Abstract
Chikungunya virus (CHIKV) is an important emerging mosquito-borne alphavirus, indigenous to tropical Africa and Asia. It can cause epidemic fever and acute illness characterized by fever and arthralgias. The epidemic cycle of this infection is similar to dengue and urban yellow fever viral infections. The generation of an efficient vaccine against CHIKV is necessary to prevent and/or control the disease manifestations of the infection. In this report, we studied immune response against a CHIKV-envelope DNA vaccine (pEnv) and the role of the CHIKV nonstructural gene 2 (nsP2) as an adjuvant for the induction of protective immune responses in a relevant mouse challenge model. When injected with the CHIKV pEnv alone, 70% of the immunized mice survived CHIKV challenge, whereas when co-injected with pEnv+pnsP2, 90% of the mice survived viral challenge. Mice also exhibited a delayed onset signs of illness, and a marked decrease in morbidity, suggesting a nsP2 mediated adjuvant effect. Co-injection of the pnsP2 adjuvant with pEnv also qualitatively and quantitatively increased antigen specific neutralizing antibody responses compared to vaccination with pEnv alone. In sum, these novel data imply that the addition of nsP2 to the pEnv vaccine enhances anti-CHIKV-Env immune responses and maybe useful to include in future CHIKV clinical vaccination strategies.
Introduction
Chikungunya virus (CHIKV) is an emerging human pathogen that causes a disease syndrome characterized by fever and arthralgias, and in severe cases a fatal hemorrhagic disease (1–5). CHIKV belongs to the genus Alphaviridae, and is geographically distributed from Africa to Southeast Asia and South America and transmitted to humans primarily by the Aedes aegypti mosquito (3, 6–9). Ae. aegypti is found in the Indian Ocean islands affected by CHIKV but is generally less abundant than Ae. albopictus (1, 10, 11). The failure to control Ae. aegypti is generally attributed more to its endemicity and athropophilicity than to insecticide resistance. Therefore, development of promising therapies or potent vaccines against CHIKV is greatly needed. One attractive approach for vaccine development is the DNA plasmid-based approach. We have previously reported that DNA vaccination with the E1, E2, and capsid genes of CHIKV-induced cellular and humoral immune responses in mice (12–15).
Recently, there has been increased interest in understanding the role of the nonstructural proteins in pathogenicity, as well as their potential to modulate antigen processing and vaccine responses against the pathogen of interest (16–18). Boural et al. (17) report a targeted ns2 vaccine was able to interrupt virus infection, while others report that chimeric proteins covering fusions of heterologous non-structural proteins from, VEE to CHIKV structural genes led to increased anti-CHIKV titers (12, 15, 17, 19, 20). Here we sought to investigate whether CHIKV NS2 protein could serve to adjuvant anti-CHIKV Env responses in a homologous manner. Such an activity would be novel and support the inclusion of broader gene sequences (notably the nonstructural proteins) in the development of effective vaccines against pathogens of interest.
In the study reported here, we explored the potential adjuvant effect of the nonstructural protein-2 (nsP2) expressing DNA plasmid (designated pnsP2) on protective immunity elicited by a CHIKV-Env expressing DNA vaccine. In these studies, addition of pnsP2 was able to markedly enhance both cellular and humoral immune responses. Furthermore, the nsP2 adjuvant increased survival in a lethal CHIKV challenge model as well as enhanced neutralizing antibody responses against several epidemic CHIKV viruses. These findings underscore further areas for CHIKV vaccine research and indicate that the nsP2 protein can function as a major immune adjuvant and also suggests that CHIKV specific antibodies are a critical component in protective immunity.
Materials and Methods
Cells, virus, and mice
HeLa (ATCC- CCL2) and Vero 76 (ATCC CRL-1587) cell lines were cultured as described previously (21). Female C57BL/6 mice, used in this study, were 6-to 8-week-old and purchased from Jackson Laboratory (Indianapolis, IN). PC-08 CHIKV strain viruses were passaged every 48 h onto Vero E6 cells until cytopathology was observed and propagated and was used for challenge experiments (21). All animals were housed in accordance with the guidelines of the National Institutes of Health (NIH-Bethesda, MD) and the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC).
Plasmid DNA construct and in vivo EP delivery
Codon and RNA optimized CHIKV-nsP2 genes were modified by inclusion of a human IgE leader peptide at the N-terminus and a HA tag to the C-terminus for immuno detection. nsP2 genes were then cloned into a pVax1 mammalian expression vector (Invitrogen, Carlsbad, CA). The pEnv utilized was the same as used in a previous study (9, 21). All plasmid constructs were amplified and purified using Qiagen maxi kit (Qiagen, Valencia, CA). Three groups of C57BL/6 mice (n=10) were immunized intramuscularly with pVax1 (Control), pEnv, and pEnv+pnsP2 respectively. Following a prime vaccination, the quadriceps muscle of C57BL/6 mice was injected with two booster doses on day 14 and day 21. For immunization, 25 μg of plasmid DNA in 50 μL volume (pVax1 and pEnv, pEnv+pnsP2 or nsP2was injected. Intramuscular injection of the DNA vaccine/adjuvants was followed by MID-EP system (CELLECTRA®; Inovio Pharmaceuticals, Blue Bell, PA) (three pulses of 0.5 Amp constant current, 1 second apart and 52 ms in length). Each animal received a total of 3 immunizations, 2 weeks apart as described previously (21). The mice were sacrificed 7 days after the last immunization, whereupon sera were collected and the spleen was isolated for immunological assays.
pCHIKV-nsP2 expression study
In vitro expression of the pCHIKV-nsP2 construct was confirmed by a T7-based coupled transcription/translation system (Promega, Madison, WI) as described previously (9). In vivo expression of nsP2 construct was confirmed after transfection of HeLa cells by indirect immunofluorescence (IFA) as described previously (9, 22). All images were captured by Zeiss LSM 510 Confocal microscope system.
Cellular responses: ELISpot assay
Ninety-six-well ELISpot plates (Millipore, Billerica, MA) were coated with anti-mouse IFN-γ capture antibody and incubated overnight at 4°C (R&D Systems; Minneapolis, MN). The following day, plates were washed with PBS and blocked for 2 h with 1% BSA. Two hundred thousand splenocytes were then added to each well and stimulated overnight with RPMI 1640 (negative control), Con A (positive control), or specific antigenic peptides (10 μg/mL) consisting of pools of 15-mer peptides, overlapping by 11 amino acids. After 24 h of stimulation, the cells were washed with PBS and incubated with biotinylated anti-mouse IFN-γ antibodies (R&D Systems, Minneapolis, MN) overnight followed by washing and incubation with streptavidin-alkaline phosphatase (R&D Systems) for 2 h. Following washing, 5-bromo-4-chloro-3′-indolylphosphate p-toluidine salt and nitro blue tetrazolium chloride (chromogen color reagent; R&D Systems) were added to each well. The plates were then rinsed with distilled water and dried, and spots were subsequently counted by an automated ELISpot reader (CTL Limited, Shaker Heights, OH).
Humoral immune responses: Antibody ELISA
Antigen-specific antibody levels, following each immunization with the pEnv or pEnv+pnsP2 were assessed by ELISA. Briefly, 96-well high-binding polystyrene plates were coated with synthesized specific peptides (2 μg/mL PBS), and incubated overnight. The plates were then washed with PBST (PBS, 0.05% Tween-20), blocked for 1 h with 3% BSA and incubated with sera (1:100) from immunized mice for 1 h at 37 °C. Bound antibody was detected using HRP-conjugated goat anti mouse IgG, IgG1, IgG2a, IgG2b, and IgG3 secondary antibodies (Research Diagnostics Inc, Flanders, NJ) (1:5000 dilution) following 1 ho incubation at 37°C. Detection was performed by addition of a chromogenic substrate solution TMB (R&D Systems). Plates were then read at 450 nm using a Biotek EL312e Bio-Kinetics reader. The IgG1/IgG2a ratios were calculated to define the T cell phenotype induced by vaccination since the IgG2a and IgG1 levels are indicative of Th1 and Th2 responses, respectively.
CHIKV viral challenge experiments
The PC-08 strain of CHIKV was cultured and quantified as previously described (21). On day 35 after immunization, 10 mice from each of the vaccination groups (pVax1, pEnv, and pEnv+pnsP2, nsP2) were challenged with 7 log10 PFU (25 μL) of the PC-08 isolate by intranasal infection (i.n.) and were evaluated daily for clinical signs of infection. The virus was administered by gradual i.n., inoculation (25–40 μL). Subsequently mice were observed for 14 days for signs of infection. Brains from mice sacrificed at day 14 were harvested, washed and fixed in 4% formaldehyde immediately. Subsequently, H and E staining was performed and slides were observed under a Leica microscope system. Ready-Set-Go mouse TNF-a kits (eBioscience, San Diego, CA) were used for detecting TNF-a cytokine levels in sera samples according to manufacturer's recommendations.
Statistical analysis
Data were analyzed using Graph Pad Prism Software (Prism Inc., San Diego, CA). Statistical comparisons among groups were performed using the Wilcoxon Matched Pairs t test. Correlations between the variables in the control and experimental groups were statistically evaluated using the Spearman rank correlation test. For all the tests, a p value less than 0.1 was considered to be significant.
Results
Cloning and expression of CHIKV-nsP2 plasmid
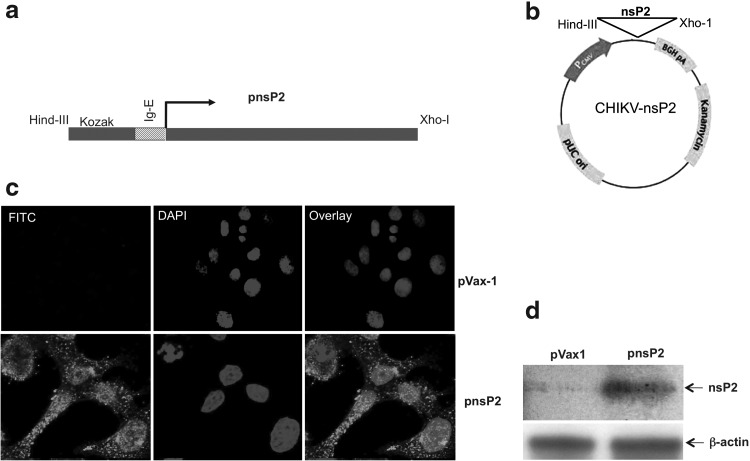
CHIKV-Env was constructed as previously described (21). The consensus sequences for CHIKV nsP2 were generated from genomic sequences from the GenBank-NCBI database. Several modifications were made in constructing the plasmid, including addition of a highly efficient IgE leader peptide sequence to facilitate expression in the pVax1 expression vector (Fig. 1a and b). To verify the expression of the nsP2 plasmid (pnsP2), an indirect immunofluorescence assay (IFA) of transfected cells was performed, using a pnsp2 antibody, which demonstrated specific cytoplasmic expression of pnsP2 (Fig. 1c, lower left panel). No expression was detected in control vector (pVax1) transfected cells (Fig 1c, upper left panel). The upper and lower middle panels demonstrate DAPI staining, while the upper and lower right panels indicate overlays of FITC+DAPI staining. Second, 35S met-labeled products were immunoprecipitated using an anti-CHIKV-nsP2 antibody followed by SDS-PAGE, demonstrating expression of the protein and thus validating its subsequent use in subsequent experiments (Fig. 1d).
FIG. 1.
Plasmid CHIKV-nsP2 construction and in vitro expression. (a) Schematic diagram of the CHIKV-nsP2 gene Including the Kozak consensus sequence and immunoglobulin E (IgE) leader; (b) pVax1 backbone vector indicating the inclusion of the nsP2 gene insert; (c) indirect immunofluorescence assay of transfected HeLa cells expressing pVax1 or pnsP2. Images indicate transfected cells viewed under Zeiss LSM510 confocal microscope, following the addition of anti-nsP2 antisera, followed by staining with a FITC-conjugated mouse secondary antibody (upper and lower left panel) or DAPI stain (upper and lower middle panels), followed by an overlayed image (FITC+DAPI shown in the upper right and lower panels); (d) immunoprecipitation of synthesized CHIKV non-structural protein 2 (nsP2), from the transfection/expression reaction mixtures, using a His-tag antibody. The image indicates that the protein product from the gene construct pnsP2 is at its predicted molecular weight.
Enhancement of humoral responses by addition of nsP2 DNA
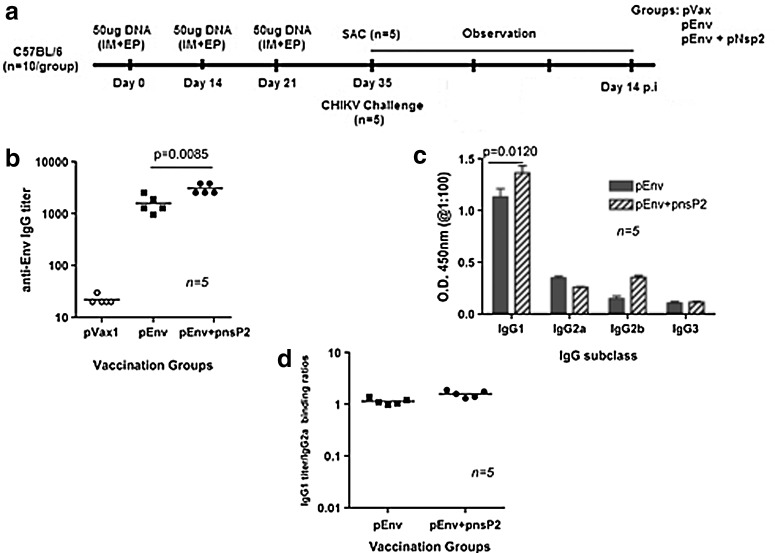
In this study, we evaluated the role of nsP2 vaccine in humoral responses. Figure 2a depicts the immunization scheme for assessing the effect of pnsP2 on Env-induced antibody responses. These analyses were performed by antigen-specific antibody binding analyses. Mice were bled on day 35 following the first immunization and an ELISA was performed. Figure 2b indicates the total IgG titer in each group against the CHIKV-Env immunogen. The total anti-CHIKV Env IgG binding titer is higher in the sera of pEnv+pnsP2 vaccinated mice than in the pEnv only vaccinated group. To further characterize the type of immune response generated, Env-specific antibody subtypes were characterized from the immunized sera. Figure 2c indicates CHIKV-Env specific IgG1, IgG2a, IgG2b, and IgG3 titers from groups that were immunized with pEnv or pEnv+pnsP2. Both groups used in this study induced IgG1 with a significant decrease in IgG2a, IgG2b, and IgG3 titers. Since the vaccine alone-vaccinated group had less antibody titers compared to the adjuvant immunized group, the IgG1: IgG2 ratios showed some variance (Fig. 2d). Furthermore, vaccination with Env+pnsP2 tended to induce higher IgG1 than IgG2. However, the differences between these two groups were not significant. These results clearly demonstrate that the pnsP2 DNA vaccine strategy resulted in active and robust CHIKV-specific immunological response when co-administered with CHIKV-Env and suggest that all vaccination regimes result in a rather balanced immune response in mice.
FIG. 2.
Immunization schema and post-vaccination antibody responses. (a) Vaccination and blood/tissue sampling time course. (b) Anti-CHIKV antibody measurement by ELISA. Sera were collected a week after the third immunization and the total IgG produced was measured in each group. Values represent the mean (±SD) of duplicate wells; p values (p<0.0085) indicate that the data are statistically significant between pEnv and pEnv+pnsP2 (i.e., the titer for pEnv + pnsP2 vaccinated animals is significantly higher. (c) IgG subtype ELISA analysis. The graph shows CHIKV-Env specific IgG1, IgG2a, IgG2b, and IgG3 antibody levels in mice that were vaccinated with pEnv or pEnv+pnsP2. Error bars represent standard deviation (±S.D) of duplicate wells. (d) IgG1 to IgG2a level binding ratios are shown for mice (n=5) immunized with pEnv or pEnv+pnsP2 intramuscularly, delivered through MID-EP. The values shown represent the mean (±S.D). The ratios were not significantly different.
pnsP2 significantly enhances cellular immune responses induced by pEnv
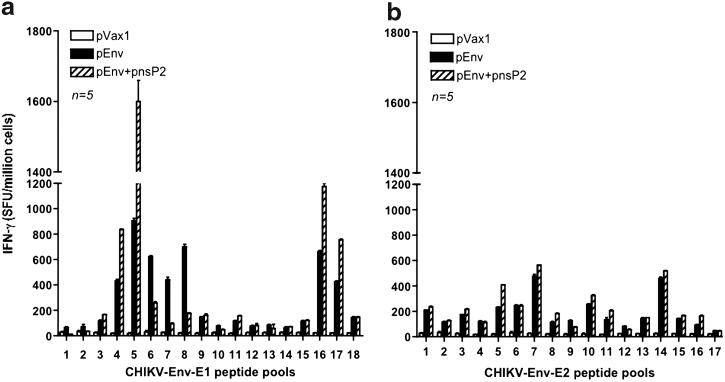
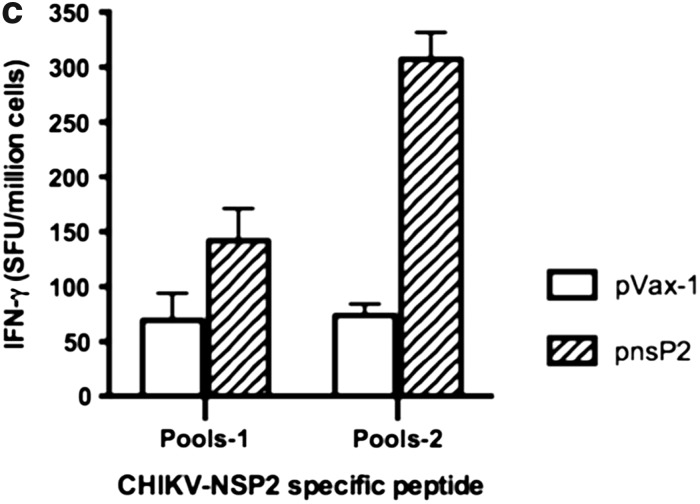
In order to examine the effects of pnsP2 on CHIKV-Env specific immune response, an IFN-g ELISpot assay was performed. One week after the third immunization, 5 of the 10 mice in the group were euthanized and the splenocytes were isolated from the immunized mice and stimulated with CHIKV- E1 and E2 specific peptide pools. The ELISpot assay was performed against a library of peptides spanning the entire E1 and E2 region of the CHIKV envelope. Seventy four 15-mer peptides with 9 amino acid overlaps, spanning the residues 1–435 of E1 protein and 1–423 of E2 protein were used. Figure 3a and b represent the average IFN-g levels secreted from 106 splenocytes that were stimulated with specific peptides. As observed, the cellular response (measured by IFN-g spot forming units (SFU) per million splenocytes), induced by the pEnv+pnsP2 DNA vaccine/adjuvant was against all regions of the protein was significantly higher than that induced by pEnv alone (1625 versus 923). Further IFN-g levels secreted form the pnsP2 immunized group also analyzed. Figure 3c shows that the pnsP2 group exhibited a significant increase in IFN-g secretion compared to the pVax1 in agreement with a role for nonstructural protein in regulating immune response.
FIG. 3.
Immunogenicity of CHIKV-nsP2. As indicated in the Materials and Methods section, C57BL/6 mice were immunized three times with 50 μg of pEnv, pEnv+pnsP2, pnsP2, or control pVax1 constructs and sacrificed 1 week following last immunization. Splenocytes were harvested, stimulated overnight with the specific peptide pools from proteins (a) Env-E1 or (b) Env-E2 and (c) Nsp2 and plates were processed and the spot forming units (SFU) were quantified by an automated ELISpot reader. For the NsP2 we used only the pools from the overlapping peptide of the protein. The raw data was normalized to SFU per million splenocytes. Values represent the mean of duplicate wells.
pnsP2+pEnv mediated anti-CHIKV immunity and enhanced protection against viral challenge
It was then determined whether there was enhanced protection against CHIKV challenge in pEnv+pnsP2 vaccinated mice compared to groups vaccinated with pEnv or pnsP2 alone. Immunized mice were challenged with 7 log10 PFU (plaque forming units) of the CHIKV isolate (PC-08) through intranasal infection one week after the third immunization. Animals were evaluated daily for up to 14 days for clinical signs of infection (i.e., lethargy and hind limb weakness). The outcome of the challenge was determined based on the common clinical signs of infection and the level of viremia (12, 21).
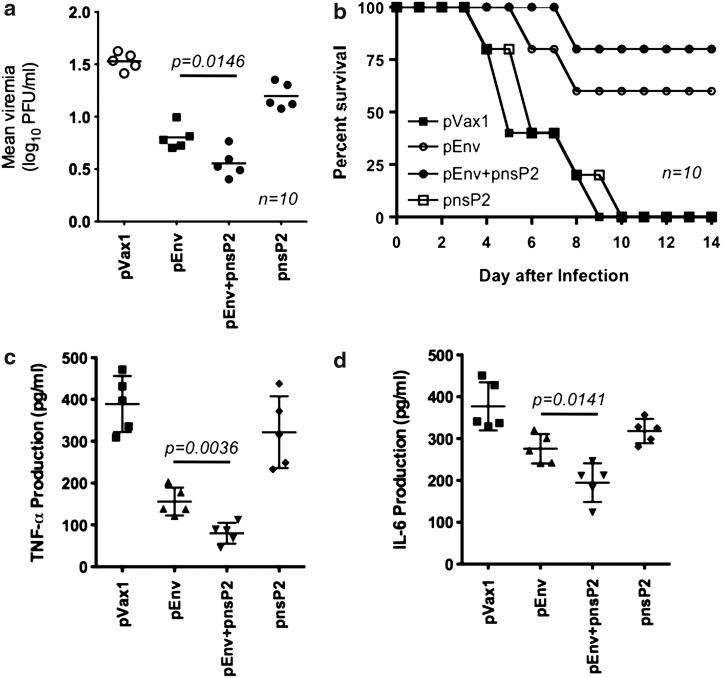
As an initial step towards evaluating the protective efficacy against viral challenge in the nsP2 DNA vaccine group, sera samples obtained from day 5 to day 10 were monitored for viremia. The mice vaccinated with the vector pVax1 control succumbed to infection and developed viremia, detectable on day 5 post inoculation (mean titer log10 1.52 PFU/mL±0.073). The viremia was similar to that in pnsP2 only vaccinated group (mean titer log10 1.13 PFU/mL±0.074). The level of viremia in pEnv only mice demonstrated a significant decrease in viremia (mean titer log10 0.764 PFU/mL±0.070), whereas a further decline was observed in pEnv+pnsP2 animals (mean titer log10 0.55 PFU/mL±0.016) (Fig. 4a). Although the viremia in the co-immunized group was only slightly lower than that of the pEnv group, there was a significantly increased rate of survival in the pEnv+pnsP2 mice (Fig. 4b). Further, a recent study suggested that the proinflamatory cytokine TNF-a and IL-6 are consider to be biomarkers of CHIKV infection (21, 23). Therefore, TNF-α and IL-6 were measured in all groups. In CHIKV challenged and infected pVax1 vaccinated mice, secretion of TNF-α and IL-6 were found to be very high compared to the pEnv immunized group. Further pEnv+pnsP2 immunized mice showed significant reduction in these two cytokines (Fig. 4c and d).
FIG. 4.
CHIKV viremia and pathogenesis in vaccinated mice. Ten mice from the control or the experimental vaccination groups were inoculated with 7log10 PFU (plaque forming units) of CHIKV by the intranasal route and subsequently examined for viremia, disease phenotype, and survival, and sacrificed for pathologic analysis. In this viral challenge model, (a) pVax1 and NsP2 immunized mice exhibited robust replication, but Env+nsP2 co-immunized mice which exhibited a 2 to 3 log reduction in mean viral tier. 50% of the mice in the pVax1 group were euthanized on day 12 post-infection, as they lost 30% of the original body weight. Significantly lower viremia (p<0.0146) in pEnv+pnsP2 vaccination group compared to pVax1 alone, pnsP2 alone groups. (b) Long-term survival of CHIKV challenged mice, demonstrating increased survival, coupled with a disease-free state, of mice in the pEnv+pnsP2 groups compared to the pVax1 or pEnv alone groups. (c and d) Proinflammatory cytokines of TNF-a and IL-6 in mouse sera after CHIKV challenge. Sera samples were evaluated on day 12-post challenge with naïve mouse sera samples used as controls. Cytokines in serum levels were determined by specific ELISAs, and results shown are geometric mean values obtained from five mice at time point.
Mice in the control group demonstrated significant clinical signs within 3 days of infection while pEnv only mice demonstrated milder clinical sings of illness. Importantly, the pEnv+pnsP2-group failed to demonstrate clinical symptoms. All mice except for one in the pEnv group survived at least to day 12, while 100% of the pEnv+pnsP2 group survived, with no symptoms, until day 12 or beyond. On day 14-post infection, all surviving mice were euthanized, with brain and kidney samples being collected and subsequently prepared for histopathology.
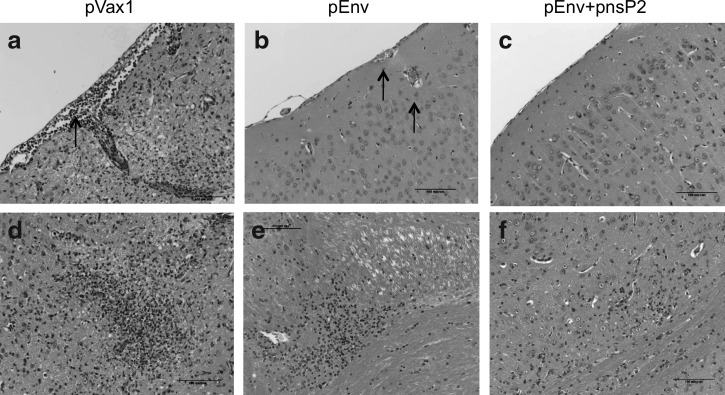
The brain sections from the pVax1 immunized mice demonstrated severe meningoencephalitis (Fig. 5a), with lymphocytic infiltration and edema observed as well. These results resemble the pathology in biopsy samples of CHIKV infected humans (21). Figure 5b and e depicts brain tissue sections from pEnv-immunized mice. Mild perivascular cuffing and edema were observed along with hyperemia of arterioles and blood capillaries. Figure 5e demonstrates infiltration of hyperplastic microglial cells and formation of a small glial nodule, indicating a low level of neuronal degeneration. However, the brain tissue sections of the pEnv+pnsP2 immunized mice (Fig. 5c and f) indicated normal brain morphology without evidence of hyperemia or edema.
FIG. 5.
DNA vaccination using CHIKV-nsP2 with EP is completely protective against lethal challenge: Immunohistopathology of brain tissue from immunized mice after CHIK viral challenge experiment. Brain tissues were collected from day 10 through 14 and were sectioned and prepared as described in the Materials and Methods section. Sections from pVax1 vaccination group: (a) infiltration of cells into the arteriole and blood capillary; (d) large focal accumulation of glial cells and formation of glial nodule. Sections from pEnv vaccination group: (b) hyperemia and mild perivascular edema; (e) mild gliosis. Sections from pEnv+pnsP2 vaccination group: (c) normal brain pathology; (f) low levels of gliosis.
Discussion
Among virally encoded molecules, nonstructural proteins are attractive targets for the development of antiviral drugs and vaccines since they are among the initial viral proteins synthesized and are assumed to be essential for the viral life cycle (5). This suggests the use of nonstructural proteins (i.e., nsP2) in a vaccine strategy against CHIKV infection. Recent reports clearly highlighted the advantages of using a nonstructural protein vaccine since a vaccine that targeted nsP2 interrupted the infection process of the virus (16, 24). More recently, three different versions of a chimeric vaccine were created and used which included the nonstructural genes of Sindibis, Venezuelan equine encephalitis viruses, as well as the structural genes of CHIKV. A higher immunogenicity and protection against CHIKV was observed with this chimeric vaccine (15, 16, 24–26). Considering the benefits of using a nonstructural gene in a vaccine cocktail, we hypothesized that the CHIKVnsP2 could function, when expressed from a DNA plasmid, as an adjuvant to elicit a more potent immune response than the previously developed and evaluated Env based vaccine.
Previously, we reported that a CHIKV Env DNA vaccine, delivered through EP, conferred an enhanced level of protection against viral challenge (21). In this study, we designed a vaccine cassette based on nsP2 specific consensus sequences with several modifications (9, 22). DNA vaccines provide a stable source of the encoded antigen, leading often to stimulation of the immune system and generation of long-lasting immunity (15, 27, 28). The major advantage of the DNA vaccine method is that both cellular and humoral immune responses are induced as the encoded antigen is processed and presented by conventional pathways (22, 27, 29, 30). These types of responses are similar to those obtained with live attenuated vaccine preparations, without many of the safety concerns (27, 31). Therefore, it was reasoned that the DNA vaccine strategy was appropriate for the potential development of an efficacious vaccine against CHIKV. As an approach to induce stronger cell-mediated immune responses to CHIKV-env, we evaluated additional vaccination regimes consisting of nonstructural protein 2 (nsP2) either vaccination with or with out Env. Previous studies have demonstrated varying efficacy of DNA vaccine plus EP strategies for individual E1, E2 compared to combined Envs (E1+E2+E3) DNA vaccination regime. A combined DNA Env regime elicited stronger cellular immune responses. In our study, the DNA-based vaccine and the DNA plus nonstructural protein adjuvant regime elicited higher levels of cell-mediated responses, both inducing a Th1-type response characterized by a predominance of IFN-γ and high cytotoxicity in mice and suggest that immune adjuvant targeting nonstructural proteins might be the appropriate approach for induction of both humoral and cellular immunity to the surface protein envelope.
In our present study, antibody binding was utilized to calculate the antibody kinetics and potency. Specifically, neutralizing antibodies are thought to be a critical immune response that mediates protection against CHIKV infection and disease. DNA vaccines on their own, however, are typically poor inducers of neutralizing antibodies. However, recent advancements such as the use of the MID-EP (Minimal Invasive Electroporation) delivery system have been demonstrated to increase the detectable neutralizing antibody response against several pathogens (21, 28, 32, 33). EP has been one of the more recent extensively evaluated methods for enhancing delivery of DNA for both therapeutic and vaccination purposes. One of the limitations of nonfacilitated DNA vaccine delivery is relatively low to moderate transfer across the cell membrane with low expression levels and associated low immunogenicity to the expressed proteins. The major advantages of EP is thought to be that it increases DNA distribution and associated expression throughout the tissue (34) and causes a concomitant local inflammatory reaction (35), both of which are reasoned to contribute to the development of a stronger immune response. Studies have demonstrated that applying EP after injection of plasmid DNA effectively enhances the immune response in a wide variety of species, nonhuman primates and humans (21, 28, 31, 32, 36, 37). These previous studies have provided the impetus for the use of EP to potentially enhance delivery and immunogenicity of the CHIKV DNA constructs evaluated in this report.
This current study suggests that the inclusion of nsP2 from CHIKV in the Env DNA vaccine preparation significantly increases both cellular and humoral immune responses in particular specific neutralizing antibodies and protection of mice against viral challenge. The protection observed, mediated by the addition of pnsP2 to pEnv, in the viral challenge studies could be attributed to the higher levels of functional antibodies and a reduction in proinflammatory cytokines (i.e., IL-6 and TNF-a). The use of adjuvants is critical for enhancing the response in immunization with DNA immunization. Several previous studies demonstrated that immune stimulatory adjuvants enhance immune responses through promoting proliferation and differentiation of B cells and antibody development. The proliferation and activation of many kinds of T cells and production of various immune stimulatory cytokines can also promoted and stimulated by adjuvant. The effect of nsP2 enhancing the level of antibody and cellular response may be related to its ability to induce protective immune responses. Therefore, we suggest that the use of this CHIKV nsP2 as a DNA adjuvant has the potential to induce a more potent neutralizing antibody targeted against CHIKV Env and, as such, may have some clinical utility. The histopathological evaluation of tissues from the brains of virally challenged immunized mice clearly showed the difference in tissue sections between the three groups evaluated in this study. The group that was co-immunized with pnsP2+pEnv showed no neuropathologic damage, whereas the CHIKV Env alone DNA immunized group demonstrated some minimal damage from the brain sections.
In summary, the data presented in this study demonstrated that immunization with a CHIKV Env DNA vaccine co-expressing nsP2 elicits a stronger humoral and cellular immune response when compared to the response obtained when immunized with CHIKV-Env DNA vaccine alone. This finding highlights the role of the early expressed nsp2 as an adjuvant in enhancing both cellular and humoral immune responses generated by a CHIKV DNA vaccine. These results have potentially important implications for the development of a successful anti-CHIKV vaccine.
Acknowledgments
We would like to acknowledge members of the Weiner laboratory for significant contributions and/or critical reading of this manuscript.
Author Disclosure Statement
Competing financial interests: DBW: has grant funding and collaborations, advising, or consulting including service on scientific review committees for commercial entities and therefore notes possible conflicts associated with this work with Pfizer, Inovio, BMS, Virxsys, Ichor, Merck, Althea, VGXI, J&J, Aldevron, and possibly others. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the article apart from those disclosed. No writing assistance was utilized in the production of this article.
References
- 1.Sourisseau M. Schilte C. Casartelli N, et al. Characterization of reemerging chikungunya virus. PLoS Pathog. 2007;3:e89. doi: 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiter P. Fontenille D. Paupy C. Aedes albopictus as an epidemic vector of chikungunya virus: Another emerging problem? Lancet Infect Dis. 2006;6:463–464. doi: 10.1016/S1473-3099(06)70531-X. [DOI] [PubMed] [Google Scholar]
- 3.Reinert JF. Harbach RE. Kitching IJ. Phylogeny and classification of Aedini (Diptera: Culicidae), based on morphological characters of all life stages. Zool J Linn Soc. 2004;142:289–368. [Google Scholar]
- 4.Grivard P. Le Roux K. Laurent P, et al. Molecular and serological diagnosis of Chikungunya virus infection. Pathol Biol (Paris) 2007;55:490–494. doi: 10.1016/j.patbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Thiboutot MM. Kannan S. Kawalekar OU, et al. Chikungunya: A potentially emerging epidemic? PLoS Negl Trop Dis. 2010;4:e623. doi: 10.1371/journal.pntd.0000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross RW. The Newala epidemic. III. The virus: Isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 1956;54:177–191. doi: 10.1017/s0022172400044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar NP. Joseph R. Kamaraj T. Jambulingam P. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol. 2008;89:1945–1948. doi: 10.1099/vir.0.83628-0. [DOI] [PubMed] [Google Scholar]
- 8.Manimunda SP. Sinigh SS. Sugunan AP, et al. Chikungunya fever, Andaman and Nicobar Islands, India. Emerg Infect Dis. 2007;13:1259–1260. doi: 10.3201/eid1308.070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muthumani K. Lankarraman KM. Laddy DJ, et al. Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine. 2008;26:5128–5134. doi: 10.1016/j.vaccine.2008.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sam IC. AbuBakar S. Chikungunya virus infection. Med J Malaysia. 2006;61:264–269. [PubMed] [Google Scholar]
- 11.Santhosh SR. Dash PK. Parida MM, et al. Comparative full genome analysis revealed E1: A226V shift in 2007 Indian Chikungunya virus isolates. Virus Res. 2008;135:36–41. doi: 10.1016/j.virusres.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Wang E, et al. Chimeric alphavirus vaccine candidates for chikungunya. Vaccine. 2008;26:5030–5039. doi: 10.1016/j.vaccine.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwari M. Parida M. Santhosh SR, et al. Assessment of immunogenic potential of Vero adapted formalin inactivated vaccine derived from novel ECSA genotype of Chikungunya virus. Vaccine. 2009;27:2513–2522. doi: 10.1016/j.vaccine.2009.02.062. [DOI] [PubMed] [Google Scholar]
- 14.Blackburn SD. Shin H. Haining WN, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akahata W. Yang ZY. Andersen H, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat Med. 2010;16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fros JJ. Liu WJ. Prow NA, et al. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J Virol. 2010;84:10877–10887. doi: 10.1128/JVI.00949-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourai M. Lucas-Hourani M. Gad HH, et al. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J Virol. 2012;86:3121–3134. doi: 10.1128/JVI.06390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang KA. Yan J. Draghia-Akli R, et al. Strong HCV NS3- and NS4A-specific cellular immune responses induced in mice and Rhesus macaques by a novel HCV genotype 1a/1b consensus DNA vaccine. Vaccine. 2008;26:6225–6231. doi: 10.1016/j.vaccine.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DY. Firth AE. Atasheva S, et al. Conservation of a packaging signal, the viral genome RNA packaging mechanism in alphavirus evolution. J Virol. 2011;85:8022–8036. doi: 10.1128/JVI.00644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frolova EI. Gorchakov R. Pereboeva L, et al. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol. 2010;84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallilankaraman K. Shedlock DJ. Bao H, et al. A DNA vaccine against Chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Negl Trop Dis. 2011;5:e928. doi: 10.1371/journal.pntd.0000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin J, et al. High antibody, cellular responses induced to HIV-1 clade C envelope following DNA vaccines delivered by electroporation. Vaccine. 2011;29:6763–6770. doi: 10.1016/j.vaccine.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng LF. Chow A. Sun YJ, et al. IL-1beta, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS ONE. 2009;4:e4261. doi: 10.1371/journal.pone.0004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartholomay LC. waterhouse RM. Mayhew GF, et al. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. 2010;330:88–90. doi: 10.1126/science.1193162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enserink M. Infectious diseases. Massive outbreak draws fresh attention to little-known virus. Science. 2006;311:1085. doi: 10.1126/science.311.5764.1085a. [DOI] [PubMed] [Google Scholar]
- 26.Higgs S. The 2005–2006 Chikungunya epidemic in the Indian Ocean. Vector Borne Zoonotic Dis. 2006;6:115–116. doi: 10.1089/vbz.2006.6.115. [DOI] [PubMed] [Google Scholar]
- 27.Kutzler MA. Weiner DB. DNA vaccines: Ready for prime time? Nat Rev Genet. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shedlock DJ. Talbott KT. Cress C, et al. A highly optimized DNA vaccine confers complete protective immunity against high-dose lethal lymphocytic choriomeningitis virus challenge. Vaccine. 2011;29:6755–6762. doi: 10.1016/j.vaccine.2010.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrow MP. Weiner DB. DNA drugs come of age. Sci Am. 2010;303:48–53. doi: 10.1038/scientificamerican0710-48. [DOI] [PubMed] [Google Scholar]
- 30.Kawalekar OU. Shedlock DJ. Weiner DB. Current strategies and limitations of HIV vaccines. Curr Opin Investig Drugs. 2010;11:192–202. [PubMed] [Google Scholar]
- 31.Boyer JD. Robinson TM. Kutzler MA, et al. Protection against simian/human immunodeficiency virus (SHIV) 89.6P in macaques after coimmunization with SHIV antigen and IL-15 plasmid. Proc Natl Acad Sci USA. 2007;104:18648–18653. doi: 10.1073/pnas.0709198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laddy DJ. Yan J. Khan AS, et al. Electroporation of synthetic DNA antigens offers protection in nonhuman primates challenged with highly pathogenic avian influenza virus. J Virol. 2009;83:4624–4630. doi: 10.1128/JVI.02335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broderick KE. Shen X. Soderholm J, et al. Prototype development and preclinical immunogenicity analysis of a novel minimally invasive electroporation device. Gene Ther. 2011;18:258–265. doi: 10.1038/gt.2010.137. [DOI] [PubMed] [Google Scholar]
- 34.Zaharoff DA. Barr RC. Li CY. Yuan F. Electromobility of plasmid DNA in tumor tissues during electric field-mediated gene delivery. Gene Ther. 2002;9:1286–1290. doi: 10.1038/sj.gt.3301799. [DOI] [PubMed] [Google Scholar]
- 35.Babiuk S. Baca-Estrada ME. Foldvari M, et al. Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J Biotechnol. 2004;110:1–10. doi: 10.1016/j.jbiotec.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Yan J, et al. Cellular immunity induced by a novel HPV18 DNA vaccine encoding an E6/E7 fusion consensus protein in mice and rhesus macaques. Vaccine. 2008;26:5210–5215. doi: 10.1016/j.vaccine.2008.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirao LA. Wu L. Khan AS, et al. Intradermal/subcutaneous immunization by electroporation improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine. 2008;26:440–448. doi: 10.1016/j.vaccine.2007.10.041. [DOI] [PubMed] [Google Scholar]