Abstract
Background
Schizophrenia (SZ) and bipolar disorder (BD) have substantial negative impact on the quality of human life. Both, microRNA (miRNA) expression profiling in SZ and BD postmortem brains [and genome-wide association studies (GWAS)] have implicated miRNAs in disease etiology. Here, we aim to determine whether significant GWAS signals observed in the Psychiatric Genetic Consortium (PGC) are enriched for miRNAs.
Method
A two-stage approach was used to determine whether association signals from PGC affect miRNAs: (i) statistical assessment of enrichment using a Simes test and sum of squares test (SST) and (ii) biological evidence that quantitative trait loci (eQTL) mapping to known miRNA genes affect their expression in an independent sample of 78 postmortem brains from the Stanley Medical Research Institute.
Results
A total of 2567 independent single nucleotide polymorphisms (SNPs) (R2 > 0.8) were mapped locally, within 1 Mb, to all known miRNAs (miRBase v. 21). We show robust enrichment for SZ- and BD-related SNPs with miRNAs using Simes (SZ: p ≤ 0.0023, BD: p ≤ 0.038), which remained significant after adjusting for background inflation in SZ (empirical p = 0.018) and approached significance in BD (empirical p = 0.07). At a false discovery rate of 10%, we identified a total of 32 eQTLs to influence miRNA expression; 11 of these overlapped with BD.
Conclusions
Our approach of integrating PGC findings with eQTL results can be used to generate specific hypotheses regarding the role of miRNAs in SZ and BD.
Keywords: Bipolar disorder, eQTLs, GWAS, miRNAs, schizophrenia
Introduction
Schizophrenia (SZ) and bipolar disorder (BD) are debilitating psychiatric disorders that affect approximately 1% of the population worldwide (McGrath et al. 2008). Since 2007, when differential expression of microRNAs (miRNAs) in postmortem brain tissue from psychiatric patients and normal controls was assessed (Perkins et al. 2007) for the first time, increasing evidence from follow-up expression studies (Beveridge et al. 2008; Kim et al. 2010; Santarelli et al. 2011; Miller et al. 2013) conducted in postmortem brain tissue of SZ and BD patients as well as recent genome-wide association studies (GWAS) (PGC, 2011; Ripke et al. 2013, 2014) have implicated miRNAs to play a role in SZ and BD etiology. miRNAs are small single-stranded RNA molecules that mainly downregulate protein-coding genes by binding to the gene’s 3′ untranslated region (UTR) (Forero et al. 2010) through a variety of mechanisms, including mRNA cleavage and degradation, translational inhibition, and shortening of the mRNA poly tail (Filipowicz et al. 2008).
In 2013, the Psychiatric Genetic Consortium (PGC) published the largest GWAS to date with an initial discovery sample of 21 856 individuals and a replication sample of 29 839 individuals (Ripke et al. 2013). Their strongest finding was located in the primary transcript of hsa-miR-137 (miR-137). Other genes, predicted as targets of miR-137, were also noted as achieving genome-wide significance; the predicted relationship between miR-137 and these genes has since been experimentally verified (Kim et al. 2012; Kwon et al. 2013; Collins et al. 2014). A second PGC study, utilizing an even larger sample size than the first, identified over 108 genome-wide significant signals, many of which also mapped to additional miRNA genes with neurodevelopmental function such as hsa-miR-640, -135a, -4304 and let-7g (Ripke et al. 2014). To our knowledge, however, no one has attempted to link aberrant miRNA expression from postmortem studies to significant genetic association signals identified in the PGC GWAS. Expression quantitative trait loci (eQTL) analysis offers a relatively straightforward approach to this task by linking genetic variants with gene expression (Westra & Franke, 2014). Moreover, specific testable hypotheses can be drawn from the significant associations found between variants and molecular features as eQTLs were shown to be also tissue- and cell-specific and to exhibit conserved effect sizes across major population groups (Stranger et al. 2012). miRNA-linked eQTLs, in particular, have been shown to be associated with the expression of complex phenotypic traits (Nicolae et al. 2010; Gamazon et al. 2012). miRNA-linked eQTLs can impact on miRNA cellular function either directly through modification of the miRNA expression (Hansen et al. 2007; Feng et al. 2009; Xu et al. 2010) or indirectly by impacting on the miRNA-binding activity to its target transcripts (Nicolae et al. 2010; Gamazon et al. 2012). In this study, we attempt to generate specific hypotheses regarding the genetic signals observed in the latest PGC GWAS and their connection to miRNAs by employing a two-stage approach. First, we ask whether GWAS signals observed in the PGC dataset are enriched for single nucleotide polymorphisms (SNPs) in linkage disequilibrium (LD, R2 > 0.8) with known miRNA precursors. Second, in a 1 Mb region flanking those miRNAs for which enrichment was detected, we ask, whether any of those specific SNPs function as eQTLs for miRNA expression. Multiple authors have noted already the enrichment of eQTLs among top GWAS findings in a wide variety of disorders, including SZ, diabetes, and specific types of cancer (Nicolae et al. 2010; Richards et al. 2012; Bacanu et al. 2014; Jiang et al. 2014; Roussos et al. 2014; Zhang et al. 2014). In this study we test two hypotheses: (1) that PGC GWAS signals are enriched for miRNA-linked eQTLs and (2) that by identifying the relationship between these genetic factors, the miRNAs and their respective gene targets, we can identify gene pathways of functional relevance to the pathophysiology of SZ and BD. Aberrant miRNA expression, especially during neurodevelopment, can modulate the expression of target protein-coding genes, which being themselves linked to the pathophysiology of SZ and BD as originally shown by PGC (PGC, 2011) could offer a novel etiological mechanism of disease development.
Method
Detection of SNP enrichment
All autosomal tag SNPs (Plink v1.07, R2 = 0.8) (Purcell et al. 2007) found locally, within a 1 Mb region flanking known miRNA precursor sequences (N = 1881, miRBase v21) (Kozomara & Griffiths-Jones, 2014) were extracted and tested for enrichment in association signals over the PGC GWAS background using the Simes and sum of squares (SST) tests (see Supplementary material). Additionally, in order to identify potential confounders that may influence the level of GWAS enrichment, we performed bioinformatic analysis of the selected tag SNPs in our dataset. SNPs were excluded from our list if they were found to be located in highly conserved regions, predicted to have detrimental protein effects (Adzhubei et al. 2013) or if they mapped to highly conserved transcription factor binding sites.
To determine whether the miRNA location affects the level of enrichment, miRNAs were categorized as intergenic and intragenic (Down & Hubbard, 2002). Intergenic miRNAs are located outside of the boundaries of protein-coding genes; intragenic miRNAs are located within the boundaries of an annotated protein-coding gene, including those found within the 5′- and 3′-UTRs, and in the exonic and intronic regions of the gene. Intergenic miRNAs have been noted to possess an independent source of transcription, whereas intragenic miRNAs are often thought to share promoter sequences with the protein-coding genes that host them (Gromak, 2012; Ramalingam et al. 2014). It is worth noting that knowledge of transcription start sites (TSS) for the intragenic miRNAs has been based on bioinformatic prediction and the full range of the predicted miRNA TSS have not yet been fully validated experimentally (Saini et al. 2007). Indeed, recent experimental work using chromatin-based assays and miRNA cloning into empty promoter sequences have suggested that some intragenic miRNAs may have TSS that are transcribed by polymerases II and III (Pol II, Pol III) independently of their host gene (Borchert et al. 2006; Monteys et al. 2010). However, the data are still inconclusive and therefore, we believe that to assess the level of SNP enrichment separately based on this distinction alone would be premature without a full-scale validation of the TSS of known miRNAs and would also lead to a substantial reduction on statistical power. We do, however, draw a distinction between eQTLs associated with intergenic v. intragenic miRNAs in our pathway enrichment analysis. Our rationale for this distinction is based on the potential confounding effect of miRNA location on its host gene; it is possible an eQTL affecting the expression of intragenic miRNA to also affect the expression of its host protein-coding gene. The distance parameters employed for determining whether a SNP is a ‘local’ or a cis-eQTL are based on previous studies in which cis-eQTLs have been identified (Gamazon et al. 2012, 2013a, b).
Data integration and genotype processing
All available genotypes were downloaded from the Stanley Online Genomics Database (Kim & Webster, 2010) and filtered with PLINK v1.02 (Supplementary material). Subjects having genotypes and miRNA expression values (Kim et al. 2010) were matched on the basis of gender, age, postmortem interval (PMI), brain pH, DSM-IV diagnosis, and gender. After applying a stringent quality control (QC) to the available genotype data, a total of 78 subjects (SZ = 27, BD = 29, controls = 22) were retained. Specific demographic information for these subjects can be found in online Supplementary Table S1. Imputation was performed on the original genotypes to make the GWAS panel used in the structural magnetic resonance imaging (sMRI) study more comparable to that used by the PGC (Supplementary material). A total of 309 531 SNPs in the sMRI GWAS was used to generate additional 332 271 genotypes meeting study criteria leading to a final count of 641 802 genotypes.
eQTL detection
eQTLs were detected using an additive model and accounting for the potential effect of age, brain pH, race, gender, DSM-IV classification, PMI and RNA integrity number (RIN) on miRNA expression. Those SNPs having missing data and/or a low minor allele frequency (MAF < 0.02) in the sMRI GWAS were not considered in the eQTL analysis. SNPs were coded as 0 (homozygous for the major allele), 1 (heterozygous) and 2 (homozygous for the minor allele) for evaluation in a linear regression model implemented by the R package Matrix EQTL (Shabalin, 2012); p values were corrected for multiple testing based on the number of SNPs within the 1 Mb region flanking of each individual miRNA that were tested in the enrichment stage at a false discovery rate (FDR) of 10%. We chose to focus exclusively on ‘local’ cis-eQTLs located within 1 Mb of the individual miRNA; by being closer to the gene of interest these have a greater chance to affect the overall level of expression than more distant (trans) eQTLs. A total of 2567 SNPs from the PGC dataset were evaluated in this manner.
Target prediction and pathway assessment
High-quality targets were predicted for all miRNAs for which a significant eQTL was identified (FDR < 0.1), using a consensus-based software approach in MiRWalk 2.0 (Dweep et al. 2011). MiRWalk 2.0 is a highly regarded software/database archive that documents predictions from 12 independent prediction algorithms, including DIANA-microTv4.0 (Maragkakis et al. 2011), DIANA-microT-CDS (Paraskevopoulou et al. 2013), miRanda-rel2010 (Betel et al. 2010), mirBridge (Tsang et al. 2010), miRDB4.0 (Wang and El Naqa, 2008), miRmap (Vejnar et al. 2013), miRNAMap (Hsu et al. 2008), doRiNA (Blin et al. 2015), PITA (Kertesz et al. 2007), PICTAR2 (Krek et al. 2005), RNAhybrid (Rehmsmeier et al. 2004), and Targetscan (Grimson et al. 2007). MiRWalk 2.0 combines the information from these platforms to build miRNA target predictions in the promoter region, coding sequences, 5′- and 3′-UTRs. A consensus-based approach to target prediction has been recently suggested to be more useful since it minimizes the occurrence of false positives (Zhang & Verbeek, 2010; Pio et al. 2014; Yu et al. 2014). Targets for each miRNA were selected if they had a seed length ≥7 bases, were located with the gene’s 3′-UTR and if the same binding site was predicted in ≥6 of the software systems that MiRWalk archives.
Using these targets, we analyzed the pathway enrichment using the human Wiki Pathways dataset, April 2014 release (Kelder et al. 2012). We first identified all genes targeted by miRNAs with significant cis-eQTLs which were then organized into gene pathways enriched for these miRNAs; second, the gene pathways were further assessed to identify pathways specifically enriched for the intragenic or intergenic miRNAs only. Pathway enrichment, at both stages, was determined using a hypergeometric test and correction for multiple testing was performed using methods outlined by Benjamini and Hochberg (Benjamini & Hochberg, 1995).
Results
Detection of SNP enrichment
Of the 2567 selected SNPs, four were shown by PolyPhen2 to be potentially impactful to mRNA/miRNA expression (score > 0.4). Furthermore, bioinformatic assessment of using SNPINFO (Xu & Taylor, 2009) indicated that the majority of SNPs had no clear predicted biological function related to the regulation of gene expression, with the exception of few being associated with conserved transcription factor binding sites (online Supplementary Table S2). We therefore believe that there was no other biological effect exerted by the SNPs on the level of enrichment other than through their relationship with the miRNAs.
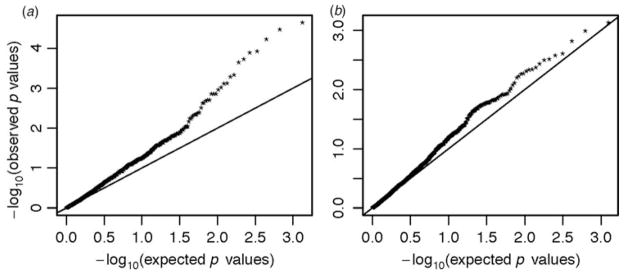
Based on the Simes test, enrichment of miRNA SNPs associated with both SZ (p = 0.0023; Fig. 1a) and to a lesser extent with BD (p = 0.038, Fig. 1b) was detected. After applying the SST, which adjusts for the overall background enrichment of GWAS signals, the enrichment for SZ remained significant (empirical p = 0.018), while the enrichment for BD was suggestive (empirical p = 0.07). Detailed description of these two statistical approaches are provided in the online Supplementary material. The types of miRNAs that these SNPs tagged were roughly 2:1 (23 intragenic: nine intergenic miRNAs; Table 1), suggesting that, the source of the signal may have also originated from the mRNA genes that harbor them. However, despite the belief that intragenic miRNAs share promoters with their host mRNAs, recent studies suggest that this may not be entirely correct with some miRNAs such as hsa-miR-128-2 contain their own transcription regulatory elements (Ozsolak et al. 2008; Monteys et al. 2010). Therefore, to maximize our power, we have chosen to analyze both sets of SNPs as a single unit; however, as there might be distinct biological implications for the genes targeted by the intergenic v. intragenic miRNAs, thus we drew a clear distinction between these two classes of miRNAs when performing the pathway analysis.
Fig. 1.
Quantile–quantile (QQ) plot of the genome-wide association studies signals enrichment among the miRNA expression quantitative trait loci in schizophrenia (SZ) and bipolar disorder (BD). The expected −log10 p values under the null hypothesis are represented on the x axis, while the observed values are represented on the y axis. (a) Shows the enrichment for SZ single nucleotide polymorphisms (SNPs) and (b) for BD SNPs.
Table 1.
A master table showing pertinent information to the detection of 32 eQTLs and the miRNAs to which they are linked
| Disorder | miRNA | miRNA origin | Chromosome | miRNA base pair location | eQTL | eQTL base pair location | Alleles | Global MAF | eQTL p value | eQTL q value |
|---|---|---|---|---|---|---|---|---|---|---|
| SZ/BD | hsa-miR-34a | Intergenic | 1 | 9151668-9151777 | rs12119878 | 9158662 | G/T | 0.1967 | 1.55 × 10−4 | 1.08 × 10−2 |
| SZ | hsa-miR-138 | Intergenic | 3 | 44114212-44114310 | rs6788142 | 44128954 | A/G | 0.1899 | 2.16 × 10−6 | 6.75 × 10−3 |
| SZ/BD | hsa-mir-206 | Intergenic | 6 | 52144349-52144434 | rs6921008 | 52610560 | C/T | 0.2274 | 1.88 × 10−4 | 8.46 × 10−2 |
| SZ/BD | hsa-mir-124-1 | Intergenic | 8 | 9903388-9903472 | rs7461939 | 9714111 | A/G | 0.4407 | 1.07 × 10−3 | 2.90 × 10−2 |
| SZ | hsa-mir-1251 | Intergenic | 12 | 97491909-97491978 | rs4762481 | 98310417 | A/G | 0.3349 | 3.64 × 10−4 | 7.28 × 10−2 |
| SZ | hsa-mir-494 | Intergenic | 14 | 101029634-101029714 | rs9324038 | 101408012 | A/G | 0.2015 | 2.55 × 10−4 | 7.53 × 10−3 |
| SZ | hsa-mir-138-2 | Intergenic | 16 | 56858518-56858601 | rs1566439 | 56990750 | A/G | 0.3806 | 3.38 × 10−3 | 8.45 × 10−2 |
| SZ/BD | hsa-mir-328 | Intergenic | 16 | 67202321-67202395 | rs28171 | 66537518 | A/G | 0.0519 | 2.92 × 10−4 | 7.00 × 10−2 |
| SZ/BD | hsa-mir-132-3p | Intergenic | 17 | 2049908-2050008 | rs17834974 | 2742577 | A/G | 0.2726 | 7.07 × 10−4 | 8.84 × 10−2 |
| SZ | hsa-mir-9-1 | Intragenic | 1 | 156420341-156420429 | rs4661040 | 156399474 | A/G | 0.2849 | 8.28 × 10−4 | 2.24 × 10−2 |
| SZ/BD | hsa-mir-205 | Intragenic | 1 | 209432133-209432242 | rs11577858 | 209404159 | A/G | 0.3478 | 5.34 × 10−4 | 6.68 × 10−2 |
| SZ | hsa-mir-664 | Intragenic | 1 | 220200538-220200619 | rs6669360 | 219474597 | A/T | 0.3636 | 1.71 × 10−3 | 5.98 × 10−2 |
| SZ | hsa-mir-153 | Intragenic | 2 | 219294111-219294200 | rs1425029 | 219729817 | A/G | 0.3736 | 1.43 × 10−3 | 5.01 × 10−2 |
| SZ | hsa-mir-217 | Intragenic | 2 | 55982967-55983076 | rs7604029 | 56042395 | C/T | 0.3231 | 7.96 × 10−4 | 7.16 × 10−2 |
| SZ | hsa-miR-135a | Intragenic | 3 | 52294219-52294308 | rs3733047 | 52837913 | A/G | 0.2452 | 1.68 × 10−6 | 1.88 × 10−3 |
| SZ | hsa-mir-218-1-3p | Intragenic | 4 | 20528275-20528384 | rs4389575 | 21020885 | A/G | 0.4599 | 6.22 × 10−4 | 1.04 × 10−2 |
| SZ | hsa-mir-874 | Intragenic | 5 | 137647572-137647649 | rs10060914 | 137497671 | A/G | 0.3766 | 1.74 × 10−4 | 1.56 × 10−2 |
| SZ | hsa-mir-103-1 | Intragenic | 5 | 168560896-168560973 | rs1422021 | 169357195 | A/G | 0.2107 | 4.38 × 10−4 | 6.58 × 10−2 |
| SZ | hsa-mir-449a | Intragenic | 5 | 55170532-55170622 | rs35930 | 54285894 | A/G | 0.2752 | 6.76 × 10−4 | 1.01 × 10−2 |
| SZ | hsa-mir-597 | Intragenic | 8 | 9741672-9741768 | rs735449 | 9245937 | G/T | 0.2388 | 8.24 × 10−4 | 2.88 × 10−2 |
| SZ/BD | hsa-mir-32 | Intragenic | 9 | 109046229-109046298 | rs10114192 | 108859784 | C/T | 0.497 | 1.69 × 10−2 | 1.18 × 10−2 |
| SZ | hsa-mir-601 | Intragenic | 9 | 123402525-123402603 | rs10739620 | 122840801 | C/G | 0.1378 | 2.12 × 10−4 | 7.42 × 10−3 |
| SZ | hsa-mir-101 | Intragenic | 9 | 4850297-4850375 | rs10974788 | 4795595 | G/T | 0.1819 | 1.69 × 10−4 | 1.18 × 10−2 |
| SZ | hsa-mir-7-1-3p | Intragenic | 9 | 83969748-83969857 | rs1502683 | 82984665 | C/T | 0.4323 | 2.69 × 10−3 | 9.42 × 10−2 |
| SZ/BD | hsa-mir-27b | Intragenic | 9 | 95085445-95085541 | rs10993280 | 94647489 | C/T | 0.4782 | 6.25 × 10−4 | 9.21 × 10−2 |
| SZ/BD | hsa-mir-148b | Intragenic | 12 | 54337216-54337314 | rs2306851 | 54854422 | C/T | 0.1372 | 7.33 × 10−3 | 9.53 × 10−2 |
| SZ/BD | hsa-mir-16-1-3p | Intragenic | 13 | 50048973-50049061 | rs7338471 | 49406928 | G/T | 0.3548 | 1.16 × 10−4 | 3.09 × 10−2 |
| SZ | hsa-mir-19b-1-3p | Intragenic | 13 | 91351192-91351278 | rs4773657 | 91963767 | C/T | 0.4692 | 4.37 × 10−4 | 5.37 × 10−2 |
| SZ/BD | hsa-mir-190a | Intragenic | 15 | 62823957-62824041 | rs12438131 | 63465635 | C/G | 0.4998 | 5.55 × 10−3 | 1.39 × 10−2 |
| SZ | hsa-mir-484 | Intragenic | 16 | 15643294-15643372 | rs6498573 | 15785516 | C/T | 0.2236 | 3.08 × 10−3 | 8.33 × 10−2 |
| SZ | hsa-mir-744 | Intragenic | 17 | 12081899-12081996 | rs1468501 | 12018627 | C/T | 0.1965 | 1.19 × 10−3 | 1.07 × 10−2 |
| SZ | hsa-mir-195 | Intragenic | 17 | 7017615-7017701 | rs11655246 | 6124519 | C/T | 0.345 | 3.65 × 10−5 | 1.61 × 10−2 |
eQTL, Expression quantitative trait loci; miRNA, microRNA; MAF, minor allele frequency; SZ, schizophrenia; BD, bipolar disorder.
eQTL mapping surrounding miRNAs
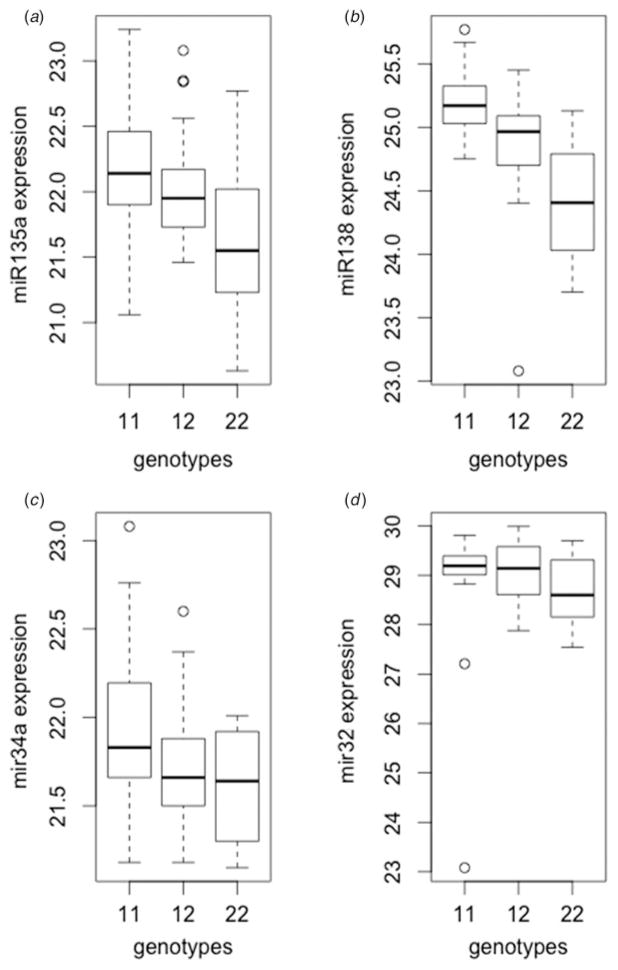
Within our enriched group of SNPs used in stage 1, we identified 116 SNPs to act as miRNA eQTLs at the nominal p ≤ 0.05 of which 32 eQTLs remained significant following FDR correction at 10%. Of these 32 miRNA eQTLs, all were found to relate to SZ and 11 overlapped with BD. Our most significant SZ eQTL, rs3733047 (p = 1.68 ×10−6, q = 0.001876; Fig. 2a) was shown to affect the expression of hsa-miR-135a, which in a recent study was shown to be differentially expressed during the normal brain development (Moreau et al. 2013). Rs3733047 is located in a transcription factor binding site for GATA2 which regulates neuronal differentiation and has been found upregulated in the prefrontal cortex of SZ subjects (Miller et al. 2013). The second most significant SZ eQTL finding (rs6788142, p = 2.16 × 10−6, q = 0.006754; Fig. 2b) affects the expression of hsa-mir-138-1 (miR-138). miR-138 has been shown to decrease the amplitude of postsynaptic currents, negatively regulating spine size in mouse and rat studies (Olsen et al. 2009). The most significant BD eQTL, rs12119878 (p = 0.001549, q = 0.010845; Fig. 2c) was shown to affect the expression of hsa-miR-34a (miR-34a). An increased level of this miRNA was recently identified in the postmortem cerebellar tissue of BD patients (Bavamian et al. 2015); increased miR-34a expression in human iPSC-derived neuronal progenitor cells impacts neuronal differentiation and morphology. The second most significant BD related finding was rs10114192 (p = 0.016858, q = 0.0118006; Fig. 2d) that affected the expression of hsa-miR-32 and this miRNA was shown to be differentially expressed in post-mortem brains exclusively between BD patients and controls and not in SZ patients (Miller et al. 2013).
Fig. 2.
Boxplots showing the relationship between single nucleotide polymorphism genotype and miRNA expression for the two most significant schizophrenia (SZ) and bipolar disorder (BD) expression quantitative trait loci (eQTLs). (a) and (b) show SZ-related eQTLs, while (c) and (d) show BD-related eQTLs.
Other notable individual eQTLs include rs10993280 (p = 0.00062592, q = 0.09213) on chromosome 9 and rs7338471 (p = 0.000116, q = 0.030872) and rs4773657 (p = 0.000437, q = 0.05369) on chromosome 13. Rs10993280 is an eQTL for the hsa-miR-23b, -24-1, and -27b cluster on chromosome 9, while rs7338471 affected the expression of the hsa-miR-15a, and -16 cluster and rs4773657 is an eQTL for the hsa-19b-1, -17, -18a, -20a, and -92a cluster on chromosome 13.
Four eQTLs, rs4389575 (p = 0.000622, q = 0.001036), rs35930 (p = 0.000676, q = 0.010133), rs4661040 (p = 0.005786, q = 0.098362) and rs1422021 (p = 0.000438, q = 0.065754) were associated with the expression of four intragenic miRNAs; these miRNAs (hsa-mir-218-1, -449a, -9-1, and -103-1) have been experimentally shown to target their respective host genes (Wilfred et al. 2007; Bazzoni et al. 2009; Lize et al. 2010; Fish et al. 2011). A list of all 32 eQTLs affecting the expression of their respective miRNAs is given in Table 1. To further ensure the validity of our results as true signals, we compared our eQTLs against the Brain eQTL Almanac (Ramasamy et al. 2014); we found 82.1% of our eQTLs to be also reported in this database.
Of the 632 human pathways currently listed on WikiPathways, 588 showed significant enrichment of the genes predicted to be targets for the 32 miRNAs (q < 0.01). The pathways that demonstrated significant enrichment for intergenic miRNAs involved: (1) apoptosis, (p = 2.00 × 10−8, q = 1.30 × 10−6), (2) B cell receptor signaling pathway (p = 8.29 × 10−7, q = 5.14 × 10−5), (3) calcium signaling in cardiac cells (p = 2.92 × 10−8, q = 1.90 × 10−6), (4) G protein signaling (p = 1.68 × 10−6, q = 1.02 × 10−4), (5) insulin signaling (p = 1.41 × 10−4, q = 5.76 × 10−3), and (6) transforming growth factor beta (TGF-β) signaling (p = 2.26 × 10−4, q = 7.70 × 10−3). The pathways that demonstrated significant enrichment for intragenic miRNAs involved: (1) B cell receptor signaling (p = 3.76 × 10−8, q = 2.22 × 10−6), (2) DNA damage dependent ataxia telangiectasia, mutated (ATM) only (p = 3.44 × 10−7, q = 1.96 × 10−5), (3) insulin signaling (p = 1.07 × 10−9, q = 6.29 × 10−8) and (4) epidermal growth factor (EGF)/epidermal growth factor receptor (EGFR) signaling (p = 1.63 × 10−11, q = 9.11 × 10−10). Table 2 provides information describing all of the significantly enriched pathways.
Table 2.
A summary table showing the enriched pathways predicted to be targeted by the 32 miRNAs
| miRNA type | miRNAs showing significant levels of target enirchment | Wiki pathway name | Number of genes in a given pathway | Average no. of predicted targets in pathway | Single most enriched miRNA | p value | q value |
|---|---|---|---|---|---|---|---|
| Intergenic | 124-1, −1251, −132-3p, −138-1,−138-2, −206, −328, −34a | Apoptosis | 84 | 55 | mir34a | 2.00 × 10−8 | 1.30 × 10−6 |
| Intergenic | 124-1, 1251, 132-3p, 138-1, 138-2, 206, 328, 34a | B cell receptor signaling pathway | 95 | 60 | mir34a | 8.29 × 10−7 | 5.14 × 10−5 |
| Intergenic | 124-1,−1251, −132-3p, −138-1, −138-2, −206, −328, −34a | Calcium regulation in the cardiac cell | 150 | 90 | mir34a | 2.92 × 10−8 | 1.90 × 10−6 |
| Intergenic | 124-1, 1251, 132-3p, 138-1, 138-2, 206, 328, 34a | G protein signaling pathways | 93 | 57 | mir34a | 1.68 × 10−6 | 1.02 × 10−4 |
| Intergenic | 124-1, 1251, 132-3p, 138-1, 138-2, 206, 328, 34a | Insulin signaling | 164 | 96 | mir34a | 1.41 × 10−4 | 5.76 × 10−3 |
| Intergenic | 124-1 −132-3p, −138-1, −138-2,−206, −328, −34a, −494 | TGF-β signaling pathway 1 | 120 | 68 | mir138-2 | 2.26 × 10−4 | 7.70 × 10−3 |
| Intragenic | 101, 103a, 135a, 16-1, 205, 218, 27b, 449a, 601, 664a, 7-1 | B cell receptor signaling pathway | 95 | 65 | mir744 | 3.76 × 10−8 | 2.22 × 10−6 |
| Intragenic | 101-3p, 103a-3p, 135a-5p,153-3p, 16-1-3p, 195-5p, 205-5p, 218-1, 27b −3p, 449a. 601, 744-5p, | DNA damage response only ATM dependent | 84 | 57 | mir744 | 3.44 × 10−7 | 1.96 × 10−5 |
| Intragenic | 101-3p, 103a-3p, 135a-3p. 153-3p, 16-1-3p, 190a-5p, 195-5p, 205-5p, 217, 218-1, 27b-3p, 449a, 597-3p, 664a-5p, 7-1-3p, 744-5p, 874-5p | Insulin signaling | 164 | 96 | mir205-5p | 1.07 × 10−9 | 6.29 × 10−8 |
| Intragenic | 101-3p, 103a-3p, 135a-5p, 153-3p, 195-5p, 205-5p, 217, 218-1, 27b-3p, 449a, 601, 744, 9-3 | EGF/EGFR signaling pathway | 145 | 93 | mir27b-3p | 1.63 × 10−11 | 9.11 × 10−10 |
miRNA, MicroRNA; TGF-β, transforming growth factor beta; ATM, ataxia telangiectasia, mutated; EGFR, epidermal growth factor receptor.
Discussion
In this paper, we present results from a two-stage study that attempts to determine whether SNPs located in close proximity to miRNAs could be enriched for a SZ and BD association signal and whether these SNPs could function as eQTLs for the miRNAs to which they are co-localized. Our approach used the enrichment we detected in the PGC results as guidance for further fine mapping of the same variants in an eQTL analysis of postmortem brain samples from SZ and BD subjects. Our goal with the eQTL analysis in this study is to generate specific testable hypotheses that address the relationship between the observed GWAS signals and the miRNAs location, suggesting the potential impact this relationship might have on the targeted mRNA gene pathways.
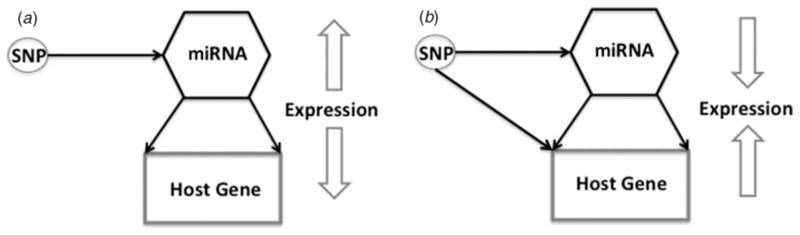
Among these eQTLs, we speculate about two potential pathways by which a GWAS variant can interact with a miRNA in SZ and/or BD (pictured in Fig. 3). We show that SZ and BD significant SNPs could directly influence miRNA expression; however, the manner in which the SNP interacts with the miRNA might differ depending on the miRNA’s origin. miRNAs in the intergenic category might be more strongly affected by an eQTL that would result in detectable levels of differential expression in postmortem expression studies because of the absence of other involved factors mediating this relationship. Of the miRNAs that fell into this group, it is important to note that hsa-miR-132, -34a, -124, -206 and -328 have been reported previously by other researchers to be associated with SZ/BD (Guo et al. 2010; Kim et al. 2010; Lai et al. 2011; Miller et al. 2013; Sun et al. 2015) in either postmortem expression studies or through genetic association. Furthermore, miRNAs within this class have been reported to be highly expressed in the brain, with miR-124 and -138 both being neurally specific and responsible for the differentiation of neural stem cells into neurons (Banerjee et al. 2009; Akerblom et al. 2012; Petri et al. 2014). Finally, hsa-miR-34a and -206, have also been noted to be responsive to mood stabilizers such as lithium and sodium valproate that provide support for their roles in SZ and BD (Hunsberger et al. 2013; Wang et al. 2014). The significantly enriched gene pathways unique to this group of miRNAs include the G protein signaling pathway and TGF-β signaling. Both of these pathways have been implicated in SZ and BD through risk genes in association studies, namely the G-protein-coupled receptors (GPCRs) (Funk et al. 2014) and ZNF804 (Umeda-Yano et al. 2013; Pietersen et al. 2014). GPCRs represent nearly half of the therapeutic drug targets in neuropsychiatric research today (Bridges & Lindsley, 2008). Both, hsa-miR-328 and some of its gene targets belonging to the TGF-β signaling pathway were also recently shown to be differentially expressed in laser-captured pyramidal neurons found in SZ subjects (Pietersen et al. 2014). Other enriched pathways of miRNAs in the intergenic class are involved in immune function and the regulation of signaling through calcium channels; both of these pathways have been implicated in SZ and BD (Hinze-Selch, 2002; de Baumont et al. 2015) with respect to etiology and treatment course.
Fig. 3.
A schematic diagram depicting the hypothesized relationship between expression quantitative trait loci, miRNAs, and mRNA genes. (a) Depiction of ‘coherent’ feed forward loop (FFL) and (b) depiction of ‘incoherent’ FFL. See text for more details. SNP, single nucleotide polymorphism.
The second way in which SZ and BD significant SNPs could influence the expression of a miRNA is by affecting both the miRNA and its host gene in a regulatory loop, although it is unclear at present whether the effect of the SNP within the loop is equally shared between the miRNA and the host or not. The potential for regulatory loops involving miRNAs is nonetheless quite interesting with respect to SZ and BD disease etiology (Guo et al. 2010; Li et al. 2014; Shenoy & Blelloch, 2014). Several types of regulatory loops have been described; a feed forward loop (FFL), for instance, involves a transcription factor (TF) that binds to a cis-regulatory element upstream of the promoter for a miRNA and its mRNA target. In ‘coherent’ FFLs where the TF activates the miRNA that represses the mRNA gene, the controlling element in the loop is the TF and the relationship between all three elements is relatively balanced. In ‘incoherent’ FFLs, however, the TF activates both the miRNA and the mRNA; the relationship becomes more difficult to parse out because the action of the miRNA compensates against the activity of the TF on the mRNA gene. Several regulatory loops involving miRNAs have been described in connection to brain development and neuronal differentiation (Zhao et al. 2009; Sun et al. 2011; Liu et al. 2013). In this study, we found eQTLs for four miRNAs (hsa-miR-218-1, -449a, -9-3, and -103-1) for which there was experimental validation of the relationship between the host and the miRNA gene and instances of FFLs involving these same miRNAs (Wilfred et al. 2007; Bazzoni et al. 2009; Lize et al. 2010; Fish et al. 2011). We believe that with additional experimental validation of the relationship between the miRNAs in this class and their predicted targets, we will see additional examples of FFLs.
The enriched pathways targeted by the intragenic miRNA group were insulin signaling, B cell receptor signaling, DNA damage response only ATM dependent, and EGF/EGFR signaling. Two of these pathways, involving insulin signaling and B cell receptor signaling, overlapped with the intergenic group of miRNAs due to sequence similarities in hsa-miR-449a and -34a. The other enriched pathways that were unique to the intragenic miRNAs were the DNA damage response only ATM dependent and the EGF/EGFR signaling pathways. Of these, the EGF/EGFR signaling pathway has the most significant implications for neuropsychiatric disorders like SZ. The ligands for this pathway, in the ErbB families and neuregulin (NRG) families, are found in abundance in GABAergic and dopaminergic neurons and in glial cells. ErbB1 ligands, in particular, have been shown to negatively regulate GABAergic development in the neocortex and influence activity of glutamate receptor channels (Nagano et al. 2007). In addition, genetic and candidate gene studies have implicated both ErbB1-4 and NRG1-6 in SZ (Barnes et al. 2012; Deng et al. 2013; Nawaz et al. 2014).
By integrating eQTL analysis with the association signal seen in the PGCGWAS dataset, we can offer general hypotheses about the role of miRNAs in SZ and BD. First, due in large part to their location and the lack of other apparent factors directly mediating expression, the influence of intergenic miRNAs may be more readily apparent and directed in SZ/BD subjects than in intragenic miRNAs. The influence of intragenic miRNAs in SZ/BD might, in turn, be easily missed if they participate in regulatory loops and focus is only given to the miRNA and not the other loop ‘participants’.
We acknowledge several limitations to this study. First, there are significant challenges posed by working with postmortem brain samples that may lead to inconsistencies in study replication and reduced power (McCullumsmith et al. 2014). Factors such as age at death, PMI, brain PH, and prior exposure to specific medicines are potential confounders that, even when attempted to adjust for, may still confound the results from different expression studies when these are compared. In addition, although in recent years the numbers of postmortem brain samples have been steadily increasing, the sample size of currently available postmortem brain studies is still small, thus reducing statistical power and replicability. Furthermore, the composition of postmortem brain samples is often a mixture of tissues and cell types making it difficult to ascertain with certainty the exact source of the finding. The use of postmortem brains, nevertheless, offers the best possible substrate for studying complex human behavior simultaneously on a genetic and molecular level and provides a way in which to generate specific testable hypotheses that animal models may not allow.
A second perceived limitation of our study is that we deliberately chose not to distinguish the level of SZ and/or BD association enrichment between intergenic and intragenic miRNAs. Instead, we chose to look at the differences between the miRNA groups in terms of the predicted targets of each group and their respective pathways. Indeed, while we cannot unequivocally state that the level of enrichment did not result from the influence of the protein coding genes that hosted the intragenic miRNAs, we would still suggest that the effect is nonetheless present and inextricably linked to miRNAs. What our study does suggest is that certain classes of miRNAs may be more directly connected to genetic factors and that these may be the ones for whom neuronal cell signaling and fate might be a key function. In that regard, our study provides a successful approach to breach the gap between purely genetic (associations) and molecular (expression) studies conducted in neuropsychiatric disorders such as SZ and BD.
Supplementary Material
Suppl Table 1. Demographic Information for the
Acknowledgments
This work was supported by the Stanley Medical Research Institute (V.I.V., no. 08R-1959), the National Institute of Mental Health (S.A.B, MH100560) and the National Institute on Alcohol Abuse and Addiction (S.A.B. and V.I.V., AA02271). The summary statistics of PGC schizophrenia were obtained from https://pgc.unc.edu/Sharing.php#SharingOpp. We are grateful to the PGC investigators who produced and analyzed these datasets.
Footnotes
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0033291715000483.
Declaration of Interest
None.
References
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics. 2013;76:7.20.1–7.20.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. Journal of Neuroscience. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacanu SA, Chen J, Sun J, Richardson K, Lai CQ, Zhao Z, O’Donovan MC, Kendler KS, Chen X. Functional SNPs are enriched for schizophrenia association signals. Molecular Psychiatry. 2014;19:276–277. doi: 10.1038/mp.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Barnes A, Isohanni M, Barnett JH, Pietilainen O, Veijola J, Miettunen J, Paunio T, Tanskanen P, Ridler K, Suckling J, Bullmore ET, Jones PB, Murray GK. Neuregulin-1 genotype is associated with structural differences in the normal human brain. Neuroimage. 2012;59:2057–2061. doi: 10.1016/j.neuroimage.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, Madison JM, Zhou F, Rueckert EH, Barker D, Perlis RH, Sur M, Haggarty SJ. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Molecular Psychiatry. 2015 doi: 10.1038/mp.2014.176. Published online: 27 January 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proceedings of the National Academy Sciences USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biology. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Human Molecular Genetics. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Blin K, Dieterich C, Wurmus R, Rajewsky N, Landthaler M, Akalin A. DoRiNA 2.0-upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Research. 2015;43:D160–D167. doi: 10.1093/nar/gku1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature Structural & Molecular Biology. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Bridges TM, Lindsley CW. G-protein-coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chemical Biology. 2008;3:530–541. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]
- Collins AL, Kim Y, Bloom RJ, Kelada SN, Sethupathy P, Sullivan PF. Transcriptional targets of the schizophrenia risk gene MIR137. Translational Psychiatry. 2014;4:e404. doi: 10.1038/tp.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Baumont A, Maschietto M, Lima L, Carraro DM, Olivieri EH, Fiorini A, Barreta LA, Palha JA, Belmonte-de-Abreu P, Moreira Filho CA, Brentani H. Innate immune response isdifferentially dysregulated between bipolar disease and schizophrenia. Schizophrenia Research. 2015;161:215–221. doi: 10.1016/j.schres.2014.10.055. [DOI] [PubMed] [Google Scholar]
- Deng C, Pan B, Engel M, Huang XF. Neuregulin-1 signalling and antipsychotic treatment: potential therapeutic targets in a schizophrenia candidate signalling pathway. Psychopharmacology (Berlin) 2013;226:201–215. doi: 10.1007/s00213-013-3003-2. [DOI] [PubMed] [Google Scholar]
- Down TA, Hubbard TJ. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Research. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk – database: prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. Journal of Biomedical Informatics. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS. Evidence for X-chromosomal schizophrenia associated with microRNA alterations. PLoS One. 2009;4:e6121. doi: 10.1371/journal.pone.0006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Reviews Genetics. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fish JE, Wythe JD, Xiao T, Bruneau BG, Stainier DY, Srivastava D, Woo S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development. 2011;138:1409–1419. doi: 10.1242/dev.060046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero DA, van der Ven K, Callaerts P, Del-Favero J. miRNA genes and the brain: implications for psychiatric disorders. Human Mutation. 2010;31:1195–1204. doi: 10.1002/humu.21344. [DOI] [PubMed] [Google Scholar]
- Funk AJ, Haroutunian V, Meador-Woodruff JH, McCullumsmith RE. Increased G protein-coupled receptor kinase (GRK) expression in the anterior cingulate cortex in schizophrenia. Schizophrenia Research. 2014;159:130–135. doi: 10.1016/j.schres.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ, Gershon ES, Kelsoe JR, Greenwood TA, Nievergelt CM, Chen C, McKinney R, Shilling PD, Schork NJ, Smith EN, Bloss CS, Nurnberger JI, Edenberg HJ, Foroud T, Koller DL, Scheftner WA, Coryell W, Rice J, Lawson WB, Nwulia EA, Hipolito M, Byerley W, McMahon FJ, Schulze TG, Berrettini WH, Potash JB, Zandi PP, Mahon PB, McInnis MG, Zöllner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Liu C. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Molecular Psychiatry. 2013a;18:340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Innocenti F, Wei R, Wang L, Zhang M, Mirkov S, Ramírez J, Huang RS, Cox NJ, Ratain MJ, Liu W. A genome-wide integrative study of microRNAs in human liver. BMC Genomics. 2013b;14:395. doi: 10.1186/1471-2164-14-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Ziliak D, Im HK, LaCroix B, Park DS, Cox NJ, Huang RS. Genetic architecture of microRNA expression: implications for the transcriptome and complex traits. American Journal of Human Genetics. 2012;90:1046–1063. doi: 10.1016/j.ajhg.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geaghan M, Cairns MJ. MicroRNA and Posttranscriptional Dysregulation in Psychiatry. Biological Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N. Intronic microRNAs: a crossroad in gene regulation. Biochemical Society Transactions. 2012;40:759–761. doi: 10.1042/BST20120023. [DOI] [PubMed] [Google Scholar]
- Guo AY, Sun J, Jia P, Zhao Z. A Novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC System Biology. 2010;4:10. doi: 10.1186/1752-0509-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T, Olsen L, Lindow M, Jakobsen KD, Ullum H, Jonsson E, Andreassen OA, Djurovic S, Melle I, Agartz I, Hall H, Timm S, Wang AG, Werge T. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS One. 2007;2:e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinze-Selch D. Infection, treatment and immune response in patients with bipolar disorder versus patients with major depression, schizophrenia or healthy controls. Bipolar Disorders. 2002;4(Suppl 1):81–83. doi: 10.1034/j.1399-5618.4.s1.32.x. [DOI] [PubMed] [Google Scholar]
- Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Research. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsberger JG, Fessler EB, Chibane FL, Leng Y, Maric D, Elkahloun AG, Chuang DM. Mood stabilizer-regulated miRNAs in neuropsychiatric and neurodegenerative diseases: identifying associations and functions. American Journal of Translational Research. 2013;5:450–464. [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Jia P, Shen B, Zhao Z. Top associated SNPs in prostate cancer are significantly enriched in cis-expression quantitative trait loci and at transcription factor binding sites. Oncotarget. 2014;5:6168–6177. doi: 10.18632/oncotarget.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Research. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nature Genetics. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophrenia Research. 2012;141:60–64. doi: 10.1016/j.schres.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophrenia Research. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Webster MJ. The stanley neuropathology consortium integrative database: a novel, web-based tool for exploring neuropathological markers in psychiatric disorders and the biological processes associated with abnormalities of those markers. Neuropsychopharmacology. 2010;35:473–482. doi: 10.1038/npp.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nature Genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Molecular Psychiatry. 2013;18:11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, Huang YH, Hsiao PC, Hsiao CK, Liu CM, Yang PC, Hwu HG, Chen WJ. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liang C, Easterbrook S, Luo J, Zhang Z. Investigating the functional implications of reinforcing feedback loops in transcriptional regulatory networks. Molecular Biosystems. 2014;10:3238–3248. doi: 10.1039/c4mb00526k. [DOI] [PubMed] [Google Scholar]
- Liu C, Teng ZQ, McQuate AL, Jobe EM, Christ CC, von Hoyningen-Huene SJ, Reyes MD, Polich ED, Xing Y, Li Y, Guo W, Zhao X. An epigenetic feedback regulatory loop involving microRNA-195 and MBD1 governs neural stem cell differentiation. PLoS One. 2013;8:e51436. doi: 10.1371/journal.pone.0051436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lize M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death and Differentiation. 2010;17:452–458. doi: 10.1038/cdd.2009.188. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG. DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Research. 2011;39:W145–W148. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullumsmith RE, Hammond JH, Shan D, Meador-Woodruff JH. Postmortem brain: an underutilized substrate for studying severe mental illness. Neuropsychopharmacology. 2014;39:65–87. doi: 10.1038/npp.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiological Reviews. 2008;30:67–76. doi: 10.1093/epirev/mxn001. [DOI] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz T, Deng S, Strathmann J, Willoughby D, Kenny P, Elsworth J, Lawrence M, Roth R, Edbauer D, Kleiman R, Wahlested C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proceedings of the National Academy of Sciences of the United States of America. 2013;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, Jornsten R, Liu Y, Brzustowicz LM. Chronological changes in microRNA expression in the developing human brain. PLoS One. 2013;8:e60480. doi: 10.1371/journal.pone.0060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Namba H, Abe Y, Aoki H, Takei N, Nawa H. In vivo administration of epidermal growth factor and its homologue attenuates developmental maturation of functional excitatory synapses in cortical GABAergic neurons. European Journal of Neuroscience. 2007;25:380–390. doi: 10.1111/j.1460-9568.2007.05297.x. [DOI] [PubMed] [Google Scholar]
- Nawaz R, Asif H, Khan A, Ishtiaq H, Shad F, Siddiqui S. Drugs targeting SNPrs35753505 of the NRG1 gene may prevent the association of neurological disorder schizophrenia in a Pakistani population. CNS Neurolologial Disorders and Drug Targets. 2014;13:1604–1614. doi: 10.2174/1871527313666140806150440. [DOI] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genetics. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS ONE. 2009;4:e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes and Development. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Research. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin JP, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biology. 2007;8:11. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R, Malmevik J, Fasching L, Akerblom M, Jakobsson J. miRNAs in brain development. Experimental Cell Research. 2014;321:84–89. doi: 10.1016/j.yexcr.2013.09.022. [DOI] [PubMed] [Google Scholar]
- Pietersen CY, Mauney SA, Kim SS, Lim MP, Rooney RJ, Goldstein JM, Petryshen TL, Seidman LJ, Shenton ME, McCarley RW, Sonntag KC, Woo TU. Molecular profiles of pyramidal neurons in the superior temporal cortex in schizophrenia. Journal of Neurogenetics. 2014;28:53–69. doi: 10.3109/01677063.2014.882918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pio G, Malerba D, D’Elia D, Ceci M. Integrating microRNA target predictions for the discovery of gene regulatory networks: a semi-supervised ensemble learning approach. BMC Bioinformatics. 2014;15(Suppl 1):S4. doi: 10.1186/1471-2105-15-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric Genetics Consortium (PGC) Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam P, Palanichamy JK, Singh A, Das P, Bhagat M, Kassab MA, Sinha S, Chattopadhyay P. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA. 2014;20:76–87. doi: 10.1261/rna.041814.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, De T, Coin L, de Silva R, Cookson MR, Singleton AB, Hardy J, Ryten M, Weale ME. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nature Neuroscience. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, Sanders AR, Purcell S, Visscher PM, Craddock N, Owen MJ, Holmans P, O’Donovan MC. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Molecular Psychiatry. 2012;17:193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Holmans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O’Neill FA, Owen MJ, Milovancevic MP, Posthuma D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Levinson DF, Gejman PV, Kendler KS, Laurent C, Mowry BJ, O’Donovan MC, Owen MJ, Pulver AE, Riley BP, Schwab SG, Wildenauer DB, Dudbridge F, Holmans P, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, O’Neill FA, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Lin K, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Powell J, Rujescu D, Van Os J, Walshe M, Weisbrod M, Wiersma D, Donnelly P, Barroso I, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, Barroso I, Deloukas P, Mathew CG, Blackwell JM, Brown MA, Corvin AP, McCarthy MI, Spencer CC, Bramon E, Corvin AP, O’Donovan MC, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, Stahl EA, Georgakopoulos A, Ruderfer DM, Charney A, Okada Y, Siminovitch KA, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Plenge RM, Raychaudhuri S, Fromer M, Purcell SM, Brennand KJ, Robakis NK, Schadt EE, Akbarian S, Sklar P. A role for noncoding variation in schizophrenia. Cell Reports. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proceedings of the National Academy of Science USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biological Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nature Reviews Molecular Cell Biology. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, Price A, Raj T, Nisbett J, Nica AC, Beazley C, Durbin R, Deloukas P, Dermitzakis ET. Patterns of cis regulatory variation in diverse human populations. PLoS Genetics. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, Ma X, Shi Y. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nature Communications. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XY, Lu J, Zhang L, Song HT, Zhao L, Fan HM, Zhong AF, Niu W, Guo ZM, Dai YH, Chen C, Ding YF, Zhang LY. Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. Journal of Clinical Neuroscience. 2015;22:570–574. doi: 10.1016/j.jocn.2014.08.018. [DOI] [PubMed] [Google Scholar]
- Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Molecular Cell. 2010;38:140–153. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda-Yano S, Hashimoto R, Yamamori H, Okada T, Yasuda Y, Ohi K, Fukumoto M, Ito A, Takeda M. The regulation of gene expression involved in TGF-beta signaling by ZNF804A, a risk gene for schizophrenia. Schizophrenia Research. 2013;146:273–278. doi: 10.1016/j.schres.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Vejnar CE, Blum M, Zdobnov EM. miRmap web: comprehensive microRNA target prediction online. Nucleic Acids Research. 2013;41:W165–W168. doi: 10.1093/nar/gkt430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang C, Huang J, Yuan C, Hong W, Chen J, Yu S, Xu L, Gao K, Fang Y. MiRNA-206 and BDNF genes interacted in bipolar I disorder. Journal of Affective Disorders. 2014;162:116–119. doi: 10.1016/j.jad.2014.03.047. [DOI] [PubMed] [Google Scholar]
- Westra HJ, Franke L. From genome to function by studying eQTLs. Biochimica et Biophysica Acta. 2014;1842:1896–1902. doi: 10.1016/j.bbadis.2014.04.024. [DOI] [PubMed] [Google Scholar]
- Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Molecular Genetics and Metabolism. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li F, Zhang B, Zhang K, Zhang F, Huang X, Sun N, Ren Y, Sui M, Liu P. MicroRNAs and target site screening reveals a pre-microRNA-30e variant associated with schizophrenia. Schizophrenia Research. 2010;119:219–227. doi: 10.1016/j.schres.2010.02.1070. [DOI] [PubMed] [Google Scholar]
- Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Research. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Kim J, Min H, Yoon S. Ensemble learning can significantly improve human microRNA target prediction. Methods. 2014;69:220–229. doi: 10.1016/j.ymeth.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gierman HJ, Levy D, Plump A, Dobrin R, Goring HH, Curran JE, Johnson MP, Blangero J, Kim SK, O’Donnell CJ, Emilsson V, Johnson AD. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics. 2014;15:532. doi: 10.1186/1471-2164-15-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Verbeek FJ. Comparison and integration of target prediction algorithms for microRNA studies. Journal of Integrative Bioinformatics. 2010;7(3):127. doi: 10.2390/biecoll-jib-2010-127. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature Structural and Molecular Biology. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl Table 1. Demographic Information for the