Abstract
Background
Minor saponins or human intestinal bacterial metabolites, such as ginsenosides Rg3, F2, Rh2, and compound K, are more pharmacologically active than major saponins, such as ginsenosides Rb1, Rb2, and Rc. In this work, enzymatic hydrolysis of ginsenoside Rb1 was studied using enzyme preparations from cultured mycelia of mushrooms.
Methods
Mycelia of Armillaria mellea, Ganoderma lucidum, Phellinus linteus, Elfvingia applanata, and Pleurotus ostreatus were cultivated in liquid media at 25°C for 2 wk. Enzyme preparations from cultured mycelia of five mushrooms were obtained by mycelia separation from cultured broth, enzyme extraction, ammonium sulfate (30–80%) precipitation, dialysis, and freeze drying, respectively. The enzyme preparations were used for enzymatic hydrolysis of ginsenoside Rb1.
Results
Among the mushrooms used in this study, the enzyme preparation from cultured mycelia of A. mellea (AMMEP) was found to convert ginsenoside Rb1 into compound K with a high yield, while those from G. lucidum, P. linteus, E. applanata, and P. ostreatus produced remarkable amounts of ginsenoside Rd from ginsenoside Rb1. The enzymatic hydrolysis pathway of ginsenoside Rb1 by AMMEP was Rb1 → Rd → F2 → compound K. The optimum reaction conditions for compound K formation from ginsenoside Rb1 were as follows: reaction time 72–96 h, pH 4.0–4.5, and temperature 45–55°C.
Conclusion
AMMEP can be used to produce the human intestinal bacterial metabolite, compound K, from ginsenoside Rb1 with a high yield and without food safety issues.
Keywords: Armillaria mellea, compound K, enzymatic hydrolysis, ginsenoside Rb1
1. Introduction
The root of Panax ginseng Meyer (Araliaceae) has been used as a folk medicine to cure diseases and maintain health for thousands of years in Asian countries such as Korea, China, and Japan. Ginseng saponins, i.e., ginsenosides, have come to be regarded as the major active components responsible for the pharmacological effects of ginseng. So far, > 150 ginsenoside derivatives have been identified from roots, leaves, fruits, flower buds, processed P. ginseng products, and other Panax spp [1]. Pharmacological effects of ginsenosides frequently depend on the type or site of the attached sugar moieties in dammarane- or oleanane-type aglycones. The major saponins in the root of P. ginseng are ginsenosides Rb1, Rb2, Rc, Rd, Re, and Rg1, which constitute > 80–90% of total ginsenosides [2].
However, these naturally occurring major ginsenosides are poorly absorbed by the human intestinal tract because of their large molecular size, low solubility, and poor permeability across the cell membrane [3], [4]. By contrast, many studies have indicated that major saponins, such as ginsenosides Rb1, Rb2, and Rc, are deglycosylated by intestinal bacteria or digestive enzymes after oral intake, and are converted to minor ginsenosides such as ginsenosides Mc1, Mc, C–O, C–Y, Rg3, F2, and compound K [5], [6]. Especially, compound K is one of the human intestinal bacterial metabolites of protopanaxadiol-type saponins, such as ginsenosides Rb1, Rb2, Rc, and Rd.
Interestingly, compound K does not exist in natural ginseng. It has various pharmacological activities such as antigenotoxic activity [7], antiallergic effect [8], and prevention of tumor invasion and metastasis [9], as well as hepatoprotective [10] and anti-inflammatory activities [11]. Various methods, such as mild acid hydrolysis [12], alkaline cleavage [13], and microbial and enzymatic degradation [14], have previously been reported for producing minor ginsenosides Rg3, F2, Rh2, and compound K from major ginsenosides. Microbial or enzymatic hydrolysis methods are considered more favorable than conventional chemical methods due to their performance under milder reaction conditions, simplicity of reaction steps, and high regio- and diastereoselectivities [15]. However, in previous studies, some attempts to produce minor ginsenosides using microbial and enzymatic methods were limited by low yields, complexity of enzyme isolation, or food safety problems.
Another approach to enhance the contents of minor ginsenosides in white ginseng, red ginseng extract, or processed ginseng products is to use mushroom mycelial fermentation. Recently, some studies reported that fermentation of ginseng extracts or ginseng extraction residue with commercial edible mushrooms such as Ganoderma lucidum, Phellinus linteus, Lentinus edodes, Grifolia frondosa, Hericium erinaceum, and Codyceps spp. could enhance the concentration of minor ginsenosides in the products. However, these studies reported that the major transformation product from major ginsenosides Rb1, Rb2, and Rc, when ginseng extracts or ginseng extraction residues were fermented with these mushrooms, was ginsenoside Rd rather than minor ginsenosides such as ginsenoside Rg3, F2, Rh2, or compound K [16], [17], [18].
In a preliminary experiment, we screened a variety of edible and/or medicinal mushrooms for activities in the production of minor ginsenosides and intestinal bacterial metabolites from ginsenoside Rb1, one of the protopanaxadiol-type saponins. From the results, the intestinal bacterial metabolite compound K was found to be produced with a high yield from ginsenoside Rb1 by enzyme preparation from the cultured mycelia of Armillaria mellea (AMMEP), whereas enzyme preparation from the cultured mycelia of G. lucidum, P. linteus, Elfvingia applanata, and Pleurotus ostreatus produced remarkable amounts of ginsenoside Rd from ginsenoside Rb1. A. mellea, known as honey mushroom, is commonly used in folk medicine as a health-promoting food in various forms and for dietary supplementation [19], [20]. In traditional Chinese medicine, A. mellea is used for treating a variety of complaints including palsy, headache, hypertension, insomnia, vertigo, and neurasthenia. It also exerts neuroprotective effects [19], [21]. Therefore, this paper describes the enzymatic formation of the human intestinal microbial metabolite compound K from ginsenoside Rb1 by AMMEP.
2. Materials and methods
2.1. Materials
Ginsenoside Rb1 was isolated from fresh P. ginseng roots according to the usual procedure [22]. The isolated compound was identified by comparing its spectral data and retention time from HPLC with those of an authentic sample. Authentic standards and standard mixtures of ginsenosides Rb1, Rb2, Rc, Rd, Rg3 (S), F2, Rh2 (S), and compound K were kindly donated by the Korea Ginseng Corporation Research Institute, Daejeon, Korea. The concentration of each ginsenoside in the standard mixture was 1,000 ppm (1.0 mg/mL). Silica gel F254 TLC plate and silica gel 60 (0.040–0.063 mm) for column chromatography were purchased from Merck Co. (Darmstadt, Germany). The compounds p-nitrophenol (PNP), PNP-α-D-glucopyranoside, PNP-β-D-glucopyranoside, PNP-α-D-galactopyranoside, PNP-β-D-galactopyranoside, PNP-α-L-arabinofuranoside, PNP-α-L-arabinopyranoside, and PNP-β-D-xylopyranoside were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). Other reagents used were of analytical grade and were procured from commercial sources.
2.2. TLC analysis
TLC was performed on silica gel 60 F254 plate with CHCl3–CH3OH–H2O (65:35:10, v/v/v, lower phase) as the developing solvent. The spots on the TLC were detected by spraying 10% (w/v) H2SO4 in ethanol and heating at 120°C for 10 min.
2.3. HPLC and LC/MS analysis
HPLC analysis was performed using an HPLC system (Waters, Millford, MA, USA) equipped with a 600E system controller, a 717 plus autosampler, and a 486 UV detector with a YMC-Pack Pro C18 RS column (250 mm × 4.6 mm, 5 μm ID; YMC Co., Ltd, Shimogyo-ku, Kyoto, Japan). The mobile phase consisted of water (A) and acetonitrile (B) at ratios of (A:B) 70:30 (0–5 min), 70:30 (5–15 min), 43:57 (15–25 min), 30:70 (25–30 min), and 70:30 (30–40 min) at a flow rate of 0.8 mL/min. Detection absorbance was 203 nm. LC/MS analysis was performed with a Waters ACQUITY UPLC system composed of a binary solvent manager and a photodiode array detector at 203 nm. Chromatographic separation was accomplished on an ACQUITY BEH C18 column (100 mm × 2.1 mm, 1.7 μm; Waters). The column temperature was 40°C. The binary gradient elution system consisted of 0.01% formic acid in water (A) and 0.01% formic acid in acetonitrile (B). Separation was achieved using the following gradient program: 0–1 min (15% B), 1–15 min (30% B), 15–20 min (38% B), 20–24 min (43% B), 27–31 min (55% B), 31–35 min (70% B), 35–38 min (90% B), and 38–43.0 min (15% B). The flow rate was set at 0.6 mL/min. MS analysis was performed on a Waters Xevo quadruple-time-of-flight MS equipped with an electrospray ionization source under negative ion mode. The conditions for MS analysis were as follows: drying gas N2 flow rate of 12 L/min, cone gas temperature of 350°C, and nebulizer pressure of 50 psi. The capillary voltage was set to 4.0 kV.
2.4. Preparation of crude enzymes from cultured mycelia of mushrooms
Strains of A. mellea, G. lucidum, P. linteus, E. applanata, and P. ostreatus were purchased from Korean Agricultural Culture Collection, Suwon, Korea, and preincubated on a potato dextrose agar (DB Difco, NJ, USA) for 6 d at 25°C. All nutrient media were sterilized at 121°C for 30 min. The strains were inoculated with germinated-malt media (11 Brixo) saccharified at 65°C with fourfold tap water for 8 h, and then cultured for 2 wk at 25°C. The scale-up production of mushroom mycelia was performed in a 4 L Erlenmeyer flask containing 1 L of germinated-malt media for 3 wk at 25–26°C with gentle shaking (120 rpm). The culture broth was centrifuged (9,000g, 20 min) to separate the mycelia from the broth. The mycelia mass was washed with distilled water to remove residual broth, and then lyophilized. Each lyophilized mycelium (100 g) was mixed with 500 mL of 0.1M sodium phosphate buffer (pH 6.0) with gentle stirring for 12 h at 4°C and then homogenized with an Omni mixer homogenizer (Omni International, NW Kennesaw, GA, USA) for 1 min at 4°C. The slurry was squeezed through a cheese cloth and the filtrate was centrifuged at 9000g for 20 min at 4°C. Solid ammonium sulfate was added to the supernatant (400 mL), initially up to 30% and eventually to 80% saturation. After centrifugation at 9,000g for 20 min at 4°C, the precipitate was dissolved in 10mM sodium acetate buffer (pH 6.0). After overnight dialysis, the solution was centrifuged at 9,000g for 20 min at 4°C, and the supernatant was lyophilized.
2.5. Enzyme assay and enzyme characterization
Activity of β-glucosidase was assayed using PNP-β-D-glucopyranoside as a substrate. A reaction mixture (1.0 mL) containing 0.1 mL of PNP-β-D-glucopyranoside (10mM), 0.1 mL of enzyme solution containing 0.02 mg of enzyme preparation, and 0.8 mL of 0.1M sodium acetate buffer (pH 4.8) was incubated for 30 min at 37°C. The reaction was terminated by adding 1.0 mL of 0.1M Na2CO3 solution. The released PNP was measured immediately using a UV–visible spectrophotometer (UV-1601; Shimadzu, Tokyo, Japan) at 400 nm. The amount of PNP released was quantified using the concentration plot of the PNP standard. One unit of β-glucosidase activity was expressed as the amount of enzyme required to release 1 μmole of PNP per min under the assay conditions.
The effect of pH on the hydrolyzing activity of PNP-β-D-glucopyranoside was assayed at 37°C in the following buffer solutions (each at 0.1M): glycine–HCl (pH 3.0), sodium acetate (pH 4.0–5.5), sodium phosphate (pH 6.0–7.0), Tris-HCl (pH 8.0–9.0), and glycine–NaOH (pH 10.0). The pH stability of the enzyme was examined by measuring residual activity after incubating it in each buffer for 4 h at 37°C. The results were expressed as a percentage of the activity obtained at the optimum pH. The effect of temperature on the hydrolyzing activity of PNP-β-D-glucopyranoside was assayed by incubating the enzyme for 4 h at various temperatures ranging from 25°C to 70°C at pH 4.8. The temperature stability of the enzyme was determined by measuring the residual activity after preincubating the enzyme in 0.1M acetate buffer (pH 4.8) for 4 h at different temperatures (20–70°C). Immediately after cooling the test samples in an ice bath, the residual activity was determined using PNP-β-D-glucopyranoside as a substrate. The results were expressed as a percentage of the activity obtained at an optimum temperature. The following substrates were also tested with the same assay method used for PNP-β-D-glucopyranoside: PNP-α-D-glucopyranoside, PNP-α-D-galactopyranoside, PNP-β-D-galactopyranoside, PNP-α-L-arabinofuranoside, PNP-α-L-arabinopyranoside, and PNP-β-D-xylopyranoside.
2.6. Enzymatic hydrolysis of ginsenoside Rb1
To investigate the hydrolysis pattern of ginsenoside Rb1 by enzyme preparations from the cultured mushroom mycelia, the reaction mixture (2.0 mL) containing 2.0 mg of ginsenoside Rb1 in 0.2 mL of methanol and each enzyme preparation containing 1.1 U of β-glucosidase activity in 1.8 mL of 0.1M sodium acetate buffer (pH 4.8) were incubated for 96 h at 37°C. The reaction mixture was extracted twice with 2.0 mL of n-butanol saturated with water. The n-butanol fraction was concentrated to dryness in vacuo, and the residue was dissolved in 1.0 mL of methanol. To investigate the time course of ginsenoside Rb1 hydrolysis by AMMEP, a 20 mL reaction mixture containing 20 mg of ginsenoside Rb1 in 2.0 mL methanol, 30 mg of AMMEP containing 11 U of β-glucosidase, and 0.1M sodium acetate buffer (pH 4.8) was incubated for 96 h at 37°C with gentle shaking. The reaction mixture (2 mL) was withdrawn at regular intervals, and extracted twice with 2.0 mL of n-butanol saturated with water. The n-butanol fraction was concentrated to dryness in vacuo. The residue was dissolved in 1.0 mL of methanol, and subjected to TLC, HPLC, and LC/MS analyses.
For enzyme characterization of the ginsenoside Rb1 hydrolysis, the reaction mixture (2.0 mL) containing 2.0 mg of ginsenoside Rb1 in 0.2 mL methanol, AMMEP containing 1.1 U of β-glucosidase, and 1.8 mL of 0.1M acetate buffer (pH 4.8) was incubated at 37°C with gentle shaking. After 24 h of incubation, each reaction mixture was extracted twice with 2.0 mL of n-butanol saturated with water. The n-butanol fraction was concentrated to dryness in vacuo, and the residue was dissolved in 1.0 mL of methanol. The effect of pH on the hydrolyzing activity of ginsenoside Rb1 was examined using ginsenoside Rb1 as a substrate at 37°C in the following buffer solutions (each at 0.1M): glycine–HCl (pH 3.0), sodium acetate (pH 4.0–5.5), sodium phosphate (pH 6.0–7.0), Tris-HCl (pH 8.0), and glycine–NaOH (pH 9.0–10.0). The effect of temperature on the hydrolyzing activities of ginsenoside Rb1 were assayed by incubating the reaction mixture for 24 h at various temperatures ranging from 30°C to 70°C at pH 4.8. To determine pH and temperature stability, the enzyme solution was preincubated in the pH range of 4.0–10.0 and in the temperature range of 30–70°C for 4 h. To determine residual enzyme activity, the reaction mixture (2.0 mL) containing 2.0 mg of ginsenoside Rb1 in 0.2 mL methanol, 0.2 mL of enzyme solution, and 1.6 mL of 0.1M acetate buffer (pH 4.8) was incubated for 24 h at 37°C with gentle shaking. The effect of the substrate concentration on the formation of compound K was examined. A reaction mixture (2.0 mL) containing various amounts of ginsenoside Rb1 (0.5–5.0 mg) in 0.2 mL of methanol and 1.5 mg of AMMEP containing 0.55 U of β-glucosidase (the weight ratio of ginsenoside Rb1 to AMMEP being 0.3–3.3:1.0) in 1.8 mL of 0.1M sodium acetate buffer (pH 4.8) was incubated for 24 h at 37°C. The concentrations of each ginsenoside in the reaction mixtures were calculated from the calibration curves of standard mixtures. All experiments were performed in triplicate, and the data were expressed as the mean ± standard deviation.
3. Results and discussion
3.1. Enzymatic hydrolysis of ginsenoside Rb1
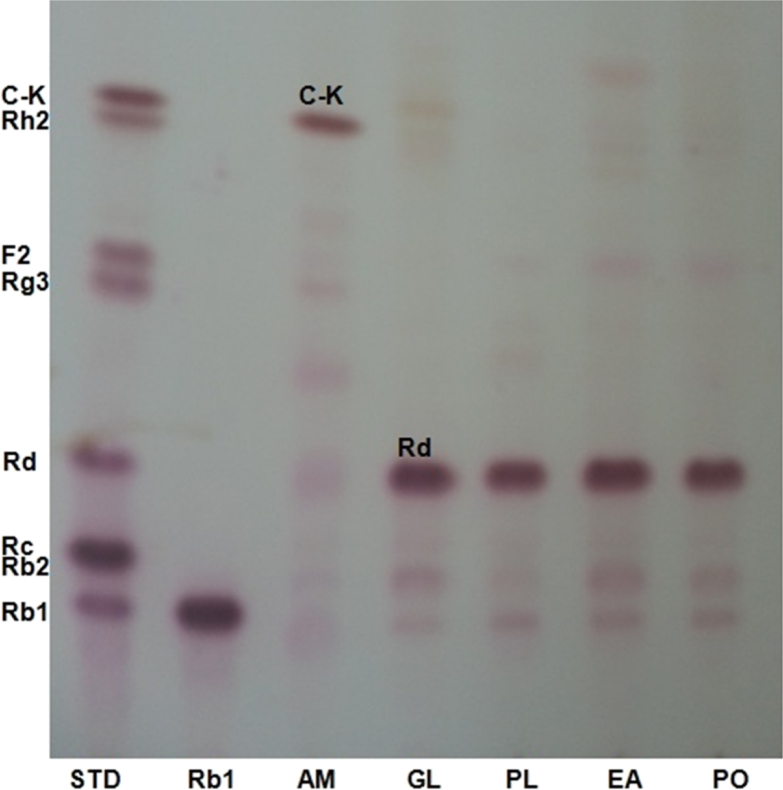
Basidiomycetes mushrooms are not only an important natural source of foods and therapeutics, but also microorganisms of great interest that secrete various hydrolytic enzymes such as cellulase, β-glucosidase, and endo- and exoglucanase [23], [24], [25]. Recently, some studies reported that fermentation of ginseng extracts or ginseng extraction residues using edible or medicinal mushrooms could enhance the contents of minor ginsenosides in their products [16], [17], [18]. Therefore, these results indicate that mushroom mycelia may convert major ginsenosides into deglycosylated minor ginsenosides by their carbohydrate hydrolases. We investigated the possibility of producing minor saponins and/or human intestinal bacterial metabolites from ginsenoside Rb1 using crude enzyme preparations from cultured mycelia of five mushrooms. From the results, we found that compound K was produced with a high yield from ginsenoside Rb1 by AMMEP, whereas enzyme preparations from G. lucidum, P. linteus, E. applanata, and P. ostreatus produced remarkable amounts of ginsenoside Rd from ginsenoside Rb1 (Fig. 1).
Fig. 1.
TLC analysis of hydrolysis of ginsenoside Rb1 by enzyme preparations from cultured mycelia of mushrooms. The reaction mixture (2.0 mL) containing 2.0 mg of ginsenoside Rb1 in 0.2 mL of methanol and each enzyme preparation containing 1.1 U β-glucosidase activity in 1.8 mL of 0.1M sodium acetate buffer (pH 4.8) was incubated for 96 h at 37°C. The reaction mixture was extracted twice with 2.0 mL of n-butanol saturated with water. The n-butanol fraction was concentrated to dryness in vacuo, and the residue was dissolved in 1.0 mL of methanol. TLC of the methanol extract was performed on a silica gel 60 F254 plate with CHCl3–CH3OH–H2O (65:35:10, lower phase). The spots on the TLC were detected by spraying 10% H2SO4 in ethanol and heating at 120°C for 10 min. AM, Armillaria mellea; EA, Elfvingia applanata; GL, Ganoderma lucidum; PL, Phellinus linteus; PO, Pleurotus ostreatus.
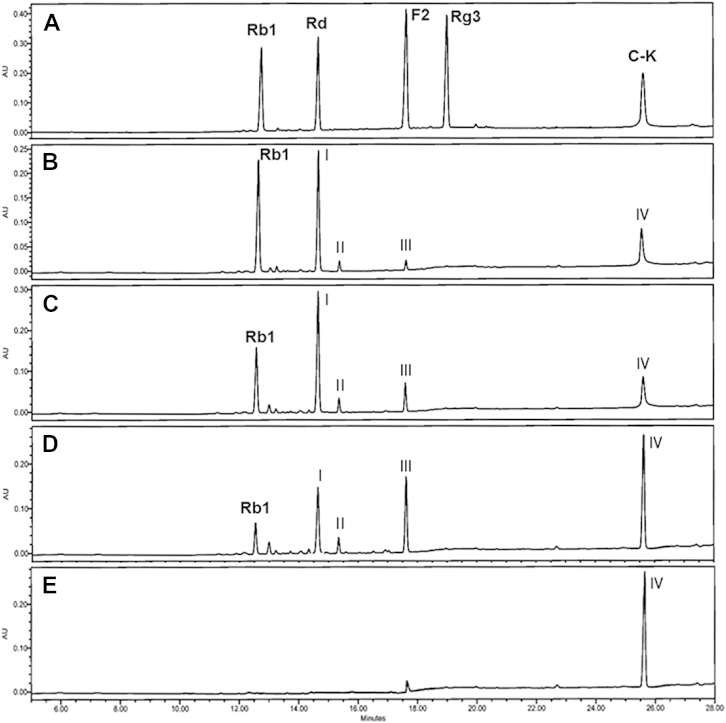
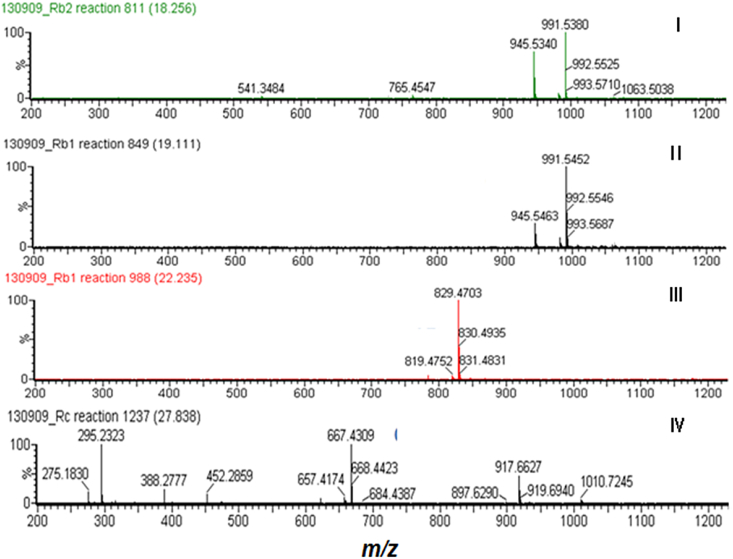
To investigate the pattern of hydrolysis of ginsenoside Rb1 by AMMEP, the sample was withdrawn at regular intervals during enzymatic hydrolysis and analyzed by HPLC (Fig. 2). HPLC profiles showed four new peaks, indicating that ginsenoside Rb1 was gradually hydrolyzed to four metabolites (Ⅰ, Ⅱ, Ⅲ, and Ⅳ). Ginsenoside Rb1 was hydrolyzed to metabolite Ⅰ in the early stage of the reaction. After 96 h, almost all of ginsenoside Rb1 and metabolites I, II, and III were hydrolyzed to metabolite IV. These results indicate that metabolites I, II, and III were intermediate metabolites, while metabolite IV was the final product. The reaction mixture was analyzed by electrospray ionization–LC/MS to determine the molecular weight of new metabolites. Peak Ⅰ in Fig. 2 appeared as a quasimolecular ion peak at m/z 991.54 [M-H + formic acid]− with a molecular ion peak [M-1]− at m/z 945.53, corresponding to the molecular formula C48H82O18 (molecular weight 946.55; Fig. 3). Thus, Peak I was attributed to ginsenoside Rd. Peak II also appeared as a quasimolecular ion peak at m/z 991.55 [M-H + formic acid]− with a molecular ion peak at m/z 945.55 [M-1]−, corresponding to the molecular formula C48H82O18 (molecular weight 946.55). Therefore, Peak II was attributed to gypenoside XVII. Peaks III and IV appeared as molecular ion peaks at m/z 829.47 [M-H + formic acid]− and m/z 667.43 [M-H + formic acid]−, corresponding to quasimolecular ions of ginsenosides F2 and compound K, respectively (Fig. 3). These results indicated that the four metabolites in Fig. 3 were ginsenoside Rd (Peak I), gypenoside XVII (Peak II), ginsenoside F2 (Peak III), and compound K (Peak IV) [26].
Fig. 2.
HPLC analysis of hydrolysis of ginsenoside Rb1 by AMMEP. The reaction conditions are the same as in Fig. 1. HPLC analysis was performed under the conditions described in the Materials and methods section. (A) Mixture of authentic ginsenosides, (B) 24-h reaction, (C) 48-h reaction, (D) 72-h reaction, and (E); 96-h reaction. AMMEP, enzyme preparation from cultured mycelia of A. mellea.
Fig. 3.
ESI–MS spectra (negative ion mode) of hydrolysis of ginsenoside Rb1 by AMMEP. AMMEP, enzyme preparation from cultured mycelia of A. mellea; ESI, electrospray ionization; I, ginsenoside Rd; II, gypenoside XVII; III, ginsenoside F2; IV, compound K.
3.2. Enzyme characterization
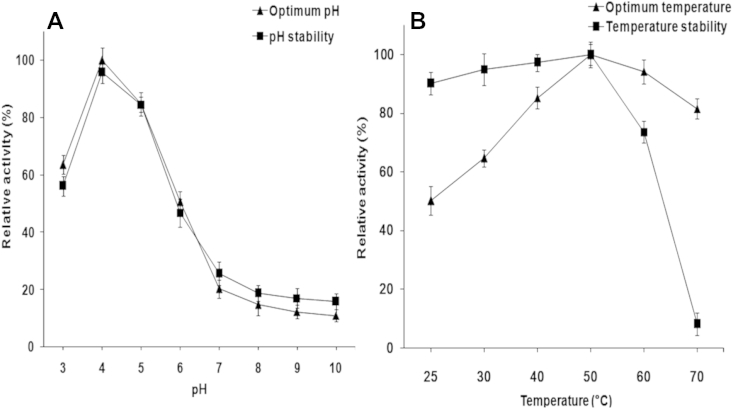
When assayed using PNP-β-D-glucopyranoside as a substrate, β-glucosidase activity in AMMEP showed optimal activity and stability at pH 4.0, and was stable between pH 4 and 5. The enzyme became vulnerable to inactivation below and above this range, and became inactivated on the extreme alkaline side of the pH profile (above pH 7.0; Fig. 4A). The optimum temperature and thermal stability of this enzyme peaked at 50°C. The enzyme was stable in a broad temperature range (25–50°C), but the activity decreased sharply above 60°C (Fig. 4B).
Fig. 4.
Hydrolysis of PNP-β-D-glucopyranoside by AMMEP. Effects of (A) pH and (B) temperature on the hydrolysis of PNP-β-D-glucopyranoside. A reaction mixture (1.0 mL) containing 0.1 mL of enzyme solution with 0.02 mg of enzyme preparation, 0.1 mL of PNP-β-D-glucopyranoside (10mM), and 0.8 mL of sodium acetate buffer (0.1M, pH 4.8) was incubated for 30 min at 37°C. The reaction was terminated by adding 1.0 mL of 0.1M Na2CO3 solution. The released PNP was measured using a UV–visible spectrophotometer at 400 nm. AMMEP, enzyme preparation from cultured mycelia of Armillaria mellea; PNP, p-nitrophenol.
The substrate specificity of AMMEP β-glucosidase was assayed using 10mM of PNP glycosides with α- and β-configuration. The results showed that AMMEP β-glucosidase had strong hydrolytic activities toward PNP-β-D-glucopyranoside and PNP-α-D-galactopyranoside. As shown in Table 1, the enzyme preparations from cultured mycelia of G. lucidum, A. applanata, and P. ostreatus also showed strong hydrolytic activities toward PNP-β-D-glucopyranoside. Interestingly, these enzyme preparations exhibited no hydrolyzing activities for converting ginsenoside Rb1 into ginsenoside F2 or compound K. These results demonstrate that β-glucosidases from G. lucidum, P. linteus, E. applanata, and P. ostreatus mycelia selectively hydrolyzed β-(1→6)-glucosidic linkage of ginsenoside Rb1, without attacking β-(1→2)-glucosidic linkage in spite of their potent β-glucosidase activities. These results were similar to those of ginsenoside Rb1-hydrolyzing β-D-glucosidases purified from China white jade snail [27] and Cladosporium fulvum [28]. The reason for this might be that the spatial configuration of ginsenoside molecule blocked the attack of enzyme to β-(1→2)-glucosidic linkage in linked at the C3-hydroxy group of aglycone [28].
Table 1.
Relative activity of enzyme preparations from mushroom mycelia on various chromogenic substrates as PNP release.1)
| Substrate | Armillaria mellea | Ganoderma lucidum | Phellinus linteus | Elfvingia applanata | Pleurotus ostreatus |
|---|---|---|---|---|---|
| PNP-α-D-glucopyranoside | 13.1 ± 1.1 | 37.5 ± 1.8 | 7.1 ± 0.5 | 40.1 ± 0.9 | 16.1 ± 0.4 |
| PNP-β-D-glucopyranoside | 100 ± 2.7 | 126 ± 4.8 | 46.5 ± 1.1 | 181 ± 1.7 | 201 ± 3.1 |
| PNP-α-D-galactopyranoside | 68.6 ± 2.6 | 350 ± 3.4 | 10.9 ± 0.2 | 181 ± 2.6 | 11.2 ± 0.9 |
| PNP-β-D-galactopyranoside | 9.6 ± 0.2 | 7.2 ± 0.8 | 50.4 ± 0.6 | 7.6 ± 0.3 | 7.9 ± 0.1 |
| PNP-α-L-arabinopyranoside | 4.4 ± 0.5 | 3.7 ± 0.3 | 0 | 6.9 ± 0.2 | 0 |
| PNP-α-L-arabinofuranoside | 6.8 ± 0.4 | 46.4 ± 1.1 | 0 | 52.4 ± 1.1 | 5.7 ± 1.2 |
| PNP-β- D-xylopyranoside | 6.4 ± 0.3 | 13.8 ± 1.2 | 4.3 ± 0.2 | 5.1 ± 0.2 | 31.8 ± 0.4 |
PNP, p-nitrophenol
The activity was assayed with 10mM of substrates at 37°C. The relative activity was based on the hydrolytic activity of PNP-β-D-glucopyranoside in the enzyme preparation from A. mellea
3.3. Effect of reaction conditions on the hydrolysis of ginsenoside Rb1
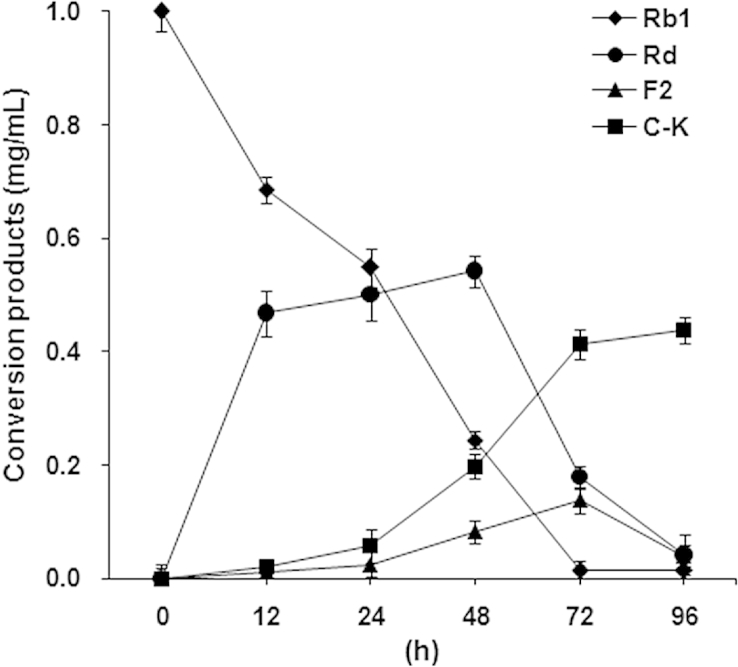
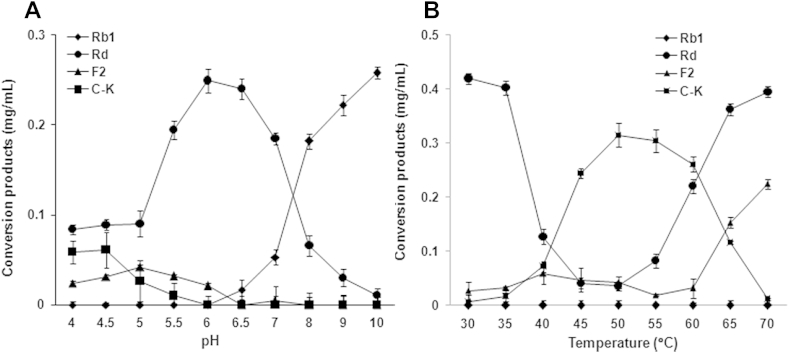
In relation to the effect of reaction time, ginsenoside Rb1 was gradually hydrolyzed into ginsenoside Rd at the early stages of the reaction (Fig. 5). However, a remarkable formation of compound K was not observed within 24 h. When the reaction time was increased from 24 h to 72 h, a significant amount of ginsenoside Rd was converted into ginsenoside F2 or compound K. After 72–96 h of incubation, ginsenosides Rd and F2 were almost completely hydrolyzed to compound K. The optimum pH range for compound K formation from ginsenoside Rb1 was 4.0–4.5 (Fig. 6A). These results indicate that ginsenoside Rb1 was converted to compound K via Rd and F2 at pH 4.0–4.5. When the pH was increased to 5.0–6.0, the rate of formation of compound K decreased. Furthermore, formation of compound K was not observed in the pH range of 6.5–10.0. From the above result, it was concluded that a weak acidic condition is ideal for the formation of compound K from ginsenoside Rb1. The influence of pH on enzymatic hydrolysis of ginsenoside Rb1 to compound K has extensively been studied with enzymes obtained from various sources, and the optimum pH for ginsenoside hydrolysis of many microbial enzymes is in the range of 3.5–7.5 [14], [29].
Fig. 5.
Time course of the hydrolysis of ginsenoside Rb1 by AMMEP. The reaction was performed under the conditions described in the Materials and methods section. AMMEP, enzyme preparation from cultured mycelia of A. mellea; C–K, compound K.
Fig. 6.
Hydrolysis of ginsenoside Rb1 by AMMEP. Effects of (A) pH and (B) temperature on the hydrolysis of ginsenoside Rb1. To determine the optimum pH and temperature, the reaction mixture (2.0 mL) containing 2.0 mg of ginsenoside Rb1 in 0.2 mL of methanol and AMMEP containing 1.1 U of β-glucosidase activity in 1.8 mL of 0.1M acetate buffer (pH 4.8) was incubated for 24 h 37°C. The optimum temperature was determined by incubating the reaction mixture for 24 h at temperatures in the range of 30–70°C. Concentrations of the hydrolysis products were analyzed by HPLC under the conditions described in the Materials and methods section. AMMEP, enzyme preparation from cultured mycelia of A. mellea; C–K, compound K.
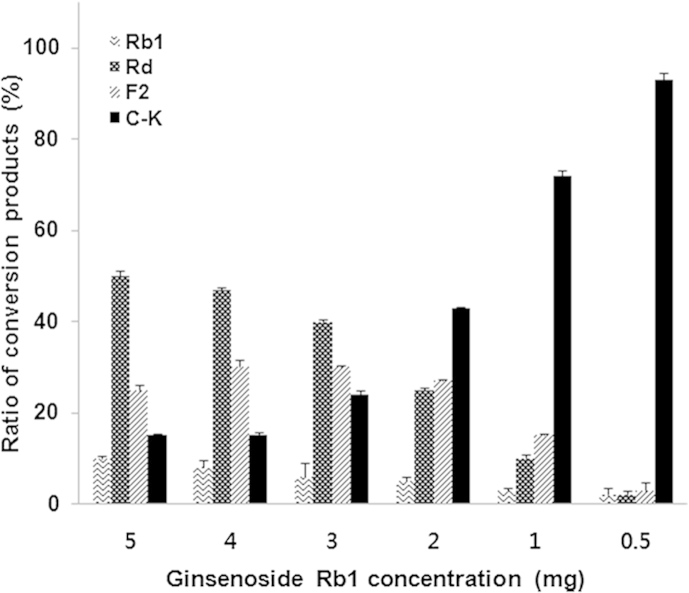
As shown in Fig. 6B, the ginsenoside Rb1-hydrolyzing activity of AMMEP β-glucosidase was greatly influenced by the reaction temperature. Ginsenoside Rb1 was incubated with AMMEP for 24 h at different temperatures (30–70°C). Initially, at 30–35°C, ginsenoside Rb1 began to hydrolyze into ginsenosides Rd and F2. As the reaction temperature was increased to 45–55°C, intermediate metabolites ginsenosides Rd and F2 were significantly hydrolyzed into compound K. When the reaction temperature was increased to 60–70°C, formation of compound K decreased. The results suggest that the optimum temperature for the formation of compound K from ginsenoside Rb1 by AMMEP β-glucosidase is in the range of 45–55°C. Generally, the optimum temperatures for ginsenoside-hydrolyzing enzymes from human intestinal bacteria and soil microorganisms are in the range of 37–45°C [14], [29]. The effect of ginsenoside Rb1 concentration on the formation of compound K was examined. The results showed that as the concentration of ginsenoside Rb1 in the reaction mixture was increased, the quantity of compound K decreased. Ginsenoside Rb1 was also completely hydrolyzed to compound K when incubated with a large quantity of AMMEP (the weight ratio of ginsenoside Rb1 to AMMEP being 1:3; Fig. 7).
Fig. 7.
Effects of ginsenoside Rb1 concentration on the formation of compound K by AMMEP. The reaction mixture (2.0 mL) containing various amounts of ginsenoside Rb1 ranging from 0.5 mg to 5.0 mg in 0.2 mL of methanol, and 1.5 mg of AMMEP (the weight ratio of ginsenoside Rb1 to AMMEP being 0.3–3.3:1.0) in 1.8 mL of 0.1M sodium acetate buffer (pH 4.8) was incubated for 24 h at 45°C. The ratio of conversion products was expressed as percentage of the HPLC peak area. AMMEP, enzyme preparation from cultured mycelia of Armillaria mellea; C–K, compound K.
3.4. Pathway of enzymatic formation of compound K from ginsenoside Rb1
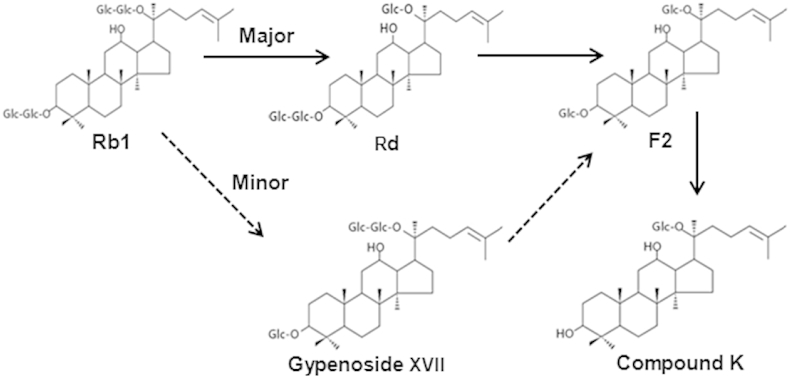
As ginsenoside Rb1 contains four molecules of glucose with β-glucosidic linkages, it can be hydrolyzed by β-glucosidase through different pathways with multiple steps. Judging from the results of TLC, HPLC, and LC/MS analyses of the hydrolysis products of ginsenoside Rb1 in this study, β-glucosidase from AMMEP first hydrolyzes the outer glucose moiety attached to the C-20 position of aglycone to produce ginsenoside Rd. Thereafter, the outer glucose moiety is attached to the C-3 position of the aglycone to produce ginsenoside F2. This is followed by detachment of the inner glucose moiety from the C-3 position to produce compound K. Therefore, we suggested that AMMEP β-glucosidase exploits the hydrolytic pathway of ginsenoside Rb1 → Rd → F2 → compound K, along with a minor pathway involving ginsenoside Rb1 → gypenoside XVII → F2 → compound K (Fig. 8).
Fig. 8.
Proposed pathway of the enzymatic hydrolysis of ginsenoside Rb1 by AMMEP. AMMEP, enzyme preparation from cultured mycelia of Armillaria mellea.
The hydrolytic pathway is determined by the stereospecific preference of the enzyme for the sugars attached to C-3 or C-20 position of the aglycone in ginsenoside Rb1 [29]. Previous studies showed that the β-glucosidases from Lactobacillus pentosus DC101 [30] and Paecilomyces bainier [31] exploit the hydrolytic pathway involving ginsenoside Rb1 → Rd → F2 → compound K. However, β-glucosidases from Fusobacterium sp [32]. exploit two hydrolytic pathways: ginsenoside Rb1 → Rd → F2 → compound K and gypenoside XVII → F2 → compound K.
In conclusion, we prepared enzyme preparations from cultured mycelia of five edible and/or medicinal mushrooms. Among them, AMMEP β-glucosidase showed potent activity in hydrolysis of ginsenoside Rb1 to compound K. The optimum conditions for compound K formation from ginsenoside Rb1 were as follows: reaction time of 72–96 h, pH of 4.0–4.5, and temperature range of 45–55°C. This result suggests that AMMEP can be used to produce the human intestinal bacterial metabolite compound K from ginsenoside Rb1 with a high yield and without food safety problems.
Conflicts of interest
All contributing authors declare no conflicts of interest.
Acknowledgments
This work was supported in part by a Korea Government Scholarship Program (J.U.).
References
- 1.Christensene L.P. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv Food Nutr Res. 2009;55:1–99. doi: 10.1016/S1043-4526(08)00401-4. [DOI] [PubMed] [Google Scholar]
- 2.Liu C.Y., Song J.G., Li P.F., Yu H.S., Jin F.X. Ginsenoside contents in three different ginseng. J Dalian Polytechnic Univ. 2011;30:79–82. [Google Scholar]
- 3.Xu Q.F., Fang X.L., Chen D.F. Pharmacokinetics and bioavailability of ginsenoside Rb1 and Rg1 from Panax notoginseng in rats. J Ethnopharmacol. 2005;84:187–192. doi: 10.1016/s0378-8741(02)00317-3. [DOI] [PubMed] [Google Scholar]
- 4.Yu K., Chen F., Li C. Absorption, disposition, and pharmacokinetics of saponins from Chinese medicinal herbs: what do we know and what do we need to know more? Curr Drug Metab. 2012;13:577–598. doi: 10.2174/1389200211209050577. [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa H. Proof of the mysterious efficacy of ginseng: basic and clinical trials: metabolic activation of ginsenoside: deglycosylation by intestinal bacteria and esterification with fatty acid. J Pharmacol Sci. 2004;95:153–157. doi: 10.1254/jphs.fmj04001x4. [DOI] [PubMed] [Google Scholar]
- 6.Kato T., Kida T., Kanaoka M., Hattori M., Kobashi K. Drug metabolism: intestinal bacterial hydrolysis is required for the appearance of compound K in rat plasma after oral administration of ginsenoside Rb1 from Panax ginseng. J Pharm Pharmacol. 1998;50:1155–1160. doi: 10.1111/j.2042-7158.1998.tb03327.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee B.H., Lee S.J., Hui J.H., Lee S.Y., Sung J.H., Huh J.D., Moon C.K. In vitro antigenotoxic activity of novel ginseng saponin metabolites formed by intestinal bacteria. Planta Med. 1998;64:500–503. doi: 10.1055/s-2006-957501. [DOI] [PubMed] [Google Scholar]
- 8.Bae E.A., Choo M.K., Park S.Y., Shin S.Y., Kim D.H. Metabolism of ginsenoside Rc by human intestinal bacteria and its related antiallergic activity. Biol Pharm Bull. 2002;25:743–747. doi: 10.1248/bpb.25.743. [DOI] [PubMed] [Google Scholar]
- 9.Nag S.A., Qin J.J., Wang W., Wang M.H., Wang H., Zhang R. Ginsenosides as anticancer agents: in vitro and in vivo activities, structure–activity relationships, and molecular mechanisms of action. Front Pharmacol. 2012;3:1–18. doi: 10.3389/fphar.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H.U., Bae E.A., Han M.J., Kim D.H. Hepatoprotective effect of ginsenoside Rb1 and compound K on tert-butyl hydroperoxide-induced liver injury. Liver Int. 2005;25:1069–1073. doi: 10.1111/j.1478-3231.2005.01068.x. [DOI] [PubMed] [Google Scholar]
- 11.Park J.S., Shin J.A., Jung J.S., Hyun J.W., Van Le T.K.V., Kim D.H., Park E.M., Kim H.S. Anti-inflammatory mechanism of compound K in activated microglia and its neuroprotective effect on experimental stroke in mice. J Pharmacol Exp. 2012;341:59–67. doi: 10.1124/jpet.111.189035. [DOI] [PubMed] [Google Scholar]
- 12.Han B.H., Park M.H., Han Y.N., Woo L.K., Sankawa U., Yahara S., Tanaka O. Degradation of ginseng saponins under mild acid condition. Planta Med. 1982;44:146–149. doi: 10.1055/s-2007-971425. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y.J., Nose M., Ogihara Y. Alkaline cleavage of ginsenosides. Chem Pharm Bull. 1987;35:1653–1655. doi: 10.1248/cpb.35.1653. [DOI] [PubMed] [Google Scholar]
- 14.Yang X.D., Yang Y.U., Ouyang D.S., Yang G.P. A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia. 2015;100:208–220. doi: 10.1016/j.fitote.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Faber K. Biotransformation in organic chemistry. 3rd ed. Springer-Verlag; Berlin: 1997. Advantages and disadvantages of biocatalysts. [Google Scholar]
- 16.Bae S.H., Lee H.S., Kim M.R., Kim S.Y., Kim J.M., Suh H.J. Changes of ginsenoside content by mushroom mycelial fermentation in red ginseng extract. J Ginseng Res. 2011;35:235–242. doi: 10.5142/jgr.2011.35.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu J.S., Lee H.J., Bae S.H., Kim S.Y., Park Y.H., Suh H.J. The bioavailability of red ginseng extract of fermented by Phellinus linteus. J Ginseng Res. 2013;37:108–116. doi: 10.5142/jgr.2013.37.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.By Hsu, Lu T.J., Chen C.H., Wang S.J., Hwang L.S. Biotransformation of ginsenoside Rd in the ginseng extraction residue by fermentation with lingzhi (Ganoderma lucidum) Food Chem. 2013;141:4186–4193. doi: 10.1016/j.foodchem.2013.06.134. [DOI] [PubMed] [Google Scholar]
- 19.Gao L.W., Li W.Y., Zhao Y.L., Wang J.W. The cultivation, bioactive components and pharmacological effects of Armillaria mellea. Afr J Biotechnol. 2009;8:7383–7440. [Google Scholar]
- 20.Lung M.Y., Chang Y.C. Antioxidant properties of the edible basidiomycete Armillaria mellea in submerged cultures. Int J Mol Sci. 2011;12:6367–6384. doi: 10.3390/ijms12106367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y.J., Wue S.Y., Chen C.C., Tsaog Y.L., Hsu N.C., Chou Y.C., Huang H.L. Armillaria mellea component armillarikin induces apoptosis in human leukemia cells. J Funct Foods. 2014;6:196–204. [Google Scholar]
- 22.Sanada S., Kondo N., Shoji J., Tanaka O., Shibata S. Studies on the saponins of ginseng. Ⅰ. Structures of ginsenoside-Ro, -Rb1, -Rb2, -Rc, and -Rd. Chem Pharm Bull. 1974;22:421–428. [Google Scholar]
- 23.Buswell J.A., Cai Y.J., Chang S.T., Peberdy J.F., Fu S.Y., Yu H.S. Lignocellulolytic enzyme profiles of edible mushroom fungi. World J Microbiol Biotechnol. 1996;12:537–542. doi: 10.1007/BF00419469. [DOI] [PubMed] [Google Scholar]
- 24.Baldrian P., Valášková V. Degradation of cellulose by basiomycetous fungi. FEBS Microbiol Rev. 2008;32:501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 25.Mfombep P.M., Senwo Z.N., Isikhuemhen O.S. Enzymatic activities and kinetic properties of β-glucosidase from selected white rot fungi. Adv Biol Chem. 2013;3:198–207. [Google Scholar]
- 26.Sun B.S., Xu M.Y., Li Z., Wang B.W., Sung C.K. UPLC-Q-TOF-MS/MS analysis for steaming times-dependent profiling of steamed Panax quinquefolius and its ginsenosides transformations induced by repetitious steaming. J Ginseng Res. 2012;36:277–290. doi: 10.5142/jgr.2012.36.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luan H., Liu X., Qi X., Hu Y., Hao D., Cui Y., Yang L. Purification and characterization of a novel stable ginsenoside Rb1-hydrolyzing β-D-glucosidase from China white jade snail. Process Biochem. 2006;41:1974–1980. [Google Scholar]
- 28.Zhao X., Gao L., Wang J., Bi H., Gao J., Du X., Zhou Y., Tai G. A novel ginsenoside Rb1-hydrolyzing β-D-glucosidase from Cladosporium fulvum. Process Biochem. 2009;44:612–618. [Google Scholar]
- 29.Park C.S., Yoo M.H., Noh K.H., Oh D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl Microbiol Biotechnol. 2010;87:9–19. doi: 10.1007/s00253-010-2567-6. [DOI] [PubMed] [Google Scholar]
- 30.Quan L.H., Cheng L.Q., Kim H.B., Kim J.H., Son N.R., Kim S.Y., Jin H.O., Yang D.C. Bioconversion of ginsenoside Rd into compound K by Lactobacillus pentosus DC101 isolated from Kimchi. J Ginseng Res. 2010;34:288–295. [Google Scholar]
- 31.Zhou W., Yan Q., Li J.Y., Zhang X.C., Zhou P. Biotransformation of Panax notoginseng saponins into ginsenoside compound K production by Paecilomyces bainier sp. 229. J Appl Microbiol. 2008;104:699–706. doi: 10.1111/j.1365-2672.2007.03586.x. [DOI] [PubMed] [Google Scholar]
- 32.Park S.Y., Bae E.A., Sung J.H., Lee S.K., Kim D.H. Purification and characterization of ginsenoside Rb1-metabolizing beta-glucosidase from Fusobacterium K-60, a human intestinal anaerobic bacterium. Biosci Biotechnol Biochem. 2001;65:1163–1169. doi: 10.1271/bbb.65.1163. [DOI] [PubMed] [Google Scholar]