Abstract
In the past decade, cumulative clinical experiences with molecular targeted therapies and immunotherapies for cancer have promoted a shift in our conceptual understanding of cancer. This view shifted from viewing solid tumors as a homogeneous mass of malignant cells to viewing tumors as heterogeneous structures that are dynamically shaped by intercellular interactions among the variety of stromal, immune, and malignant cells present within the tumor microenvironment. As in any dynamic system, identifying how cells communicate to maintain homeostasis and how this communication is altered during oncogenesis are key hurdles for developing therapies to restore normal tissue homeostasis. Here, I discuss tissues as dynamic systems, using the mammary gland as an example, and the evolutionary concepts applied to oncogenesis. Drawing from these concepts, I present 2 competing hypotheses for how intercellular communication might be altered during oncogenesis. As an initial test of these competing hypotheses, a recent secretome comparison between normal human mammary and HER2+ breast cancer cell lines suggested that the particular proteins secreted by the malignant cells reflect a convergent evolutionary path associated with oncogenesis in a specific anatomical niche, despite arising in different individuals. Overall, this study illustrates the emerging power of secretome proteomics to probe, in an unbiased way, how intercellular communication changes during oncogenesis.
Keywords: cell-to-cell communication, dynamic systems, somatic evolution, standard evolutionary theory, secretome
Introduction
The clinical management of metastatic breast cancer has been revolutionized following the introduction of molecular targeted therapies that promise both specificity and efficacy in treatment.1 While molecular targeted therapies can exhibit a remarkable efficacy in a subset of patients, achieving a durable clinical response remains a challenge (see ref. 22). The preclinical development of many of these molecular targeted therapies, such as herceptin, focused on their intrinsic activity to induce cell death in malignant cells, as commonly assayed in vitro or using xenograft studies. However in the case of herceptin, clinical activity also depends on engaging effector mechanisms external to the malignant cell, such as innate immunity by targeting HER2-expressing cancer cells for natural killer (NK) cell cytolysis3,4 or adaptive immunity.5 Herceptin engages adaptive immunity by downregulating HER2, thus enhancing antigen presentation by increasing MHC class I expression6 and exposing HER2+ cancer cells to the cytotoxic activity of HER2-specific CD8+ T lymphocytes.7 Moreover, the presence of an adaptive type 1 immune response within the tumor microenvironment is an independent predictor of survival in breast cancer.8-11 Collectively, these data paint a more complicated picture of the tumor than a mass of heterogeneous malignant clones. The emerging view is that the development of effective treatments for cancer will require us to consider the cancer as a dynamic system, in which malignant cells interact with a variety of other cell types present within the tumor microenvironment to maintain a disease state. The emergence of immune checkpoint inhibitors for cancer immunotherapy reinforces this perspective. For instance, regression of tumors is associated with local proliferation of CD8+ T cells within the tumor following blockade of the programmed cell death pathway,12 whereas progression exhibits at least 3 different phenotypes:13 (1) a non-functional immune response in which CD8+ T cells are present within the tumor but seem to be non-functional, (2) immunological ignorance with little to no immune infiltration into the tumor, and (3) an excluded infiltrate where immune cells are present solely around the outer edge of the tumor cell mass. In this commentary, I will summarize a number of ideas that support this view of cancer as a dynamic system and describe an experimental approach to eavesdrop on how communication among cells becomes altered during oncogenesis.
Tissues as dynamic systems
As a dynamic system, multiple cell types collectively maintain tissue organization and function even if they are subjected to external perturbations. Regulatory control networks that detect deviations from homeostasis and coordinate a cellular response to restore homeostasis through cellular communication facilitate the maintenance of cellular composition and function. The modes of cellular communication involved in restoring homeostasis include the embedding of biochemical cues within the extracellular matrix, cell-to-cell contact, and release of soluble signals. In addition to direct cell-to-cell communication, cell-to-cell contact can establish barriers that limit the entry of cells into a tissue and create diffusion gradients of soluble signals. For example, increased directional migration and proliferation during wound healing is a biological response that is orchestrated in part by a disruption in the gradient of the growth factor heregulin.14 In response to changing functional requirements, natural biological processes also promote the reorganization of tissue structures. For instance, developmental stages are associated with the coordinated expression of distinct proteins that promote homotypic interactions between cells, called adherens junctions, and facilitate the spatial organization of tissue structures.15 Once established, the functional requirements for a tissue may change periodically during an organism's lifespan. For example, the mammary epithelium reorganizes during distinct stages of the ovarian cycle in preparation for lactation.16-18 Conversely, pathogens may destabilize homotypic interactions between adjacent cells and thereby destabilize the epithelial monolayer to gain entry.19,20 Although many of the components involved in cellular communication are known, identifying the regulatory networks that control the collective cellular response within tissues remains a challenge.21 Moreover, the importance of a specific component in regulating the cellular response is highly dependent on context.
Focusing on the mammary gland as an example, a milk duct is composed of a number of different cell types (Fig. 1A). Luminal epithelial cells respond to female hormones; basal myoepithelial cells express smooth muscle contractile proteins. During lactation, the luminal epithelial cells produce milk and myoepithelial cells aid in milk expulsion. Although the cartoon in Figure 1A presents a static view, the mammary gland, like other reproductive organs, is a dynamic structure. During the ovarian cycle, plasma gonadotropins and endocrine hormones vary in concentration and coordinate the remodeling of the ovary, endometrium in the uterus, and the mammary gland in preparation for reproduction (see Fig. 1B), as reviewed in refs. 22-24. The mammary gland undergoes a cycle of cell proliferation, followed by cell differentiation, and finally an involution phase that involves programmed cell death at the end of the cycle. To summarize these dynamics, we can consider this process to be controlled by global triggers in the form of endocrine hormones and by local modes of cell-to-cell communication that help control the overall structure and function of this tissue as new cells join this system (see Fig. 1C). Secretion of proteins is one way in which cells communicate. For instance, cytokine signaling, traditionally considered a mechanism of communication among immune cells, also plays a role in mammary gland development during pregnancy.25 Secretome proteomics is a powerful and unbiased, yet underemployed, tool to eavesdrop on these local modes of communication.26 Ultimately, understanding how these local mechanisms for cell-to-cell communication help maintain the integrity of dynamic tissues, like the mammary gland, and how these modes for communication become altered during oncogenesis is central for designing ways to restore or maintain a healthy state.
Figure 1.
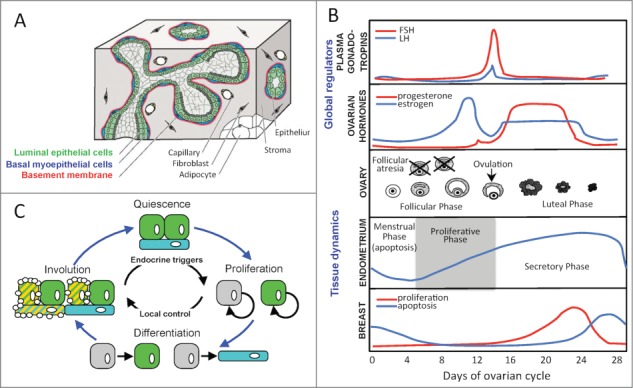
The mammary gland as a dynamic system. (A) A cartoon of the spatial organization of different cell types within the mammary gland, as similarly depicted in ref. 46. Luminal epithelial cells (green shading), basal myoepithelial cells (blue shading), and the basement membrane (red shading) are the main cellular components of an epithelial tree composed of ducts and terminal alveolar buds that remodel to produce milk during lactation. The epithelial tree is surrounded by a fibrous tissue structure, shown in gray, that contains stromal cells, fibroblasts, adipocytes, and blood capillaries. (B) The coordinated dynamics associated with plasma gonadotropins and ovarian hormones levels and with reorganization of the ovary, uterine endometrium, and breast during the phases of the ovarian cycle, as depicted in ref. 22. (C) A graphical depiction of the reorganization of the mammary gland during the reproductive cycle. Endocrine hormones initiate cellular proliferation whereas cellular differentiation into different subtypes is controlled by local contextual signals. In the absence of a reproductive event, an involution stage, which involves programmed cell death to reduce the tissue structure, precedes the return to a quiescent phase.
Cancer as a process of somatic evolution
Viewing cancer through an evolutionary lens may help us understand how these local models for cell-to-cell communication becomes altered during oncogenesis. Evolutionary processes share a number of key traits, which are collectively referred to as standard evolutionary theory (SET) or the Modern Synthesis.27 First, genetic alterations create variants that dominate a heterogeneous population based on their ability to survive and reproduce, thereby passing these genetic alterations on to progeny. The majority of genetic alterations have a neutral effect on survival or reproduction, while detrimental variants are ultimately removed from the population. Next, the influence of non-genetic and environmental factors in determining which particular variants survive is represented by a fitness landscape. Finally, the dynamics associated with evolutionary change is proportional to the mutation rate and the time associated with a reproductive generation.
In the field of cancer biology, there has been a resurgence in thinking about cancer as an evolutionary process.28,29 In short, the contemporary interest in somatic evolution has been driven by results from sequencing cancer genomes and clinical results using molecular-targeted therapies. Consistent with SET, mutagens—such as UV radiation, oncogenic viruses, or carcinogens contained within tobacco smoke—introduce genetic alterations that are passed along to progeny through somatic cell division.30 Implied by neutral theory, most of the mutations that are retained have a neutral effect on survival or proliferation.31 Mutations that provide an advantage, endowing the cell with an improved ability to persist and proliferate, are infrequent. Along these lines, Gray and Druker noted that “individual breast cancers typically carry a few consistent and functionally characterized abnormalities, along with tens to thousands of other changes that are rare or unique to the individual tumor and about which little is known.”32 Following from the “gene-centric” view commonly associated with SET,33 we could think of this fitness advantage as intrinsic properties of the cell, like the ability to survive in a nutrient poor environment, an inability to respond to cues that initiate programmed cell death, or a cell proliferation program that is stuck in the “on” state. We collectively know a lot about how these intrinsic properties change in cancer cells, as embodied in the hallmarks of cancer.34 However, despite the importance of intercellular communication within a tissue, we know much less about how cell-to-cell communication is changed during oncogenesis.
This last point highlights a potential key difference between SET and somatic evolution. SET treats the fitness landscape, or simply the environment, as a background condition, which implies a unidirectional influence of the environment on the genetic variants. Mathematically, SET applied to tissues can be summarized by 2 coupled differential equations.35 The first equation, dC/dt = g(C, E), describes the rate of change of a particular cellular variant (C) that depends on the evolutionary process (g), which is a function of 2 state variables: C and the environment, E. The second equation, dE/dt = f (E), describes the rate of change of the environment that depends solely on an autonomous function (f ) of the environment. In contrast, individual cells help create the fitness landscape in tissues, which implies that individual cells can have a significant influence on the nature of the fitness landscape and that the influence is bidirectional. This bidirectional influence suggests that cells can construct a “niche” to co-direct their own evolution by systematically reshaping the fitness landscape.36,37 In mathematical terms, niche construction theory implies that the differential equation for the environment, dE/dt = f’(C, E), depends on both the cellular and environmental states. The ability to regenerate a functional heart by seeding decellularized tissue scaffolds with neonatal cardiocytes is an example of how a normal tissue niche influences cell fate and phenotype.38
To test whether niche construction is a key trait of somatic evolution, we can formulate the relationship between malignant cells and the fitness landscape in terms of 2 alternative hypotheses (see Fig. 2). To formulate these hypotheses, we assume that an evolutionary process shapes the biochemical cues secreted by malignant cell lines. Mutations alter the genome by changing copy numbers or the sequence of promoter or coding regions. These altered sequences then alter the levels of expression and the cellular disposition of the resulting protein products. By generating genetic variants, mutagens would randomly alter the profile of proteins secreted by malignant cells, which could be viewed as a form of phenotypic drift. While the levels of each secreted protein could constitute a unique phenotype dimension within the fitness landscape, we will consider a fitness landscape with only 2 phenotype dimensions to visualize the hypotheses. The two hypotheses correspond to how the fitness landscape could potentially shape the profile of proteins secreted by malignant cells. The first hypothesis, or null hypothesis, is that the secreted proteins derived from a particular malignant clone do not influence the clone's fitness and that these alterations are neutral and independent from any intrinsic fitness effects. This implies that if altering cell-to-cell communication is not important for oncogenesis, the biochemical cues secreted by cancer cells that arise should be different from each other and different from those of normal epithelial cells derived from the same tissue, as there is no fitness gradient and mutagens promote phenotypic divergence. This hypothesis is shown graphically in the left panel of Figure 2. Alternatively, if altering cell-to-cell communication in a particular way is important for oncogenesis within a specific anatomical niche, then the biochemical cues secreted by cancer cells that arise in different patients but originate in the same anatomical tissue should share phenotypic commonalities and represent a convergent evolutionary path, as depicted in the right panel in Figure 2.1
Figure 2.
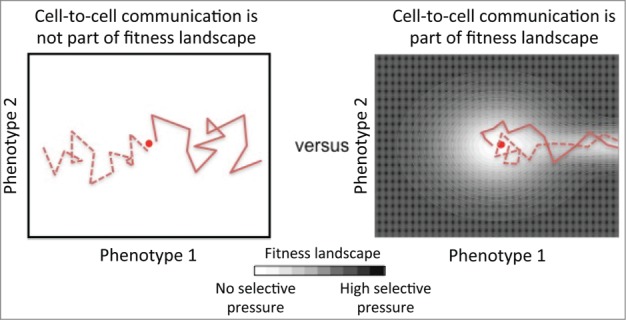
A simplified graphical depiction of 2 competing hypotheses regarding whether cell-to-cell communication is part of the fitness landscape. The left panel represents a null hypothesis that cell-to-cell communication is not a component of the fitness landscape and that malignant clones diverge from the origin in a way that is consistent with standard evolutionary theory. The right panel represents an alternative hypothesis that cell-to-cell communication is a component of the fitness landscape and that this communication must be altered in a particular way for malignant cells to dominate this tissue niche. The secretome of a normal cell is represented by the red dot at the center of the 2 panels. The dotted and solid red lines depict the evolutionary paths associated with oncogenesis of 2 malignant clones that arise within the same anatomical niche. The extent of the selective pressure exerted on a cell is depicted using black-white shading, where white indicates no selective pressure and black indicates high selective pressure.
Conclusion
In a recent study, we tested these competing hypotheses by comparing the proteins secreted, or the secretome, for 2 similar human breast carcinoma cell lines to that of a normal human mammary epithelial (HME) cell line using a mass spectrometry-based proteomics workflow.39 Interestingly, we found that the secretomes for the 2 breast carcinoma cells lines were similar to each other yet distinct from the secretome of a normal HME cell line, despite the fact that all 3 lines have been adapted to in vitro culture. Specifically, we found that 50% of the identified proteins (19 proteins) were common between the secretomes of the 2 breast carcinoma cell lines and that these proteins target similar canonical pathways (7 of the top 10 enriched pathways were common). In contrast, the secretome of the normal HME cell line shared only a single protein in common with both breast cancer cell lines and 2 additional proteins were common to each of the breast cancer cell lines individually. Moreover, no canonical pathways were identified as enriched in the secretome of the normal HME cell line. While further studies will be necessary to clarify how these secreted proteins promote oncogenesis, the results are consistent with a convergent evolutionary path associated with oncogenesis; that is, altering cell-to-cell communication in a particular way is important for oncogenesis in the mammary gland. Employing a similar approach in a variety of cancer and “normal” cell lines derived from different anatomical niches may inform the importance of biochemical nodes within intracellular signaling networks in specific tissues, such as the role of specific cytokines in coordinating antitumor immunity.
Using mass spectrometry-based proteomics as a discovery tool, we were also able to identify secretome proteins for which the biological roles as mediators of cell-to-cell communication are not well known. Specifically, in 2 different secretome studies we found that the majority of secreted proteins are associated with extracellular vesicles called exosomes.39,40 Exosomes are an emerging area of interest as they may provide a more complicated mode of communication between cells. Specifically, exosomes have been reported to transport proteins and coding and non-coding RNA between cells.41,42 Results of our pathway enrichment analysis suggest that these exosomes alter the metabolic profile of immune cells that enter the tumor microenvironment by delivering a collection of metabolic enzymes and alter antigen presentation. Moreover, the size of exosomes also suggests that their diffusivity within tissues may help create spatial gradients.
While the technical challenges associated with a mass spectrometry approach place limitations on the experimental design,26 these unbiased methods can be used in conjunction with more focused methods, such as reverse protein arrays, as described in ref. 43, or possibly multispectral imaging of immunohistochemically labeled tumor tissue, as described in refs. 44 and 45. Collectively, combining different approaches can be used to compensate for the limitations of the different technologies and provide a more comprehensive view of how cell-to-cell communication within tissues becomes altered during oncogenesis. Identifying common mechanisms, or convergent evolutionary paths, for subverting these intercellular communication networks will help in the rational design of therapies to restore normal tissue homeostasis. Moreover, this information can be used to stratify patients into different treatment protocols based on the specific network alterations that underpin their disease state, such as the stratification of patients with diabetes into type 1 and type 2 phenotypes.
Footnotes
There is a third possibility that the phenotypes of cancer cells are similar to normal cells. This could occur when a mutation occurs in an oncogene or tumor suppressor gene only thereby increasing a malignant cells intrinsic fitness with no change in secreted proteins. In human breast cancer, the quote from Joe Gray and Brian Druker (see ref.32) argues against this possibility. Although this is a scenario recreated in genetically engineered mouse models of cancer. Alternatively, similar phenotypes could also occur when any change in secreted proteins provides a significant fitness disadvantage. In short, observing that malignant cells have the same profile of secreted proteins as normal cells would be inconclusive for inferring whether altering cell-to-cell communication is important for oncogenesis.
Funding
This work was supported by grants from the National Science Foundation (CAREER 1053490) and the National Cancer Institute (R15CA132124). The content is solely the responsibility of the author and does not necessarily represent the official views of the NSF, the National Cancer Institute, or the NIH.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Schlotter CM, Vogt U, Allgayer H, Brandt B. Molecular targeted therapies for breast cancer treatment. Breast Cancer Res 2008; 10:211; PMID:18671839; http://dx.doi.org/ 10.1186/bcr2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiszman GL, Jasnis MA. Molecular mechanisms of trastuzumab resistance in HER2 over- expressing breast cancer. Int J Breast Cancer 2011; 2011:352182; PMID: 22295219; http://dx.doi.org/ 10.4061/2011/352182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnould L, Gelly M, Penault-Llorca F, Benoit L, Bonnetain F, Migeon C, Cabaret V, Fermeaux V, Bertheau P, Garnier J, et al.. Trastuzumab-based treatment of HER2-positive breast cancer: an antibody-dependent cellular cytotoxicity mechanism? Br J Cancer 2006; 94:259-67; PMID:16404427; http://dx.doi.org/ 10.1038/sj.bjc.6602930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaime-Ramirez AC, Mundy-Bosse BL, Kondadasula S, Jones NB, Roda JM, Mani A, Parihar R, Karpa V, Papenfuss TL, LaPerle KM, et al.. IL-12 enhances the antitumor actions of trastuzumab via NK cell IFN-γ production. J Immunol 2011; 186:3401-9; PMID:21321106; http://dx.doi.org/ 10.4049/jimmunol.1000328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellati F, Napoletano C, Ruscito I, Liberati M, Panici PB, Nuti M. Cellular adaptive immune system plays a crucial role in trastuzumab clinical efficacy. J Clin Oncol 2010; 28(21):e369-70; PMID:20479394; http://dx.doi.org/ 10.1200/JCO.2010.28.6922 [DOI] [PubMed] [Google Scholar]
- 6.Herrmann F, Lehr HA, Drexler I, Sutter G, Hengstler J, Wollscheid U, Seliger B. HER-2/neu-mediated regulation of components of the MHC class I antigen-processing pathway. Cancer Res 2004; 64:215-20; PMID:14729627; http://dx.doi.org/ 10.1158/0008-5472.CAN-2522-2 [DOI] [PubMed] [Google Scholar]
- 7.zum Büschenfeld CM, Hermann C, Schmidt B, Peschel C, Bernhard H. Antihuman epidermal growth factor receptor 2 (HER2) monoclonal antibody trastuzumab enhances cytolytic activity of class I-restricted HER2-specific T lymphocytes against HER2-overexpressing tumor cells. Cancer Res 2002; 62:2244-7 [PubMed] [Google Scholar]
- 8.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, Earl HM, Poole CJ, Hiller L, Dunn JA, et al.. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol 2014; 25:1536-43; PMID:24915873; http://dx.doi.org/ 10.1093/annonc/mdu191 [DOI] [PubMed] [Google Scholar]
- 9.Klinke DJ. Induction of Wnt-inducible signaling protein-1 correlates with invasive breast cancer oncogenesis and reduced type 1 cell-mediated cytotoxic immunity: a retrospective study. PLoS Comput Biol 2014; 10:e1003409; PMID:24426833; http://dx.doi.org/ 10.1371/journal.pcbi.1003409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C, et al.. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol 2014; 25:1544-50; PMID:24608200; http://dx.doi.org/ 10.1093/annonc/mdu112 [DOI] [PubMed] [Google Scholar]
- 11.Perez EA, Thompson EA, Ballman KV, Anderson SK, Asmann YW, Kalari KR, Eckel-Passow JE, Dueck AC, Tenner KS, Jen J, et al.. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the north central cancer treatment group n9831 adjuvant trastuzumab trial. J Clin Oncol 2015; 33:701-8; PMID:25605861; http://dx.doi.org/ 10.1200/JCO.2014.57.6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al.. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014; 515:568-71; PMID:25428505; http://dx.doi.org/ 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al.. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014; 515:563-7; PMID:25428504; http://dx.doi.org/ 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vermeer PD, Einwalter LA, Moninger TO, Rokhlina T, Kern JA, Zabner J, Welsh MJ. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature 2003; 422:322-6; PMID:12646923; http://dx.doi.org/ 10.1038/nature01440 [DOI] [PubMed] [Google Scholar]
- 15.Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol 2011; 192:907-17; PMID:21422226; http://dx.doi.org/ 10.1083/jcb.201009141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen DA, Parlow AF, Neville MC. Hormonal regulation of tight junction closure in the mouse mammary epithelium during the transition from pregnancy to lactation. J Endocrinol 2001; 170:347-56; PMID:11479131; http://dx.doi.org/ 10.1677/joe.0.1700347 [DOI] [PubMed] [Google Scholar]
- 17.Oakes SR, Rogers RL, Naylor MJ, Ormandy CJ. Prolactin regulation of mammary gland development. J Mammary Gland Biol Neoplasia 2008; 13:13-28; PMID:18219564; http://dx.doi.org/ 10.1007/s10911-008-9069-5 [DOI] [PubMed] [Google Scholar]
- 18.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development 2008; 135:995-1003; PMID:18296651; http://dx.doi.org/ 10.1242/dev.005439 [DOI] [PubMed] [Google Scholar]
- 19.Frank CF, Hostetter MK. Cleavage of E-cadherin: a mechanism for disruption of the intestinal epithelial barrier by Candida albicans. Transl Res 2007; 149:211-22; PMID:17383595; http://dx.doi.org/ 10.1016/j.trsl.2006.11.006 [DOI] [PubMed] [Google Scholar]
- 20.Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci 2007; 120:1944-52; PMID: 17504810; http://dx.doi.org/ 10.1242/jcs.03455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell 2013; 152:945-56; PMID:23452846; http://dx.doi.org/ 10.1016/j.cell.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medh RD, Thompson EB. Hormonal regulation of physiological cell turnover and apoptosis. Cell Tissue Res 2000; 301:101-24; PMID:10928284; http://dx.doi.org/ 10.1007/s004419900159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hovey RC, Trott JF, Vonderhaar BK. Establishing a framework for the functional mammary gland: from endocrinology to morphology. J Mammary Gland Biol Neoplasia 2002; 7:17-38; PMID:12160083; http://dx.doi.org/ 10.1023/A:1015766322258 [DOI] [PubMed] [Google Scholar]
- 24.Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol 2005; 6:715-25; PMID:16231422; http://dx.doi.org/ 10.1038/nrm1714 [DOI] [PubMed] [Google Scholar]
- 25.Watson CJ, Oliver CH, Khaled WT. Cytokine signalling in mammary gland development. J Reprod Immunol 2011; 88:124-29; PMID:21255846; http://dx.doi.org/ 10.1016/j.jri.2010.11.006 [DOI] [PubMed] [Google Scholar]
- 26.Tian R. Exploring intercellular signaling by proteomic approaches. Proteomics 2014; 14:498-512; PMID:24214021; http://dx.doi.org/ 10.1002/pmic.201300259 [DOI] [PubMed] [Google Scholar]
- 27.Pigliucci M. Biology's last paradigm shift. The transition from natural theology to Darwinism. Paradigmi 2012; 3:45-58 [Google Scholar]
- 28.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer 2006; 6:924-35; PMID:17109012; http://dx.doi.org/ 10.1038/nrc2013 [DOI] [PubMed] [Google Scholar]
- 29.Klinke DJ. An evolutionary perspective on anti-tumor immunity. Front Oncol 2012; 2:202; PMID:23336100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al.. Signatures of mutational processes in human cancer. Nature 2013; 500:415-21; PMID:23945592; http://dx.doi.org/ 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet 2010; 11:265-89; PMID:20565254; http://dx.doi.org/ 10.1146/annurev-genom-082908-150129 [DOI] [PubMed] [Google Scholar]
- 32.Gray J, Druker B. Genomics: the breast cancer landscape. Nature 2012; 486:328-9; PMID:22722187; http://dx.doi.org/ 10.1038/486328a [DOI] [PubMed] [Google Scholar]
- 33.Laland K, Uller T, Feldman M, Sterelny K, Muller GB, Moczek A, Jablonka E, Odling-Smee J, Wray GA, Hoekstra HE, et al.. Does evolutionary theory need a rethink? Nature 2014; 514:161-4; PMID:25297418; http://dx.doi.org/ 10.1038/514161a [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 35.Lewontin RC. The Organism as the subject and object of evolution. Scientia 1983; 118:63-82 [Google Scholar]
- 36.Odling-Smee FJ, Laland KN, Feldman MW. Niche Construction The Neglected Process in Evolution. Princeton University Press; 2003. [Google Scholar]
- 37.Scott-Phillips TC, Laland KN, Shuker DM, Dickins TE, West SA. The niche construction perspective: a critical appraisal. Evolution 2014; 68:1231-43; PMID:24325256; http://dx.doi.org/ 10.1111/evo.12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med 2008; 14:213-21; PMID:18193059; http://dx.doi.org/ 10.1038/nm1684 [DOI] [PubMed] [Google Scholar]
- 39.Klinke DJ, Kulkarni YM, Wu Y, Byrne-Hoffman C. Inferring alterations in cell-to- cell communication in HER2+ breast cancer using secretome profiling of three cell models. Biotechnol Bioeng 2014; 111:1853-63; PMID:24752654; http://dx.doi.org/ 10.1002/bit.25238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulkarni YM, Chambers E, McGray AJ, Ware JS, Bramson JL, Klinke DJ. A quantitative systems approach to identify paracrine mechanisms that locally suppress immune response to interleukin-12 in the B16 melanoma model. Integr Biol (Camb) 2012; 4:925-36; PMID:22777646; http://dx.doi.org/ 10.1039/c2ib20053h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bobrie A, Thery C. Unraveling the physiological functions of exosome secretion by tumors. Oncoimmunology 2013; 2:e22565; PMID:23483742; http://dx.doi.org/ 10.4161/onci.22565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 2009; 9:581-93; PMID:19498381; http://dx.doi.org/ 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- 43.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al.. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012; 487:500-4; PMID:22763439; http://dx.doi.org/ 10.1038/nature11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blenman KR, Lee PP. Quantitative and spatial image analysis of tumor and draining lymph nodes using immunohistochemistry and high-resolution multispectral imaging to predict metastasis. Methods Mol Biol 2014; 1102:601-21; PMID:24259001; http://dx.doi.org/ 10.1007/978-1-62703-727-3_32 [DOI] [PubMed] [Google Scholar]
- 45.Webb JR, Milne K, Watson P, Deleeuw RJ, Nelson BH. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin Cancer Res 2014; 20:434-44; PMID:24190978; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-1877 [DOI] [PubMed] [Google Scholar]
- 46.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol 2006; 22:287-309; PMID:16824016; http://dx.doi.org/ 10.1146/annurev.cellbio.22.010305.104315 [DOI] [PMC free article] [PubMed] [Google Scholar]


