Abstract
Background
Rhizobacteria play an important role in plant defense and could be promising sources of biocontrol agents. This study aimed to screen antagonistic bacteria and develop a biocontrol system for root rot complex of Panax notoginseng.
Methods
Pure-culture methods were used to isolate bacteria from the rhizosphere soil of notoginseng plants. The identification of isolates was based on the analysis of 16S ribosomal RNA (rRNA) sequences.
Results
A total of 279 bacteria were obtained from rhizosphere soils of healthy and root-rot notoginseng plants, and uncultivated soil. Among all the isolates, 88 showed antagonistic activity to at least one of three phytopathogenic fungi, Fusarium oxysporum, Fusarium solani, and Phoma herbarum mainly causing root rot disease of P. notoginseng. Based on the 16S rRNA sequencing, the antagonistic bacteria were characterized into four clusters, Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetesi. The genus Bacillus was the most frequently isolated, and Bacillus siamensis (Hs02), Bacillus atrophaeus (Hs09) showed strong antagonistic activity to the three pathogens. The distribution pattern differed in soil types, genera Achromobacter, Acidovorax, Brevibacterium, Brevundimonas, Flavimonas, and Streptomyces were only found in rhizosphere of healthy plants, while Delftia, Leclercia, Brevibacillus, Microbacterium, Pantoea, Rhizobium, and Stenotrophomonas only exist in soil of diseased plant, and Acinetobacter only exist in uncultivated soil.
Conclusion
The results suggest that diverse bacteria exist in the P. notoginseng rhizosphere soil, with differences in community in the same field, and antagonistic isolates may be good potential biological control agent for the notoginseng root-rot diseases caused by F. oxysporum, Fusarium solani, and Panax herbarum.
Keywords: antagonistic activities, diversity, Panax notoginseng, rhizobacteria
1. Introduction
Panax notoginseng F. H. Chen, known as Sanqi or Tianqi in Chinese, is a well-known traditional Chinese medicine [1], widely used for promotion of blood circulation, removal of blood stasis, induction of blood clotting, relief of swelling, alleviation of pain, and cure of coronary heart disease and cardiovascular disease [2]. Roots of P. notoginseng have been used as a variety of raw materials in Chinese medicinal products in China [3]. It has been mainly cultivated for 400 years in the Southwest regions of China, especially in Wenshan, Yunnan Province [4].
P. notoginseng should be grown in the field for at least 3 y to obtain high-quality raw roots [5]. However, the long period planting conditions make P. notoginseng vulnerable to attacks by many soil-borne pathogens including fungi, bacteria, and nematodes [5], [6], [7], [8], [9], [10], [11], [12], [13], [14]. Soil-borne pathogens of P. notoginseng have been reported by fungi including Fusarium oxysporum, Fusarium solani, Phoma herbarum, Alternaria tenuis, Alternaria panax, Cylindrocarpon destructans, Cylindrocarpon didynum, Phytophthora cactorum, Rhizoctonia solani, and by bacterial pathogens including Pseudomonas sp., Ralstonia sp., and by parasitic nematodes, such as Ditylenchus sp., Rhabditis elegans, and Meloidogyne spp. [15], [16]. In this case, the control of soil-borne diseases mainly relies on chemical pesticides, fungicides, and crop rotation. Chemical pesticides and fungicides are less effective on the soil-borne diseases, and lead to reduction of P. notoginseng quality. Meanwhile, pesticides may be toxic to crops, humans, animals [17], [18]. However, a 15–20 y replanting interval leads to the lack of appropriate fields, resulting in searching for a new field or/and transferring to a less appropriate field to grow P. notoginseng.
It is obvious that pesticides and less appropriate cultivation soil are not suitable to control the qualities of P. notoginseng required by the good agriculture practice (GAP). Friendly approaches are urgently needed to effectively manage or solve the questions. Biological control, a bioeffector method with other living organisms to control pests (insects, mites, weeds, and plant diseases) [19], has been considered as effective approaches. Soil bacteria, especially rhizospheric ones with antagonistic properties, demonstrate biological control effectiveness to some plant diseases, and are the most potential for development of biological control agents (BCAs) [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]. However, little is known about the bacterial diversity, distribution, and ecological effects in the cultivation soil of P. notoginseng. In this study, we developed the investigation of rhizobacteria of 3-y-old P. notoginseng from Wenshan, Yunnan Province, by culture-dependent methods. The bacterial isolates were also challenged by three pathogens, F. oxysporum, F. solani, and P. herbarum, which are associated with the root rot disease of P. notoginseng.
2. Materials and methods
2.1. Soil sample collection and isolation of soil bacteria
Soil samples were collected from a 3-y-old P. notoginseng plantation in Wenshan, Yunnan Province, in July 2014. Ten healthy and 10 root-rot notoginseng plants were uprooted. Soil was collected around 3 cm from the main roots, and rhizosphere soil was gently stripped from the roots. Root-adjacent soil and rhizospheric soil were mixed together, recorded as healthy plant soil and diseased plant soil, respectively. Uncultivated soil sample was obtained without planting notoginseng at the same field. All the soil samples were placed into sterile plastic bags, transferred to the laboratory in 24 h, and kept at 4°C before treatment.
Bacterial isolation were developed using serial dilution spread plate method. Ten grams of soil was mixed with 90 mL of sterile phosphate buffered saline (PBS, pH 7.4) and stirred for 30 min at 200 rpm with a magnetic stirrer. The soil suspension was left to stand for 10 min at room temperature to allow settling of large particles, tenfold serial diluted in PBS (from 10−2 to 10−5). Then, 80 μL of the first to fourth and fifth diluents were transferred to petri dishes with LB agar medium (10.0 g peptone, 5.0 g yeast extract, 10.0 g NaCl, and 13.0 g agar, 1.0 L distilled water, pH 7.2) and nutrition agar (NA) medium (3.0 g beef extract, 5.0 g peptone, 5.0 g NaCl, 13.0 g agar, 1.0 L distilled water, pH 7.0). The plates were incubation at 28°C, and bacterial colonies were selected and purified according to their morphological characteristics.
2.2. Screening of antagonistic bacteria against fungal pathogens
Three fungal pathogens F. oxysporum, F. solani, and P. herbarum were isolated from the rotten root of P. notoginseng, and their pathogenicity was verified [15], [16]. The target fungi were cultured on potato dextrose agar (PDA) medium (200.0 g fresh potato, 20.0 g starch, 13.0 g agar, 1.0 L distilled water, pH not adjusted). The antagonism of all bacterial isolates was checked with respect to the ability to suppress fungal growth. Antifungal bioassay was performed with the dual culture and agar well diffusion plate on PDA.
In dual culture tests, a 5-mm mycelial disk of pathogenic fungus, collected from the edge of actively growing colonies, was placed into the center of plates containing fresh PDA. Bacterial isolates were grown around the target fungus with a distance of 3.0 cm (Fig. 1A, 1B). The dual culture plates were incubation at 28°C, and checked every 12 h after inoculation. All treatments were tested in duplicate.
Fig. 1.
Antagonistic assay of bacterial isolates (A) Fusarium solani, (B) Phoma herbarum, and (C) Fusarium oxysporum.
In agar well plate tests, a 200-μL fresh culture of pathogenic fungus with concentration of 108 spores/mL was mixed with 250 mL PDA and evenly distributed into 10 petri dishes (90 mm). On each plate, four wells of 5 mm in diameter were made (Fig. 1C). Bacterial isolates were cultured in nutrient broth medium at 28°C, 135 rpm for 72 h. The bacterial suspension was adjusted to the final cell concentration of 107 cfu/mL with nutrient broth medium. Next, 200 μL of suspension was added to each well, and the same volume of nutrient broth was used as control. All treatments were tested in duplicate.
2.3. Phylogenetic analysis
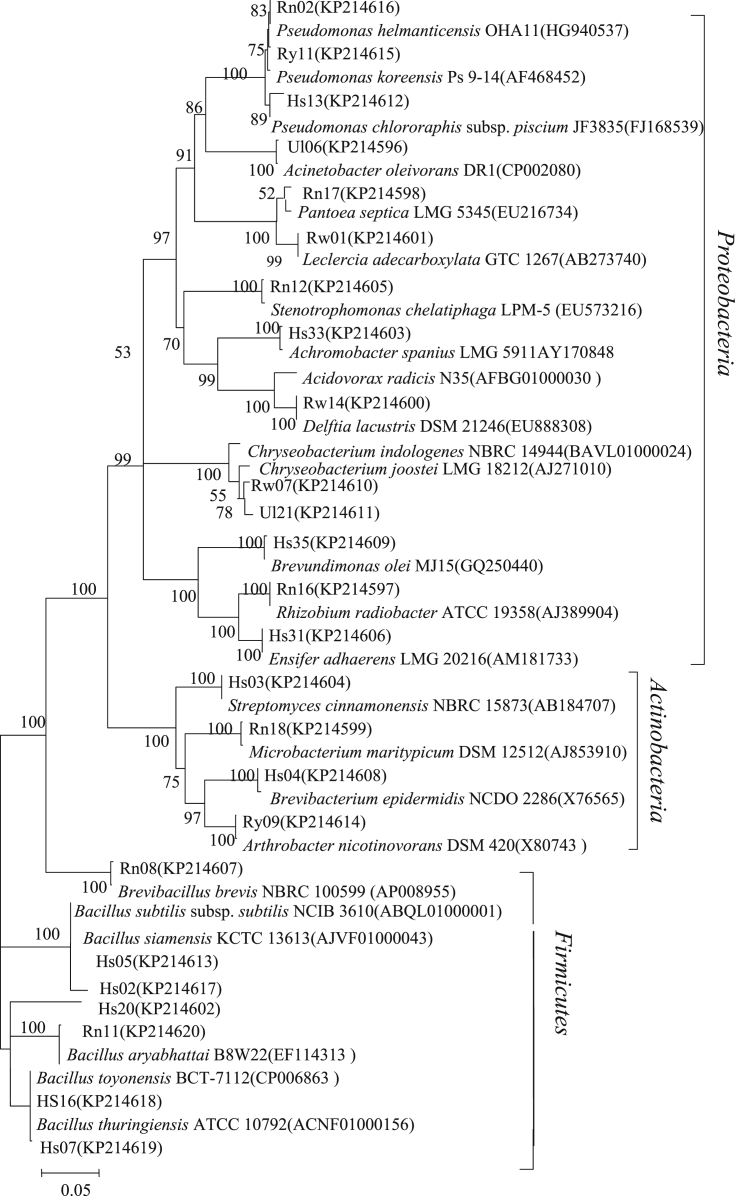
The genomic DNA of bacteria was extracted using a bacterial genomic DNA extraction kit (BioTeke Corporation, China, Cat#: DP2001) and 16S ribosomal RNA (rRNA) genes were amplified by PCR using the primer pair of PA (5′-AGAGTTTGATCCTGGCTCAG-3′) and PB (5′-AAGGAGGTGATCCAGCCGCA-3′) [30]. The PCR reaction was performed in 50 mL reaction mixture containing 1 mL of DNA, 1 μL forward primer (10 mM), 4 μL reverse primer (10 mM), 5 μL reaction buffer (10 ×), 4 μL dNTP (each 2.5 mM), 0.5 μL of Taq DNA polymerase (500 U), and 37.5 μL sterile double-distilled water. The PCR cycling protocol consisted of an initial denaturation at 94 °C for 4 min, followed by 32 cycles of 94 °C for 1 min, 56 °C for 1 min and 72 °C for 1 min, and a final elongation step of 72 °C for 10 min. As a negative control, the DNA was replaced by sterile double-distilled water. The PCR amplified products were separated by agarose gel electrophoresis, and sequenced on an ABI Prism 3730 sequencer at Sangon Biotech (Shanghai, China). The sequences of the isolates were searched in EzBioCloud (http://www.ezbiocloud.net/). The approximate phylogenetic affiliations and 16S rRNA gene sequence similarities were determined according to Altschul et al [31]. Sequences chimera checking were performed by the program CHIMERA CHECK of the Ribosomal Database Project (RDP) [32], and sequences with a potential chimeric structure were excluded. The alignments of 16S rRNA genes sequences were performed using Clustal X [33]. The 16S rRNA sequences were used to construct a phylogenetic tree with the Kimura 2-parameter model and MEGA (version 5.05) by bootstrap analysis of 1,000 replications [34], [35]. The partial 16S rRNA gene sequences obtained for rhizosphere antagonistic bacteria have been deposited in GenBank with accession numbers: KP214596–KP214641.
3. Results
3.1. Number of bacteria in different soil samples
A total of 279 bacterial isolates were obtained from healthy soil, diseased soil, and uncultivated soil. The distribution is 132 isolates (47.3%) in diseased soil, 77 isolates (27.6%) in healthy soil, and 70 isolates (25.1%) in uncultivated soil (Table 1). Bacteria in diseased soil are much richer than that in healthy and uncultivated soil.
Table 1.
Number of rhizospheric bacteria in different soil samples of Panax notoginseng
| No. of bacteria | Soil sample |
Sum | ||
|---|---|---|---|---|
| Healthy plant soil | Root rot plant soil | Uncultivated soil | ||
| Total | 77 | 132 | 70 | 279 |
| Antagonistic bacteria | 37 | 24 | 27 | 88 |
| Percent of antagonistic bacteria (%) | 48.1 | 18.2 | 38.6 | 31.5 |
3.2. Antagonistic soil bacteria associated with P. notoginseng
All the soil bacterial isolates were evaluated for their antagonistic activity to three fungal pathogens, F. oxysporum, F. solani, and P. herbarum. Eighty-eight isolates (31.5% of the total) displayed antagonistic activities against at least one of fungal pathogens (Table 1). The large number of bacterial antagonists was isolated from healthy plant soil which offered 37 strains (48.1% of 77 isolates from healthy plant soil), followed by uncultivated soil (27, 38.6% of 70 isolates from uncultivated land), diseased soil (24, 18.2% of 132 isolates from diseased soil).
Among the 88 antagonists, 33 displayed antagonistic activity only against one of three fungal pathogens (Table 2), which included four strains obtained from healthy plant soil toward F. oxysporum and 19 toward F. solani, and 10 toward P. herbarum.
Table 2.
The number of rhizobacteria obtained from different soil of Panax notoginseng with antagonisitic activities toward three host plant pathogens of root rot disease
| Pathogens | No. of rhizosphere antagonistic bacteria |
Sum | ||
|---|---|---|---|---|
| Healthy plant soil | Root rot plant soil | Uncultivated soil | ||
| Fo | 4 | 0 | 0 | 4 |
| Fs | 8 | 7 | 4 | 19 |
| Ph | 4 | 5 | 1 | 10 |
| Fo & Fs | 1 | 1 | 4 | 6 |
| Fo & Ph | 1 | 1 | 0 | 2 |
| Fs & Ph | 9 | 5 | 10 | 24 |
| Fo, Fs, & Ph | 10 | 5 | 8 | 23 |
| Total | 37 | 24 | 27 | 88 |
Fo, Fusarium oxysporum; Fs, Fusarium solani; Ph, Phoma herbarum.
There were 32 bacterial isolates showing antagonistic activities against two of three pathogens (Table 2). Among them, six isolates had antagonistic activity to F. oxysporum and F. solani, two isolates against F. oxysporum and P. herbarum, and 24 isolates against F. solani and P. herbarum.
Furthermore, there were 23 isolates exhibiting different antagonistic activities against all the three fungal pathogens (Table 2).
3.3. Phylogeny of bacterial antagonists from P. notoginseng
The molecular analysis revealed that the 88 strains belonged to four bacterial groups, Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria (Table 3, Fig. 2). Over half of the soil antagonistic bacteria (46 isolates, 52.3% of total) were accommodated in the Firmicutes group. In this group, Bacillus spp. represented the majority, with 42 isolates (91.3%). Phylogenetic analysis based on the 16S rRNA gene sequences indicated that most active Bacillus isolates were closely related to the species Bacillus thuringiensis (12 isolates, 28.6%), Bacillus aryabhattai (9 isolates, 21.4%) and Bacillus siamensis (5 isolates, 11.9%) with the sequence similarities of 99.9–100.0%, 98.9–100.0%, and 99.1–100.0%, respectively. Other 16 Bacillus isolates were assigned to nine species according to their sequence similarities: Bacillus subtilis subsp. subtilis (2 isolates), Bacillus simplex (1), Bacullus anthracis (1), B. atrophaeus (2), Bacillus cereus (4), Bacillus licheniformis (1), Bacillus safensis (1), Bacillus toyonensis (3), and Bacillus acidiceler (1). The four remaining Firmicutes were respectively assigned to Paenibacillus chitinolyticus (1), Paenibacillus jamilae (2), and Brevibacillus brevis (1) with similarity > 99.4%. Twenty-six isolates belonging to Proteobacteria were assigned to 10 genera: Achromobacter, Acinetobacter, Acidovorax, Brevundimonas, Delftia, Ensifer, Leclercia, Pseudomonas, Rhizobium, and Stenotrophomonas. Pseudomonas included nine species: Pseudomonas baetica (1 isolates), Pseudomonas helmanticensis (1), Pseudomonas hunanensis (2), Pseudomonas koreensis (2), Pseudomonas libanensis (1), Pseudomonas moorei (1), Flavimonas oryzihabitans (1), Pseudomonas chlororaphis subsp. aurantiaca (1), and Pseudomonas chlororaphis subsp. piscium (2) with sequence similarities of 98.3–100.0%.
Table 3.
Antagonistic activities of rhizospheric bacteria towards Fusarium oxysporum (Fo), Fusarium solani (Fs), and Panax herbarum (Ph) in different soil types from Panax notoginseng and their closest phylogenetic affiliation (based on partial 16S ribosomal RNA gene sequences)
| Isolate(Accession No.) | Closest NCBI library strain & accession No. | Antagonistic activities |
Similarity(%) | Origin of strains | ||
|---|---|---|---|---|---|---|
| Fo | Fs | Ph | ||||
| Hs02(KP214617) | Bacillus siamensis KCTC 13613(AJVF01000043) | +++ | +++ | +++ | 100.0 | Hs |
| Hs03(KP214604) | Streptomyces cinnamonensis NBRC 15873(AB184707) | + | ++ | ++ | 100.0 | Hs |
| Hs04(KP214608) | Brevibacterium epidermidis NCDO 2286(X76565) | − | − | + | 100.0 | Hs |
| Hs05(KP214613) | Bacillus subtilis subsp. subtilis NCIB 3610(ABQL01000001) | − | + | − | 99.7 | Hs |
| Hs07(KP214619) | Bacillus toyonensis BCT-7112(CP006863) | − | + | − | 100.0 | Hs |
| Hs08(KP214632) | Bacillus safensis FO-36b(ASJD01000027) | − | + | ++ | 99.9 | Hs |
| Hs09(KP214630) | Bacillus atrophaeus JCM 9070(AB021181) | +++ | +++ | +++ | 99.9 | Hs |
| Hs10(KP214629) | Pseudomonas hunanensis LV(JX545210) | − | + | − | 98.6 | Hs |
| Hs11(KP214626) | Pseudomonas baetica a390(FM201274) | − | + | − | 99.6 | Hs |
| Hs13(KP214612) | Pseudomonas chlororaphis subsp. piscium JF3835(FJ168539) | + | + | ++ | 98.1 | Hs |
| Hs14(KP214624) | Paenibacillus jamilae CECT 5266(AJ271157) | + | + | ++ | 100.0 | Hs |
| Hs16(KP214618) | Bacillus thuringiensis ATCC 10792(ACNF01000156) | − | + | − | 99.9 | Hs |
| Hs18(KP214628) | Pseudomonas libanensis CIP 105460(AF057645) | − | + | ++ | 99.9 | Hs |
| Hs20(KP214602) | Acidovorax radicis N35(AFBG01000030 ) | − | + | − | 97.3 | Hs |
| Hs22(KP214611) | Bacillus cereus ATCC 14579(AE016877) | − | + | − | 100.0 | Hs |
| Hs23(KP214640) | Arthrobacter pascens DSM 20545(X80740 ) | − | − | + | 99.6 | Hs |
| Hs24(KP214631) | Flavimonas oryzihabitans IAM 1568(D84004) | − | − | ++ | 98.3 | Hs |
| Hs25(KP214622) | Chryseobacterium vrystaatense LMG 22846(AJ871397) | − | + | + | 97.2 | Hs |
| Hs26(KP214627 ) | Pseudomonas moorei RW10(AM293566) | + | + | − | 99.5 | Hs |
| Hs31(KP214606) | Ensifer adhaerens LMG 20216(AM181733) | + | − | + | 100.0 | Hs |
| Hs33(KP214603) | Achromobacter spanius LMG 5911(AY170848) | + | − | − | 99.9 | Hs |
| Hs35(KP214609) | Brevundimonas olei MJ15(GQ250440) | + | − | − | 99.7 | Hs |
| Rw01(KP214601) | Leclercia adecarboxylata GTC 1267(AB273740) | + | + | + | 99.9 | Rw |
| Rw04(KP214633) | Bacillus licheniformis ATCC 14580(AE017333) | +++ | +++ | +++ | 98.9 | Rw |
| Rw07(KP214610) | Chryseobacterium joostei LMG 18212(AJ271010) | + | + | + | 98.6 | Rw |
| Rw12(KP214634) | Bacillus sonorensis NBRC 101234(AYTN01000016) | − | − | ++ | 98.8 | Rw |
| Rw14(KP214600) | Delftia lacustris DSM 21246(EU888308) | ++ | + | − | 99.3 | Rw |
| Ry07(KP214638) | Arthrobacter ureafaciens DSM 20126(X80744) | − | + | ++ | 99.5 | Ry |
| Ry09(KP214614) | Arthrobacter nicotinovorans DSM 420(X80743 ) | − | + | ++ | 100.0 | Ry |
| Ry11(KP214615) | Pseudomonas koreensis Ps 9-14(AF468452) | ++ | − | ++ | 99.6 | Ry |
| Rn02(KP214616) | Pseudomonas helmanticensis OHA11(HG940537) | − | + | ++ | 99.7 | Rn |
| Rn06(KP214623) | Paenibacillus chitinolyticus IFO 15660(AB021183) | − | ++ | − | 99.9 | Rn |
| Rn08(KP214607) | Brevibacillus brevis NBRC 100599 (AP008955) | +++ | +++ | +++ | 99.4 | Rn |
| Rn11(KP214620) | Bacillus aryabhattai B8W22(EF114313) | + | + | + | 100.0 | Rn |
| Rn12(KP214605) | Stenotrophomonas chelatiphaga LPM-5 (EU573216) | − | + | − | 98.2 | Rn |
| Rn13(KP214639 ) | Arthrobacter arilaitensis Re117(FQ311875) | − | + | − | 100.0 | Rn |
| Rn16(KP214597) | Rhizobium radiobacter ATCC 19358(AJ389904) | − | +++ | +++ | 100.0 | Rn |
| Rn17(KP214598) | Pantoea septica LMG 5345(EU216734) | − | + | − | 98.8 | Rn |
| Rn18(KP214599) | Microbacterium maritypicum DSM 12512(AJ853910) | − | − | ++ | 99.4 | Rn |
| Ul06(KP214596) | Acinetobacter calcoaceticus DSM 30006(AIEC01000170 ) | + | + | + | 99.9 | Ul |
| Ul07(KP214625) | Pseudomonas chlororaphis subsp. aurantiaca NCIB 10068(DQ682655) | − | + | + | 99.7 | Ul |
| Ul09(KP214637) | Bacillus anthracis ATCC 14578(AB190217) | + | + | + | 100.0 | Ul |
| Ul10(KP214636) | Bacillus acidiceler CBD 119(DQ374637) | + | + | − | 99.9 | Ul |
| Ul11(KP214635) | Bacillus simplex NBRC 15720(AB363738) | − | + | − | 100.0 | Ul |
| Ul16(KP214621) | Acinetobacter oleivorans DR1(CP002080) | + | + | − | 100.0 | Ul |
| Ul21(KP214641) | Chryseobacterium contaminans C26(KF652079) | − | + | − | 99.3 | Ul |
+++, highly active; ++ medially active; +, showing active; −, not active; Hs, rhizosphere soil of healthy plants; NCBI, National Center of Biotechnology Information; Rw, Ry Rn, rhizosphere soil of root-rotten plants; Ul, uncultivated soil.
Fig. 2.
Neighbor-joining tree of partial rhizospheric antagonistic bacteria obtained from five different soil types (healthy plant soil, root rot plant soil, uncultivated soil) of Panax notoginseng and their closest relatives based on the 16S ribosomal RNA gene sequences. The significance of each branch is indicated by a bootstrap value calculated for 1,000 subsets. The scale bar represents 0.05 substitutions per base position. Accession numbers are given in parenthesis. Only values above 50% were shown. The rhizospheric antagonistic bacteria of P. notoginseng were encoded as Hs01-37, Ry01-15, Rw01-14, Rn01-19, and Ul01-27.
In Actinobacteria, 11 isolates were assigned to nine species of five genera (Arthrobacter, Microbacterium, Brevibacterium, Pantoea, and Streptomyces) based on their similarities of 98.8–100.0%. Five isolates in Bacteroidetes were phylogenetically related to Chryseobacterium vrystaatense (1), Chryseobacterium joostei (2), Chryseobacterium contaminans (1), and Chryseobacterium stationis (1) with a similarity > 98.0%.
3.4. Distribution of antagonists in different soil types
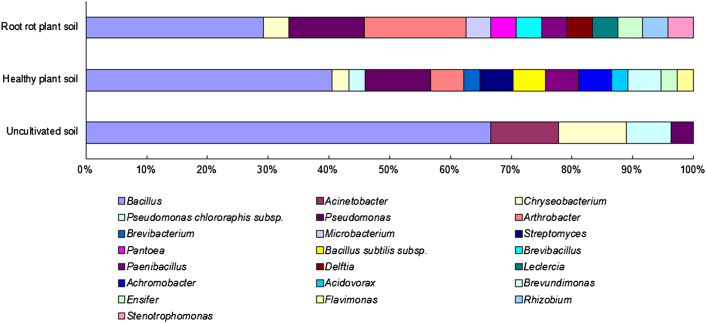
The distribution of active isolates obtained from healthy plant soil, diseased plant soil, and uncultivated soil of P. notoginseng is presented in Fig. 2. At the phylum level, isolates in Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes were widely distributed in all types of the soils. Firmicutes was dominant and accounted for 45.9%, 29.2%, and 66.7% in healthy plant soil, diseased plant soil, and uncultivated soil, respectively. The isolates in Bacteroidetes are much less than that in the other three phyla. Analysis at the genera level showed that Bacillus and Pseudomonas were present in all soil types and represented the majority of antagonists, especially Bacillus spp. which accounted for 45.9% of antagonists in healthy plant soil, 29.2% in diseased plant soil, and 66.7% in uncultivated soil. Arthrobacter was distributed in soils with P. notoginseng, and not found in the uncultivated soil (Fig. 3).
Fig. 3.
Comparative taxonomic distribution of rhizospheric antagonistic bacteria from different soil types of P. notoginseng. The different color show to different taxa.
For all isolated genera, Acidovorax, Brevibacterium, and Flavimonas were exclusively found in healthy plant soil, whereas Delftia, Leclercia, Brevibacillus, Microbacterium, Pantoea, Rhizobium, and Stenotrophomonas were only present in diseased plant soil. Acinetobacter was only found in uncultivated soil (Fig. 3).
4. Discussion
Soil-plant-microorganisms shape a complex soil ecosystem, and soil microorganisms are regarded as an important and essential component of soil quality due to their crucial activities in many ecosystem processes [36], [37], [38]. Soil bacteria exist in almost every soil type. Some soil bacteria are developed as biocontrol agents (BCAs), an environment-friendly approach to control pests (insects, mites, weeds, and plant diseases) [19]. Panax plants, P. ginseng, P. notoginseng, and Panax quniquefolius, are perennial plants and mainly cultivated in artificial shads for several years. Cultivation can be affected by diseases caused by soil-borne and foliar pathogens [16], [39], [40], [41], [42]. In recent years, using antagonistic microorganisms to control ginseng diseases is increasing [9], [41], [42], but few researches on P. notoginseng [43]. In our study, we screened 88 antagonistic strains out of 279 soil bacterial isolates of P. notoginseng with three pathogens as targets, and analyzed their phylogenetic diversity and distribution in healthy plant soil, diseased plant soil, and uncultivated soil.
Phylogenetic analysis indicated that soil antagonistic bacteria of P. notoginseng were assigned into four bacterial groups: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. In four bacterial groups, Firmicutes, especially Bacillus species, represented the majority of the active isolates. This result is similar to that of endophytic bacteria [15]. Meanwhile, the member of Proteobacteria showed high diversity in taxonomy, and were assigned into 10 genera, 19 species. Actinobacteria and Proteobacteria were also discovered from two types of soil associated to P. notoginseng. Most of the Bacillus species exist in the rhizosphere soil of P. notoginseng. This is the same as other results described in medical plants [43]. Additionally, species in Arthrobacter, Brevibacterium, Microbacterium, Streptomyces, Pantoea, Brevibacillus, Paenibacillus, Delftia, Leclercia, Achromobacter, Brevundimonas, Ensifer, Stenotrophomonas, and Pseudomonas detected as antagonistic endophytic bacteria from P. notoginseng have not been reported from cultivation soil of P. notoginseng [15]. Antagonistic bacteria existed in all types of tested soil in the same field of P. notoginseng, but the number and species of antagonists are different (Table 1). There are 23 species in 11 genera in healthy plant soil, 14 species in four genera in uncultivated soil, and 20 species in 13 genera in diseased soil, respectively. The biodiversity of antagonistic bacteria in diseased soil is much lower than that in healthy plant soil and uncultivated soil (Table 1). It is unclear what affect the patterns of distribution and diversity of soil bacteria as the soil properties and agromanagement approaches are not different in the same plantation, especially for healthy plant soil and diseased soil. Further studies might focus on the interaction between fungal pathogens and soil bacteria.
More than half of soil antagonistic bacteria of P. notogingseng (55 strains, 62.5% of antagonistic bacteria) showed antagonistic activities against two or three pathogens (Table 4). Most of these antagonists were assigned into the genus Bacillus. Bacillus spp. have been frequently reported as the major rhizobacteria for diverse host plants and used to suppress pathogens. In this study, Bacillus siamensis, Bacillus thuringiensis, and Bacillus aryabhattai were the most dominant and widespread species within different rhizosphere soil types of P. notoginseng, inferring that the three species can offer a promising way to screen biocontrol Bacillus strains for P. notoginseng. Moreover, other Bacillus spp. isolates, such as Bacillus atrophaeus, Bacillus toyonensis, Bacillus licheniformis, and Bacillus subtilis subsp. subtilis, with broad-spectrum antagonisms also are promising candidates to resist root rot disease of P. notoginseng. Further studies should be taken to evaluate their antagonistic ability in pot and field condition.
Table 4.
Species affiliations of rhizobacteria from Panax notoginseng with antagonistic activities toward Fusarium oxysporum (Fo), Fusarium solani (Fs), and Panax Herbarum (Ph)
| Phylogenetic species | Antagonistic activities |
||||||
|---|---|---|---|---|---|---|---|
| Fo | Fs | Ph | Fo & Fs | Fo & Ph | Fs & Ph | Fo, Fs, & Ph | |
| Bacillus siamensis | 1 | 4 | |||||
| Bacillus atrophaeus | 1 | ||||||
| Bacillus cereus | 2 | 2 | |||||
| Bacillus safensis | 1 | ||||||
| Bacillus thuringiensis | 2 | 1 | 8 | 1 | |||
| Bacillus toyonensis | 2 | 1 | |||||
| Bacillus licheniformis | 1 | ||||||
| Bacillus sonorensis | 1 | ||||||
| Bacillus simplex | 1 | ||||||
| Bacillus acidiceler | 1 | ||||||
| Bacillus anthracis | 1 | ||||||
| Bacillus aryabhattai | 1 | 2 | 4 | 2 | |||
| Bacillus subtilis subsp. subtilis | 1 | 1 | |||||
| Brevibacterium epidermidis | 1 | ||||||
| Brevundimonas olei | 2 | ||||||
| Brevibacillus brevis | 1 | ||||||
| Paenibacillus chitinolyticus | 1 | ||||||
| Paenibacillus jamilae | 2 | ||||||
| Ensifer adhaerens | 1 | 1 | |||||
| Chryseobacterium vrystaatense | 1 | ||||||
| Chryseobacterium indologenes | 1 | ||||||
| Coryseobacterium contaminans | 2 | ||||||
| Corynebacterium stationis | 1 | ||||||
| Flavimonas oryzihabitans | 1 | ||||||
| Pseudomonas hunanensis | 1 | 1 | |||||
| Pseudomonas libanensis | 1 | ||||||
| Pseudomonas moorei | 1 | ||||||
| Pseudomonas baetica | 1 | ||||||
| Pseudomonas koreensis | 1 | 1 | |||||
| Pseudomonas helmanticensis | 1 | ||||||
| Pseudomonas chlororaphis subsp. piscium | 2 | ||||||
| Pseudomonas chlororaphis subsp. aurantiaca | 1 | ||||||
| Streptomyces cinnamonensis | 2 | ||||||
| Stenotrophomonas chelatiphaga | 1 | ||||||
| Arthrobacter nitroguajacolicus | 1 | ||||||
| Arthrobacter nicotinovorans | 1 | 1 | |||||
| Arthrobacter ureafaciens | 1 | ||||||
| Arthrobacter arilaitensis | 1 | ||||||
| Arthrobacter pascens | 1 | ||||||
| Achromobacter spanius | 2 | ||||||
| Acidovorax radicis | 1 | ||||||
| Leclercia adecarboxylata | 1 | ||||||
| Delftia lacustris | 1 | ||||||
| Microbacterium maritypicum | 1 | ||||||
| Pantoea septica | 1 | ||||||
| Rhizobium radiobacter | 1 | ||||||
| Acinetobacter calcoaceticus | 1 | 1 | |||||
| Acinetobacter oleivorans | 1 | ||||||
| Total | 4 | 19 | 10 | 6 | 2 | 24 | 23 |
In conclusion, this investigation provides the first evidence of bacterial differences in healthy plant soil and diseased plant soil of P. notoginseng, although antagonistic bacteria are harbored in all types of tested soil. This will provide some clues for us to understand the interaction among soil bacteria, pathogenic fungi, and plant.
Conflicts of interest
The authors have no conflicts of interest with any parties or individuals.
Acknowledgments
This work was partly supported by the grants from the National Natural Science Foundation of China (Nos. 31100009 and 41361075), Yunnan Natural Science Foundation (No. 2013FA015), and Foundation of Yunnan Educational Committee (No. ZD2013008).
Contributor Information
Li-Xing Zhao, Email: zlx70@163.com.
Hui-Lin Guan, Email: ghl0871@163.com.
References
- 1.Park H.J., Kim D.H., Park S.J., Kim J.M., Ryu J.H. Ginseng in traditional herbal prescriptions. J Ginseng Res. 2012;36:225–241. doi: 10.5142/jgr.2012.36.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J.H. Cardiovascular diseases and Panax ginseng: a review on molecular mechanisms and medical applications. J Ginseng Res. 2012;36:16–26. doi: 10.5142/jgr.2012.36.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong T.T., Cui X.M., Song Z.H., Zhao K.J., Ji Z.N., Lo C.K., Tsim K.W. Chemical assessment of roots of P. notoginseng in China: regional and seasonal variations in its active constituents. J Agric Food Chem. 2003;51:4617–4623. doi: 10.1021/jf034229k. [DOI] [PubMed] [Google Scholar]
- 4.Wei J.X., Du Y.C. Yunnan Science and Technology Press; Kunming: 1996. Modern science research and application of P. notoginseng. [Google Scholar]
- 5.Guo H.B., Cui X.M., An N., Cai G.P. Sanchi ginseng (P. notoginseng (Burkill) F. H. Chen) in China: distribution, cultivation and variations. Genet Resour Crop Evol. 2006;57:453–460. [Google Scholar]
- 6.Chen Y.J., Wang Y.Y., Feng G.Q., Li Z.Y. Relationship between root rot of P. notoginseng and ecological conditions. Yunnan Agric Sci Technol. 2001;6:33–35. [Google Scholar]
- 7.Wang C.L., Cui X.M., Li Z.Y., He C.F., Yu S.F., Luo W.F. Studies on relationship between root rot on P. notoginseng (Burk). F. H. Chen and its environmental conditions. Chin J Chin Materia Medica. 1998;23:714–716. [PubMed] [Google Scholar]
- 8.Kim J.H., Jeon Y.H., Park H., Lee B.D., Cho D.H., Park B.Y., Khan Z., Kim Y.H. The root lesion nematode, Pratylenchus subpenetrans, on ginseng (Panax ginseng) in Korea. Nematology. 2006;8:637–639. [Google Scholar]
- 9.Lee S.K. Fusarium species associated with ginseng (Panax ginseng) and their role in the root-rot of ginseng plants. Res Plant Dis. 2004;10:248–259. [Google Scholar]
- 10.Lee J.H., Yu Y.H., Kim Y.H., Ohh S.H., Park W.M. Morphological characteristics and pathogenicity of Alternaria isolates causing leaf and stem blights and black root rot of Korea ginseng. Korea J Plant Pathol. 1990;6:13–20. [Google Scholar]
- 11.Jeon Y.H., Park H., Lee B.D., Yu Y.H., Chang S.P., Kim S.G., Hwang I., Kim Y.H. First description of crown gall disease on ginseng. Plant Pathol J. 2008;24:207–210. [Google Scholar]
- 12.Ohh S.H., Lee S.K., Lee J.H., Han S.C. New root rot disease of Panax ginseng due to Ditylenchus destructor Thorne. Korea J Plant Prot. 1983;22:181–185. [Google Scholar]
- 13.Ohh S.H., Yu Y.H., Cho D.H., Lee J.H., Kim Y.H. Effect of chemical treatments on population changes of Ditylenchus destructor and responses of Panax ginseng. Korea J Plant Prot. 1986;25:169–173. [Google Scholar]
- 14.Yu Y.H., Ohh S.H. Research on ginseng diseases in Korea. Korea J Ginseng Sci. 1993;17:61–68. [Google Scholar]
- 15.Ma L., Cao Y.H., Cheng M.H., Huang Y., Mo M.H., Wang Y., Yang J.Z., Yang F.X. Phylogenetic diversity of bacterial endophytes of P. notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Antonie van Leeuwenhoek. 2013;103:299–312. doi: 10.1007/s10482-012-9810-3. [DOI] [PubMed] [Google Scholar]
- 16.Miao Z.Q., Li S.D., Liu X.Z., Chen Y.J., Li Y.H., Wang Y., Xia Z.Y., Zhang K.Q. The causal microorganisms of P. notoginseng root rot disease. Scientia Agricultura Sinica. 2006;39:1371–1378. [Google Scholar]
- 17.Handelsman J., Stabb E.V. Biocontrol of soil borne plant pathogens. Plant Cell. 1996;8:1855–1869. doi: 10.1105/tpc.8.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma C.Z., Li S.D., Gu Z.R., Chen Y.J., Zhou W., Wang Y., Liu Y.Z., Xia Z.Y., Li Y.H. Measures of integrated control of root rot complex of continuous cropping P. notoginseng and their control efficacy. Acta Agriculture Shanghai. 2006;22:63–68. [Google Scholar]
- 19.Flint M.L., Dreistadt S.H., Clark J.K. University of California Press; Oakland: 1998. Natural Enemies Handbook: the illustrated guide to biological pest control. [Google Scholar]
- 20.Hameeda B., Rupela O.P., Reddy G. Antagonistic activity of bacteria inhabiting composts against soil-borne plant pathogenic fungi. Indian J Microbiol. 2006;46:389–396. [Google Scholar]
- 21.Berg G., Opelt J., Zachow C., Lottmann J., Götz M., Costa R., Smalla K. The rhizosphere effect on bacteria antagonistic towards the pathogenic fungus Verticillium differs depending on plant species and site. FEMS Microbiol Ecol. 2006;56:250–261. doi: 10.1111/j.1574-6941.2005.00025.x. [DOI] [PubMed] [Google Scholar]
- 22.Patkowska E., Konopiński M. Antagonistic activity of selected bacteria occurring in the soil after root chicory cultivation. Plant Soil Environ. 2014;60:320–324. [Google Scholar]
- 23.Landa B.B., Hervás A., Bettiol W., Jiménez-Díaz R.M. Antagonistic activity of bacteria from the chickpea rhizosphere against Fusarium oxysporum f. sp. Ciceris. Phytoparasitica. 1997;25:305–318. [Google Scholar]
- 24.Raaijmakers J.M., Paulitz T.C., Steinberg C., Alabouvette C., Yvan M.L. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009;321:341–361. [Google Scholar]
- 25.Whipps J.M. Ecological considerations involved in commercial development of biological control agents for soil-borne diseases. In: JD van Elsas, Trevors J.T., Wellington E.M.H., editors. Modern Soil Microbiology. Marcel Dekker Inc; New York: 1997. pp. 525–546. [Google Scholar]
- 26.Huang Y., Ma L., Fang D.H., Xi J.Q., Zhu M.L., Mo M.H., Zhang K.Q., Jie Y.P. Isolation and characterisation of rhizosphere bacteria active against Meloidogyne incognita, Phytophthora nicotianae and the root knot-black shank complex in tobacco. Pest Manag Sci. 2015;71:415–422. doi: 10.1002/ps.3820. [DOI] [PubMed] [Google Scholar]
- 27.Adam M., Heuer H., Hallmann J. Bacterial antagonists of fungal pathogens also control root-knot nematodes by induced systemic resistance of tomato plants. PLoS One. 2014;9:e90402. doi: 10.1371/journal.pone.0090402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas D., Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 29.Essghaier B., Fardeau M.L., Cayol J.L., Hajlaoui M.R., Boudabous A., Jijakli H., Sadfi-Zouaoui N. Biological control of grey mould in strawberry fruits by halophilic bacteria. J Appl Microbiol. 2009;106:833–846. doi: 10.1111/j.1365-2672.2008.04053.x. [DOI] [PubMed] [Google Scholar]
- 30.Qin S., Li J., Chen H.H., Zhao G.Z., Zhu W.Y., Jiang C.L., Xu L.H., Li W.J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol. 2009;75:6176–6186. doi: 10.1128/AEM.01034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maidak B.L., Olsen G.J., Larsen N., Overbeek R., Mc-Caughey M.J., Woese C.R. The RDP (ribosomal database project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgens D.G. The Clustal X windows interface, flexible strategies for multiple sequences alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garbeva P., van Veen J.A., van Elsas J.D. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- 37.Atkinson A., Watson C.A. The beneficial rhizosphere: a dynamic entity. Appl Soil Ecol. 2000;48:99–104. [Google Scholar]
- 38.Kent A.D., Triplett E.W. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu Rev Microbiol. 2002;56:211–236. doi: 10.1146/annurev.micro.56.012302.161120. [DOI] [PubMed] [Google Scholar]
- 39.Song J.Y., Seo M.W., Kim S.I., Nam M.H., Lim H.S., Kim H.G. Genetic diversity and pathogenicity of Cylindrocarpon destructans isolates obtained from Korean Panax ginseng. Mycobiology. 2014;42:174–180. doi: 10.5941/MYCO.2014.42.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahman M., Punja Z.K. Biological control of damping-off on American ginseng (Panax quinquefolius) by Clonostachys rosea f. catenulate (= Gliocladium catenulatum) Can J Plant Pathol. 2007;29:203–207. [Google Scholar]
- 41.Ryu H., Park H., Suh D.S., Jung G.H., Park J., Lee B.D. Biological control of Colletotrichum panacicola on Panax ginseng by Bacillus subtilis HK-CSM-1. J Ginseng Res. 2014;38:215–219. doi: 10.1016/j.jgr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song M., Yun H.Y., Kim Y.H. Antagonistic Bacillus species as a biological control of ginseng root rot caused by Fusarium cf. incarnatum. J Ginseng Res. 2014;38:136–145. doi: 10.1016/j.jgr.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo R., Liu X., Li S., Miao Z. In vitro inhibition of fungal root-rot pathogens of Panax notoginseng by rhizobacteria. Plant Pathol J. 2009;25:70–76. [Google Scholar]