artyukova E.V., Lauve L.S., Zhuravlev Y.N., Reunova G.D. The genetic variability of Panax ginseng callus lines. BioTechnologia. 2001;1:19–26. [Google Scholar]
- 46.Zhuravlev Y.N., Reunova G.D., Kozyrenko M.M., Artyukova E.V., Muzarok T.I. Genetic variation of wild ginseng populations (RAPD analysis) Mol Biol. 1998;32:910–914. [in Russian] [Google Scholar]
- 47.Komatsu K., Zhu S., Fushimi H., Qui T.K., Cai S., Kadota S. Phylogenetic analysis based on 18S rRNA gene and matK gene sequences of Panax vietnamensis and five related species. Planta Med. 2001;67:461–465. doi: 10.1055/s-2001-15821. [DOI] [PubMed] [Google Scholar]
- 48.Grushvitskii I.V. Nauka; Leningrad: 1961. Ginseng: the aspects of biology. [Google Scholar]
- 49.Gurzenkov N.N., Kolyada A.S. Study of the karyotype of Panax ginseng C.A. Meyer (Araliaceae) In: Gursenkov N.N., Moskaliuk T.A., Chernyshev V.D., editors. Biological researches of the Gornotaezhnaya station. Rossiiskaia akademiia nauk; Ussuriysk: 1996. pp. 101–105. [Google Scholar]
- 50.Raven P.H. The bases of angiosperm phylogeny: cytology. Ann Missouri Bot Gard. 1975;62:724–764. [Google Scholar]
- 51.Yi T., Lowry P.P., Plunkett G.M. Chromosomal evolution in Araliaceae and close relatives. Taxon. 2004;19:987–1005. [Google Scholar]
- 52.Bulgakov V.P., Lauve L.S., Tchernoded G.K., Khodakovskaya M.V., Zhuravlev Y.N. Chromosome variation in ginseng cells transformed with the rolC plant oncogene. Russ J Genet. 2000;36:150–156. [PubMed] [Google Scholar]
- 53.Kunakh V.A. Genome variation of plant somatic cells, II: variation in nature. Biopolim Kletka. 1995;11:5–40. [Google Scholar]
- 54.Lauve L.S., Burundukova O.L., Muzarok T.I., Zhuravlev Y.N. Chromosome numbers of Panax ginseng (Araliaceae) Bot J. 2008;93:158–161. [Google Scholar]
- 55.Kunakh V.A., Mozhilevskaya L.P., Adonin V.I., Gubar S.I. Productivity and genetic structure of Panax ginseng C.A. Meyer cell populations during the in vitro cultivation. BioTechnologia. 2003;3:25–35. [Google Scholar]
- 56.Dorofeev V.Y., Karnachuk R.A., Pulkina S.V., Komleva E.V., Dubina V.B., Medvedeva J.V. Atragene speciosa Weinm culture in vitro: the cytogenetic analysis and formation of triterpenoid glycosides and flavonoids. Vestnik Tomskogo Univ. 2009;3:37–41. [Google Scholar]
- 57.Shakhbazov V.G., Shestopalova N.G. Some peculiarities of nucleoli in cells of onion seed. Dokl USSR. 1971;196:58–64. [Google Scholar]
- 58.Korshikov, Tkachev Y.A. The nucleus–nucleolus characteristics of seed progeny for Crimean pine (Pinus pallasiana D. Don) from wild population and planting of anthropogenic contaminated territories. Ind Bot. 2011;11:157–161. [Google Scholar]
- 59.Burundukova O.L., Ivanov L.A., Ivanova L.A., Kiselev K.V., Makhan’kov V.V., Lauve L.S., Khrolenko Y.A., Burkovskaya E.V., Velivetskaya T.A., Ignatiev A.V. Morphofunctional principles determining the changes in the adaptation strategy of ginseng (Panax ginseng C.A. Meyer) during its domestication. Dokl Academii Nauk. 2012;446:584–597. doi: 10.1134/S0012496612050079. [DOI] [PubMed] [Google Scholar]
- 60.Ngezahayo F., Wang X.L., Yu X.M., Jiang L.L., Chu Y.J., Shen B.H., Yan Z.K., Liu B. Habitat-induced reciprocal transformation in the root phenotype of Oriental ginseng is associated with alteration in DNA methylation. Chinese Sci Bull. 2011;56:1685–1690. [Google Scholar]
- 61.Alvenson A.J., Kolnick L. Intragenomic nucleotide polymorphism among small subunit (18S) rDNA paralogs in the diatom genus Skeletonema (Bacillariophyta) J Phycol. 2005;41:1248–1257. [Google Scholar]
- 62.Chelomina G.N., Rozhkovan K.V., Ivanov S.A., Bulgakov V.P. Multiplicity of alleles of nuclear 18S rRNA gene of Amur sturgeons: genes and pseudogenes? Dokl Biochem Biophys. 2008;420:115–118. doi: 10.1134/s1607672908030058. [DOI] [PubMed] [Google Scholar]
- 63.Chelomina G.N., Rozhkovan K.V., Rachek E.I., Zhuravlev Y.N. Increased genetic diversity of 18S rDNA in genomes of F1 hybrids of sturgeons Acipenser schrenckii × A. baerii and A. schrenckii × Huso dauricus. Dokl Biol Sci. 2008;421:271–274. doi: 10.1134/s0012496608040157. [DOI] [PubMed] [Google Scholar]
- 64.Rittie L., Perbal B. Enzymes used in molecular biology: a useful guide. J Cell Commun Signal. 2008;2:25–45. doi: 10.1007/s12079-008-0026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J. W.H. Freeman and Company; New York: 1999. DNA replication, repair, and recombination. Molecular cell biology; pp. 454–481. [Google Scholar]
- 66.Cronn R., Cedroni M., Haselkorn T., Grover C., Wendel J.F. PCR-mediated recombination in amplification products derived from polyploid cotton. Theor Appl Genet. 2002;104:482–489. doi: 10.1007/s001220100741. [DOI] [PubMed] [Google Scholar]