ABSTRACT
In primary acute lymphoblastic leukemia cells exhibiting de novo resistance to glucocorticoids, we recently discovered decreased promoter methylation of caspase 1 (CASP1) and NLR family, pyrin domain containing 3 (NLRP3), which resulted in increased transcription, constitutive NALP3 (NACHT, LRR and PYD domains-containing protein 3) inflammasome activation, and caspase 1-mediated cleavage of the glucocorticoid receptor. This revealed a novel mechanism of glucocorticoid resistance that was recapitulated in model systems.
KEYWORDS: CASP1, glucocorticoids, inflammasome, NALP3, NR3C1
Glucocorticoids are a widely prescribed class of medications that are used to treat a broad spectrum of inflammatory, allergic, autoimmune, and malignant diseases including leukemias and lymphomas, and as such are an essential component of curative treatment of acute lymphoblastic leukemia (ALL). Children whose leukemia cells show relative in vitro resistance to glucocorticoids have a poorer prognosis than those showing good in vitro sensitivity.1 Glucocorticoids act by binding to the glucocorticoid receptor (encoded by the NR3C1, nuclear receptor subfamily 3 gene) in the cytoplasm, leading to translocation of the receptor to the nucleus where it binds to specific DNA motifs (glucocorticoid response elements, GREs)2 and acts as a transcription factor mediating the activation or repression of gene transcription.
In a recent genome wide association study of 444 patients diagnosed with pediatric ALL we analyzed gene expression data and information on patient tumor cell response to glucocorticoids (i.e., LC50) and showed that 2 of the most differentially expressed genes in glucocorticoid-sensitive and -resistant pediatric acute lymphoblastic leukemia cells were key components in the NALP3 inflammasome pathway (CASP1 and NLRP3).3 We found that mRNA expression of CASP1 and NLRP3 was 1.6-fold and 2.4-fold higher, respectively, in glucocorticoid-resistant ALL cells compared to glucocorticoid-sensitive ALL cells. Moreover, in patients who eventually experienced disease recurrence, CASP1 and NLRP3 expression was higher in leukemia cells analyzed at relapse than in leukemia cells analyzed at the initial diagnosis. To understand the mechanism of this differential expression, we focused on known gene regulatory mechanisms including DNA methylation because it is well established that methylation of promoter regions results in transcriptional silencing of the corresponding genes.4 Our genome-wide analysis of DNA methylation revealed a highly significant relationship between lower levels of CASP1 promoter methylation and elevated CASP1 mRNA expression, and, concordantly, a highly significant relationship between lower levels of NLRP3 promoter methylation and higher NLRP3 mRNA expression in glucocorticoid-resistant ALL cells compared to glucocorticoid-sensitive ALL cells. Based on a published report of CASP1-mediated androgen receptor5 cleavage, we used bioinformatics to identify 2 similar CASP1 cleavage 4-residue motifs (LLID and IKQE) in the transactivation domain of the glucocorticoid receptor and experimentally confirmed their clinical relevance (Fig. 1). When overexpressed and activated in human leukemia cells lines, CASP1 increased glucocorticoid resistance by cleaving and inactivating the receptor, preferably at the LLID site; this could be mitigated by either downregulation of CASP1 expression or by overexpression of the cowpox virus protein CrmA, a known inhibitor of CASP1.6
Figure 1.
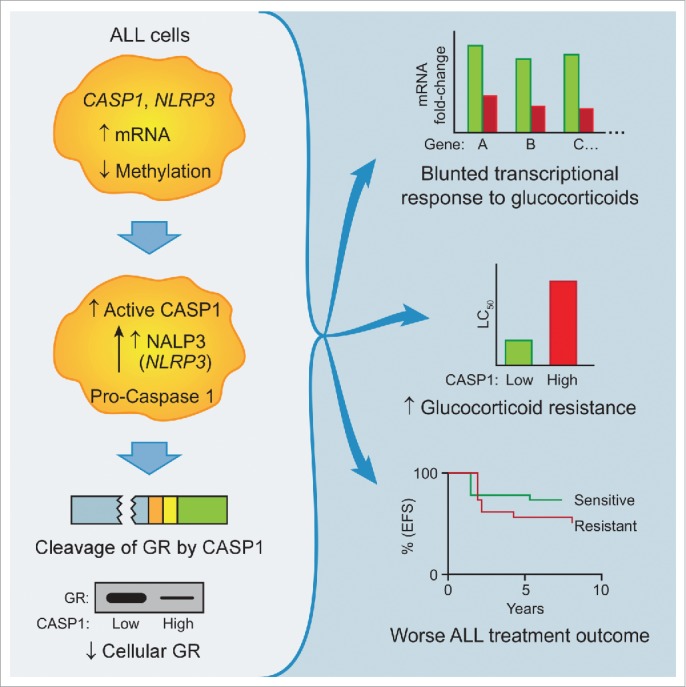
Caspase 1 induces glucocorticoid resistance via glucocorticoid receptor cleavage. Schematic illustration showing that increased expression of inflammasome components caspase 1 (CASP1) and NLR family, pyrin domain containing 3 (NLRP3) via hypomethylation of promoter regions leads to glucocorticoid receptor cleavage as a result of increased CASP1 activity. Decreased levels of functional glucocorticoid receptor (GR) lead to a blunted transcriptional response to glucocorticoids, glucocorticoid resistance, and inferior treatment outcomes.
A potential extension of our findings could be the development of new therapeutic interventions to mitigate glucocorticoid resistance by inhibiting caspase 1 in leukemia cells that have high expression of CASP1 and NLRP3. Based on our findings, we hypothesize that small-molecule inhibitors of CASP1 may be a viable therapeutic strategy to overcome this mechanism of glucocorticoid resistance in ALL. To demonstrate this pathway as a potential target for reversing glucocorticoid resistance in ALL patients, we overexpressed a known CASP1 inhibitor (CrmA) or small hairpin RNA (shRNA) to destroy either CASP1 catalytic activity or the CASP1 message. These distinct inhibition methods both resulted in enhanced glucocorticoid sensitivity and blockage of glucocorticoid receptor cleavage. An alternative strategy may be to boost the pool of available glucocorticoid receptors in CASP1 overexpressing cells by increasing its half-life. We have performed experiments to test this option and showed that increasing the level of wild-type glucocorticoid receptor by overexpression of the wild-type receptor protein in high-CASP1 leukemia cells was insufficient to overcome CASP1-induced glucocorticoid resistance. This is consistent with the susceptibility of this newly introduced wild-type glucocorticoid receptor to CASP1 cleavage. In contrast, overexpression of a mutant form of the receptor, engineered to be resistant to CASP1-cleavage by substitution of alanine residues at the 2 cleavage sites, resulted in substantially reduced CASP1-induced glucocorticoid resistance. Thus, analogous to this experiment, a potential therapeutic strategy that aims to increase the levels of wild-type endogenous glucocorticoid receptor is unlikely to be successful. Preferable strategies would likely be masking the CASP1-cleavage sites7 or blocking CASP1 activity with a targeted small molecule.
The potential for future investigation into this newly discovered link between inflammatory and anti-inflammatory mechanisms is clear. This previously unrecognized mechanism of blocking the anti-inflammatory activities of endogenous glucocorticoids by amplification of CASP1 activity further enhances the inflammatory effects. Our studies open the way for small-molecule screens for CASP1 inhibitory compounds that can lower glucocorticoid resistance and for clinical trials involving prospective screening of ALL patients for CASP1 and NLRP3 expression to optimize their treatment, in addition to evolving studies of known CASP1 inhibitors (e.g., VX-765, VRT-043198).8
Overall, our work identifies a new mechanism of glucocorticoid resistance in pediatric leukemia patients; however, the story may not end there. As glucocorticoids are some of the most highly prescribed medications, this same mechanism may hold true for rheumatoid arthritis, asthma, ulcerative colitis, and many other medical conditions. It remains to be seen whether this mechanism might be operative in the regulation of other nuclear hormone receptors such as the androgen receptor, estrogen receptor, or vitamin D receptor, although some preliminary evidence exists for the androgen receptor.5 In any case, the regulatory link between inflammatory pathways (NALP3, CASP1) and anti-inflammatory pathways (GR, NR3C1) is likely conserved across species and the operational mode of the inflammasome shutting down the opposing anti-inflammatory pathway is compatible with evolutionary selective pressure for a strong inflammatory response to pathogens or damage.9,10
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
W.E.E., S.W.P. and E.J.B. are named as co-inventors on a pending patent application that relates to the subject matter of the article, which was filed by St. Jude Children's Research Hospital.
References
- 1.Den Boer ML, Harms DO, Pieters R, Kazemier KM, Gobel U, Körholz D, Graubner U, Haas RJ, Jorch N, Spaar HJ, et al. Patient stratification based on prednisolone-vincristine-asparaginase resistance profiles in children with acute lymphoblastic leukemia. J Clin Oncol 2003; 21:3262–8; PMID:12947061; http://dx.doi.org/ 10.1200/JCO.2003.11.031 [DOI] [PubMed] [Google Scholar]
- 2.Nicolaides NC, Galata Z, Kino T, Chrousos GP, harmandari E. The human glucocorticoid receptor: molecular basis of biologic function. Steroids 2010; 75; 1–12; PMID:19818358; http://dx.doi.org/ 10.1016/j.steroids.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paugh SW, Bonten EJ, Savic D, Ramsey LB, Thierfelder WE, Gurung P, Malireddi RK, Actis M, Mayasundari A, Min J, et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat Genet 2015; 47(6):607–14; PMID:25938942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubeler D. Function and information content of DNA methylation. Nature 2015; 517:321–6; PMID:25592537; http://dx.doi.org/ 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- 5.Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem 1998; 273:9158–67; PMID:9535906; http://dx.doi.org/ 10.1074/jbc.273.15.9158 [DOI] [PubMed] [Google Scholar]
- 6.Komiyama T, Ray CA, Pickup DJ, Howard AD, Thornberry NA, Peterson EP, Salvesen G. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J Biol Chem 1994; 269:19331–7; PMID:8034697 [PubMed] [Google Scholar]
- 7.Luchini A, Espina V, Liotta LA. Protein painting reveals solvent-excluded drug targets hidden within native protein-protein interfaces. Nat Commun 2014; 5:4413; PMID:25048602; http://dx.doi.org/ 10.1038/ncomms5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boxer MB, Shen M, Auld DS, Wells JA, Thomas CJ. A small molecule inhibitor of Caspase 1. in Probe Reports from the NIH Molecular Libraries Program (Bethesda (MD: ), 2010) [Google Scholar]
- 9.Raj T, Kuchroo M, Replogle JM, Raychaudhuri S, Stranger BE, De Jager PL. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am J Hum Genet 2013; 92:517–29; PMID:23522783; http://dx.doi.org/ 10.1016/j.ajhg.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admetlla A, Pattini L, Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet 2011; 7:e1002355; PMID:22072984; http://dx.doi.org/ 10.1371/journal.pgen.1002355 [DOI] [PMC free article] [PubMed] [Google Scholar]


