Summary
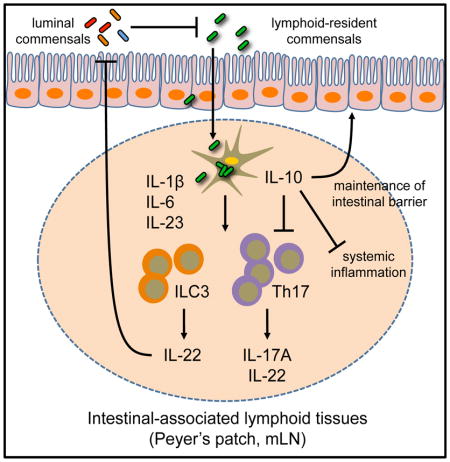
Physical separation between the mammalian immune system and commensal bacteria is necessary to limit chronic inflammation. However, selective species of commensal bacteria can reside within intestinal-associated lymphoid tissues of healthy mammals. Here, we demonstrate that lymphoid tissue-resident commensal bacteria (LRC) colonized murine dendritic cells and modulate their cytokine production. In germ-free and antibiotic-treated mice, LRCs colonized intestinal-associated lymphoid tissues and induced multiple members of the IL-10 cytokine family, including dendritic cell-derived IL-10 and group 3 innate lymphoid cell (ILC3)-derived IL-22. Notably, IL-10 limited the development of pro-inflammatory Th17 cell responses, and IL-22 production enhanced LRC colonization in the steady state. Furthermore, LRC colonization protected mice from lethal intestinal damage in an IL-10-IL-10R-dependent manner. Collectively, our data reveal a unique host-commensal bacteria dialogue whereby selective subsets of commensal bacteria interact with dendritic cells to facilitate tissue-specific responses that are mutually beneficial for both the host and the microbe.
Graphical abstract
Introduction
Trillions of microorganisms constitutively colonize the mammalian gastrointestinal (GI) tract and are essential to aid in nutrient metabolism and resistance to pathogen infection, as well as the development and maturation of the immune system (Belkaid and Hand, 2014; Honda and Littman, 2012; Hooper et al., 2012). In the healthy intestine, most commensal bacteria are restricted to the lumen of the GI tract or found associated with the surface of the intestinal epithelium and remain physically separated from the immune cells that populate the lamina propria and intestinal-associated lymphoid tissues including isolated lymphoid follicles (ILF), Peyer’s patches (PP) and the mesenteric lymph nodes (mLN) (Chow et al., 2010; Hooper et al., 2012; Hooper and Macpherson, 2010). Physical separation of commensal bacteria from immune cells is achieved by multiple physical and biochemical mechanisms that include epithelial cells that line the intestine, tight junction proteins, and secretion of anti-microbial peptides, mucus and immunoglobulin A (Chow et al., 2010; Hooper et al., 2012; Hooper and Macpherson, 2010). Anatomical segregation of commensal bacteria from the immune system, often referred to as the “demilitarized zone”, is essential to prevent pathologic immune responses against commensal bacteria (Hooper et al., 2012; Hooper and Macpherson, 2010). Consistent with this, translocation of commensal bacteria across the intestinal epithelium can lead to the generation of pro-inflammatory immune cell responses and are often associated with the pathogenesis of multiple chronic diseases, such as inflammatory bowel disease, metabolic disorders and HIV/AIDS (Brenchley and Douek, 2012; Manichanh et al., 2012).
Recent studies suggest that a unique subset of commensal bacteria can colonize the interior of intestinal-associated lymphoid tissues of healthy mammals (Fung et al., 2014; Kunisawa and Kiyono, 2012; Obata et al., 2010; Sonnenberg et al., 2012). Using 16S rDNA sequencing and fluorescence in situ hybridization (FISH), one report identified the presence of multiple species of commensal bacteria in the interior of ILFs, PPs and the mLN of healthy mice, non-human primates and humans (Obata et al., 2010). These bacteria include Alcaligenes spp., Achromobacter spp., Bordetella spp. and Ochrobactrum spp. 16S rDNA for many of these lymphoid tissue-resident commensal bacteria (LRCs) were found associated with CD11c+ dendritic cells (DCs), suggesting a role for DCs in lymphoid tissue colonization. In a subsequent study, it was demonstrated that interleukin (IL)-22 and group 3 innate lymphoid cells (ILC3) are important in preventing systemic dissemination of one LRC, Alcaligenes xylosoxidans, and subsequent induction of systemic inflammation (Sonnenberg et al., 2012). These data highlight that intestinal-associated lymphoid tissues are potential sites for colonization by commensal bacteria and innate immune pathways maintain anatomical containment between LRCs and the systemic immune system. Despite these findings, the mechanisms by which commensal bacteria colonize immune cell-rich lymphoid compartments associated with the intestine of healthy mammals, and the functional significance of this colonization to the host remain undefined.
In this report, we demonstrate that LRCs can colonize murine DCs, modulate DC cytokine production in a viability-dependent manner and promote local Th17 cell and ILC3 responses in intestinal-associated lymphoid tissues in vivo. Further, our data indicate that ILC3-derived IL-22 enhanced LRC colonization of intestinal-associated lymphoid tissues, and LRC-induced IL-10 limited pro-inflammatory responses in the steady state and could protect mice in a model of intestinal damage. Collectively, these data demonstrate that LRCs engage in a mutualistic dialogue with mammalian hosts by eliciting members of the IL-10 cytokine family.
Results
Lymphoid tissue-resident commensal bacteria colonize murine dendritic cells
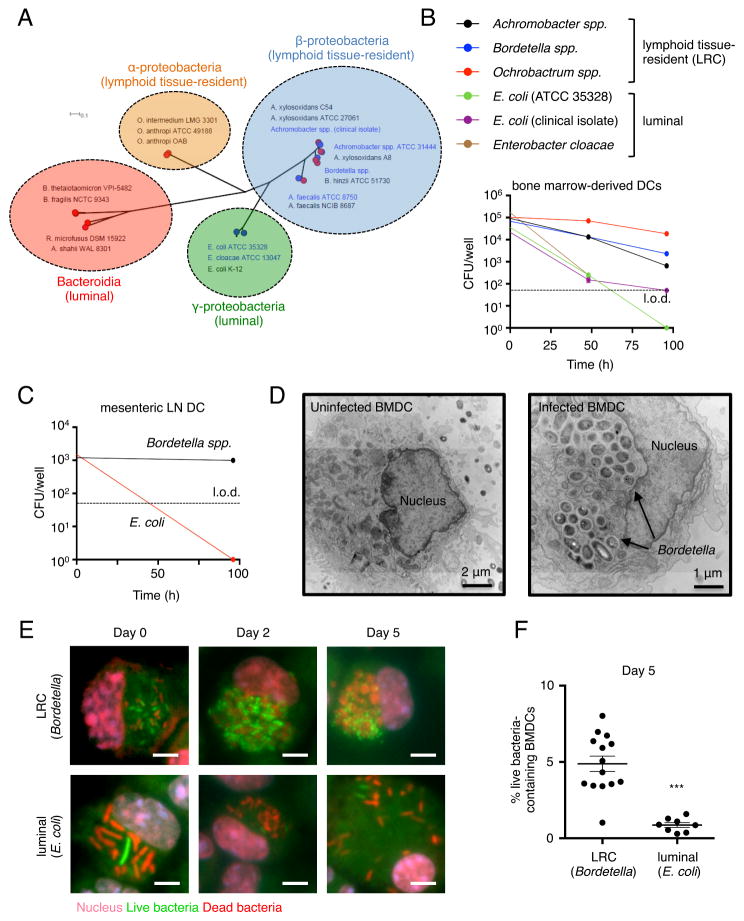
16S rDNA for several groups of bacteria have been detected in the interior of PPs and the mLN of healthy mice (Obata et al., 2010; Sonnenberg et al., 2012). These LRCs, including Achromobacter spp., Alcaligenes spp., Bordetella spp. and Ochrobactrum spp., belong exclusively to the α- and β-proteobacteria groups, which are not highly represented in sequence analyses of the luminal contents of the intestine (Belkaid and Hand, 2014; Honda and Littman, 2012; Hooper et al., 2012). We constructed a phylogenetic tree using in-house sequenced and publically available reference genomes of α-, β-, γ-proteobacteria and Bacteroidia members (Table S1). Phylogenetic analyses indicated that members of the α-proteobacteria and β-proteobacteria LRCs form two distinct clades from γ-proteobacteria and Bacteroidia members, which are typically found in the lumen of the intestine and not associated with intestinal lymphoid tissues in healthy mammals (Figure 1A).
Figure 1. Lymphoid tissue-resident commensal bacteria colonize bone marrow-derived and primary dendritic cells.
(A) Maximum-likelihood tree of 47 bacterial genomes based on 40 universal single copy marker genes. Red symbols indicate published reference genomes. Blue symbols and text indicate in-house sequenced genomes. Scale bar indicates number of substitutions per site. (B) Bone marrow-derived dendritic cells (BMDCs) were co-cultured with representative LRCs or luminal-resident commensals and bacterial survival was measured at 0, 48 and 96 hours. l.o.d., limit of detection. Data are representative of at least 2 independent experiments. (C) Primary DCs isolated from the mesenteric lymph node (mLN) of C57BL/6 mice were co-cultured with the mouse-derived LRC, Bordetella spp. or luminal-resident commensal, E. coli ATCC 35328, and bacterial survival was measured at 0 and 96 hours. l.o.d., limit of detection. Data representative of 2 independent experiments (D) Bordetella-colonized BMDCs were analyzed at day 5 post co-culture by transmission electron microscopy. Images are representative of 10 individual bacteria-containing DCs. (E) Bordetella- and E. coli-colonized BMDCs were analyzed at day 0, 2 and 5 post-co-culture by immunofluorescence. Scale bar – 5 μm. Data representative of 2 independent experiments. (F) Percentage of BMDCs containing live (green) bacteria was quantified across 7–16 distinct fields of view. Infection efficiency (BMDCs containing live or dead bacteria/total BMDCs) on day 0 was approximately 15% for Bordetella spp. and 30% for E. coli. Data are represented as mean ± SEM. Statistics shown were performed using unpaired, two-tailed, student’s t test. ***, p < 0.0001. ND, not detectable. See also Figure S1 and Table S1.
16S rDNA for LRCs have been detected in CD11c+ cells from PPs and the mLN of healthy mice (Obata et al., 2010), suggesting LRCs may colonize DCs. To test this, bone marrow-derived DCs (BMDCs) were co-cultured with bacteria predicted to be LRCs or representative intestinal microbes not predicted to be LRCs (non-LRCs), and the ability of each bacterium to survive in DCs was determined using a gentamicin protection assay. LRCs and non-LRCs were both internalized by DCs, but the numbers of viable non-LRCs declined rapidly within the first 3 days of culture, while viable LRCs persisted at high levels on days 2 and 4 (Figure 1B). LRCs but not non-LRCs were also able to colonize primary DCs isolated from the mLN and spleen (Figure 1C and S1A). Transmission electron microscopy demonstrated intracellular colonization of DCs by LRCs as noted by the presence of dense bacterial clusters within intracellular vesicles (Figure 1D), and an immunofluorescence assay confirmed the presence of intracellular live LRCs on days 2 and 5 post colonization (Figure 1E–F). Together, these data suggest that, in contrast to luminal-resident commensal bacteria, LRCs have the ability to efficiently colonize and persist in murine DCs.
Lymphoid tissue-resident commensal bacteria modulate dendritic cell cytokine production in a viability-dependent manner
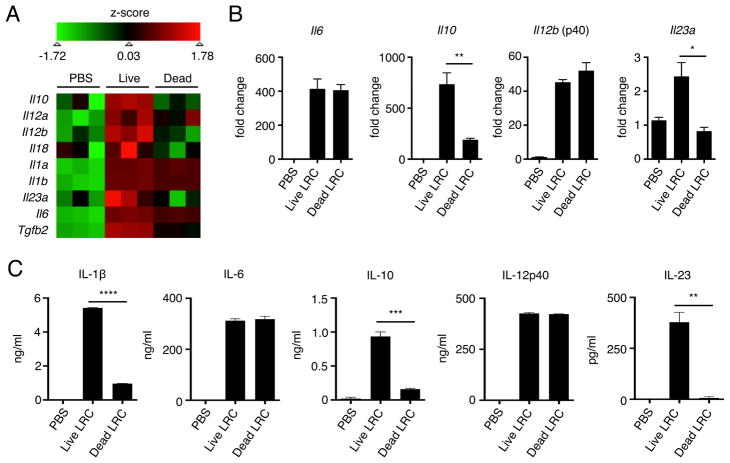
Based on the ability of LRCs to colonize and persist in murine DCs, we sought to characterize DC cytokine induction following exposure to live versus heat-killed LRCs. To investigate this, genome-wide transcriptional profiling was performed on DCs treated with PBS, live or heat-killed Bordetella spp., a model LRC isolated from our mouse colony, at 4 days post-colonization. Analysis of canonical DC-derived cytokines revealed that DCs colonized with live LRCs expressed greater levels of both pro- and anti-inflammatory cytokines Il6, Il23a, Il18, Il12a, Il12b, Il10 and Tgfb2 as compared with PBS-treated DCs (Figure 2A). Notably, the induction of mRNA and protein for several of these cytokines, including IL-1β, IL-6, IL-10, IL-12p40 and IL-23, by live LRCs was detected as early as 24 hours post co-culture (Figure 2B and 2C), and the induction of IL-10 and IL-23 mRNA (Figure 2A and 2B) and was diminished in DCs exposed to only heat-killed LRCs, suggesting that viability-dependent factors expressed by LRCs induce selective DC cytokine responses. Primary splenic DCs exposed to live LRCs also produced IL-1β, IL-6, IL-10 and IL-12p40 (Figure S1B). Efficient induction of both viability- (IL-1β, IL-10, IL-23) and non-viability-dependent cytokines (IL-6, IL-12p40) required MYD88, suggesting a role for Toll-like receptors (TLRs) in recognition of LRCs (Figure S2A). Consistent with this, production of IL-1β and IL-23 was significantly reduced in Tlr4−/− but not Tlr2−/− BMDCs (Figure S2B). Collectively, these data indicate that LRC colonization of DCs induces distinct pro- and anti-inflammatory cytokine responses in a viability- and TLR-dependent manner.
Figure 2. Lymphoid tissue-resident commensal bacteria modulate cytokine responses in bone marrow-derived dendritic cells in a viability-dependent manner.
(A) Genome-wide transcriptional profiling was performed on BMDCs co-cultured with live or heat-killed Bordetella spp. for 4 days. Numbers in colored legend represent Z scores. (B) BMDCs co-cultured with live or heat-killed Bordetella spp. for 24 hours were analyzed for cytokine gene expression by qPCR. (C) BMDCs co-cultured with live or heat-killed Bordetella spp. for 24 hours were analyzed for cytokine protein secretion by ELISA. qPCR and ELISA data were representative of at least 2 independent experiments. Data are represented as mean ± SEM. Statistics shown in B and C were performed using unpaired, two-tailed, student’s t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. See also Figure S2.
Lymphoid tissue-resident commensal bacteria promote tissue-specific Th17 cell responses
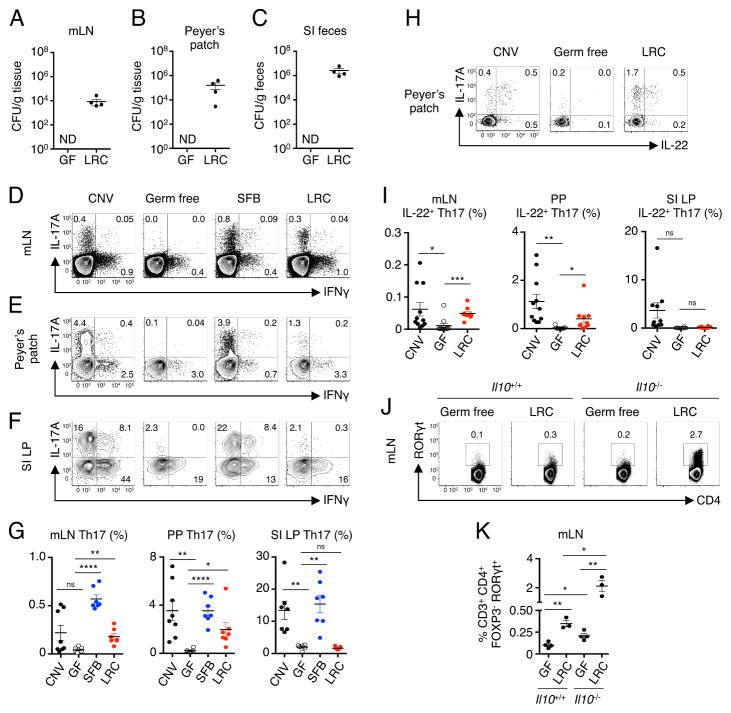
To test whether these LRCs can stably colonize and survive in intestinal-associated lymphoid tissues, germ-free (GF) mice were monocolonized with Bordetella spp., a model LRC isolated from our mouse colony. At day 10 post-inoculation, viable LRCs were consistently detected in the mLN (Figure 3A), PPs (Figure 3B) and fecal contents of the intestinal lumen (Figure 3C), but not the spleen and liver. However, colonization by LRCs was not associated with macroscopic or microscopic inflammation in the intestine at 2 weeks post colonization (Figure S3A). These data support the hypothesis that LRCs can colonize and persist in intestinal-associated lymphoid tissues of healthy mammals.
Figure 3. Colonization of intestinal-associated lymphoid tissues by lymphoid tissue-resident commensal bacteria modulates local Th17 cell responses.
(A) Mesenteric lymph nodes (mLN) homogenates, (B) Peyer’s patches (PP) homogenates and (C) small intestinal lumen contents from GF and Bordetella spp. (LRC)-monocolonized mice were cultured determine bacterial CFUs. Data in (A) and (B) are representative of at least 2 independent experiments. (D) mLN, (E) PP and (F) small intestine lamina propria (SI LP) of conventionally-housed (CNV), GF, SFB-monocolonized and LRC-monocolonized mice were analyzed for frequencies of IL-17A+ and IFNγ+ CD4+ T cells by flow cytometry. (G) Frequencies of Th17 cells in CNV, GF, SFB-monocolonized and LRC-monocolonized mice. Values represent frequencies of IL-17A+ cells among CD4+ T cells. One-way ANOVA, mLN and SI LP - ****p < 0.0001; PP - **p < 0.01 (H and I) PPs of CNV, GF, SFB-monocolonized and LRC-monocolonized mice were analyzed for frequencies of IL-22-producing Th17 cells by flow cytometry. Values represent frequencies of IL-22+ cells of IL-17A+ CD4+ T cells. One way ANOVA, mLN - ****p < 0.0001; PP - **p < 0.01; SI LP - *p < 0.05. Data are pooled from 2 independent experiments for a total of 6–8 mice per group. (J and K) Frequencies of Th17 cells (gated as CD3+CD4+RORγt+FOXP3−) in the mLN of LRC-monocolonized Il10+/+ or Il10−/− mice. Data representative of 2 independent experiments with 2–5 mice per group using Il10−/− monocolonized mice or C57BL/6 monocolonized mice with anti-IL-10R treatment (500 μg/mouse i.p. every 3 days, analyzed on day 7). One-way ANOVA, ****p < 0.0001. Cells in all flow cytometry plots are gated as live, CD3+ and CD4+. Data are represented as mean ± SEM. Statistics shown in panels G, I and K were performed using one-way ANOVA with unpaired, two-tailed, student’s t test with no correction for multiple comparisons. *, p < 0.05; **, p < 0.01; ***, p < 0.001. ND, not detectable. See also Figure S3.
Based on the ability of live LRCs to elicit IL-1β, IL-6, IL-23 and TGFβ, we hypothesized that LRCs may promote Th17 cell responses in colonized lymphoid tissues. To test this, we analyzed CD4+ T cells in the PP, mLN and small intestine lamina propria (SI LP) of conventionally-housed (CNV), GF, LRC-monocolonized mice, as well as mice monocolonized with segmented filamentous bacteria (SFB), an epithelial-associated commensal bacterium which has previously been shown to induce robust Th17 cell responses (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). Consistent with published reports, GF mice lacked Th17 cells in the mLN (Figure 3D), PP (Figure 3E) and SI LP (Figure 3F) compared to CNV mice, and SFB was sufficient to promote robust Th17 cell responses in these tissues. In contrast, LRCs promoted Th17 cell responses in the mLN and PP (Figure 3D, 3E and 3G) but failed to promote Th17 cell responses in the SI LP, suggesting tissue-specific modulation of the immune system (Figure 3F–G). Analysis of LRC-elicited Th17 cells in the PP revealed co-expression of the IL-10 family cytokine, IL-22 (Figure 3H and 3I). Accumulation of Th17 cells was selective, as LRCs did not significantly promote IFNγ-producing Th1 cell responses in the mLN, PP or SI LP (Figure S3B). Although significant Th17 cell responses were observed in the PP and mLN of LRC-monocolonized mice, the magnitude of these responses was lower than that observed in SFB-monocolonized mice (Figure 3D–F), suggesting that LRCs are less efficient at inducing Th17 cell responses or are actively supressing Th17 cell responses. Since LRCs could promote IL-10 by DCs in vitro (Figures 2A–C and S1B) and IL-10 has been shown to suppress Th17 cell responses (Liu et al., 2011; McGeachy et al., 2007; Ouyang et al., 2011), we hypothesized that IL-10 may restrain LRC-induced Th17 cells. Consistent with this hypothesis, Th17 cells were significantly increased in the mLN (Figure 3J and 3K) and PP (Figure S3C and S3D) of LRC-monocolonized Il10−/− mice compared to LRC-monocolonized Il10+/+ mice. Thus, LRCs selectively promote local Th17 cell responses in intestinal-associated lymphoid tissues, and the magnitude of these responses is limited by co-induction of IL-10.
Lymphoid tissue-resident commensal bacteria promote tissue-specific ILC3 responses
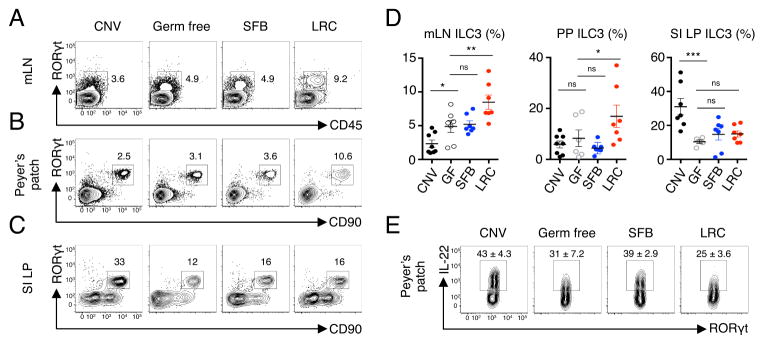
Based on the ability of live LRCs to elicit DC-derived IL-1β, IL-6 and IL-23, we hypothesized that LRCs may also promote ILC3 responses in intestinal-associated lymphoid tissues. To test this, we measured ILC3 frequencies in the PP, mLN and SI LP of CNV, GF, SFB-monocolonized and LRC-monocolonized mice. Consistent with published reports, GF mice have reduced frequencies of ILC3 in the SI LP as compared to CNV mice (Klose et al., 2013; Satoh-Takayama et al., 2008). Monocolonization by SFB did not significantly alter ILC3 frequencies in the PP, mLN and SI LP as compared to GF mice (Figure 4A–D). In contrast, LRC-monocolonized mice revealed a significant increase in ILC3 frequencies in the PP and mLN (Figure 4A, 4B and 4D) but not in the SI LP as compared to GF mice (Figure 4C and 4D). However, total numbers of ILC3s in the mLN did not change following LRC monocolonization (Figure S3E). Notably, ILC3s in the PP of LRC-monocolonized mice produced IL-22 (Figure 4E). Thus, LRCs modulate tissue-specific Th17 cell and ILC3 responses in healthy mammalian lymphoid tissues.
Figure 4. Colonization of intestinal-associated lymphoid tissues by lymphoid tissue-resident commensal bacteria promotes local ILC3 responses.
(A) mLN, (B) PP and (C) SI LP of CNV, GF, SFB-monocolonized and Bordetella (LRC)-monocolonized mice were analyzed for frequencies of ILC3 cells by flow cytometry. Cells in (A) were gated as live, lineage− (CD3−, CD5−, CD8α, CD11b−, B220−, NK1.1−). Cells in (B) and (C) were gated as live, CD45+ and lineage− . (D) Quantification of ILC3 frequencies in the mLN, PP and SI LP of CNV, GF, SFB-monocolonized and LRC-monocolonized mice. mLN ILC3 values represent frequencies of CD45+RORγt+ of lineage- cells. PP and SI LP ILC3 values represent frequencies of CD90+RORγt+ of lineage− cells. One-way ANOVA, mLN - ****p < 0.0001; PP - *p < 0.05; SI LP - **p < 0.01. (E) PPs of CNV, GF, SFB-monocolonized and LRC-monocolonized mice were analyzed for frequencies of IL-22+ ILC3 cells by flow cytometry. Cells are gated as live, lineage−, CD90+ and RORγt+. Data are pooled from 2 independent experiments for a total of 6–8 mice per group. Data are represented as mean ± SEM. Statistics shown in panel D were performed using one-way ANOVA with uncorrected Fisher’s LSD test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Lymphoid tissue-resident commensal bacteria promote ILC3-derived IL-22 responses that enhance tissue colonization
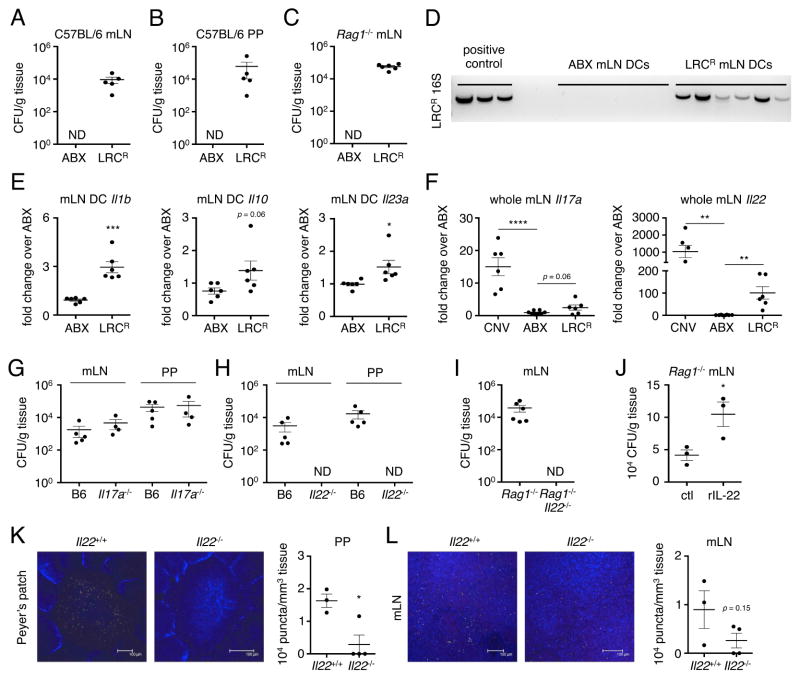
ILC3 and IL-22 responses induce anti-microbial proteins in the intestine (Sonnenberg et al., 2011; Sonnenberg and Artis, 2015), suggesting that LRCs may induce immune responses that limit its own colonization or the colonization of other bacteria. To test these possibilities, we established a mouse model of LRC colonization by treating conventional C57BL/6 mice with a limited cocktail of antibiotics (ABX) followed by inoculation with an antibiotic-resistant LRC, Achromobacter (LRCR). We confirmed that, similar to our results using LRC-monocolonized mice, LRCR could be cultured from the mLN, PPs and intestinal lumen of LRCR-colonized C57BL/6 mice (Figure 5A–B). Furthermore, analyses of ILC3 responses in LRCR-colonized C57BL/6 mice revealed a significant increase in IL-22+ ILC3 frequencies as compared to ABX-treated controls (Figure S4A). Live LRCR were also detected in the mLN of LRCR-colonized Rag1−/− mice (Figure 5C), allowing us to interrogate selective interactions between LRCs and the innate immune system. To investigate whether DC colonization and cytokine modulation by LRCs also occur in the absence an adaptive immune system in vivo, we measured LRCR 16S rDNA and cytokine gene expression in CD11c+ cells isolated from the mLN of LRCR-colonized Rag1−/− mice. LRCR 16S rDNA was detected in CD11c+ cells from LRCR-colonized but not in control ABX-treated mice (Figure 5D). Furthermore, CD11c+ cells from LRCR-colonized mice expressed increased levels of Il1b, Il10 and Il23a as compared to CD11c+ cells from control ABX-treated mice (Figure 5E). Induction of Il1b and Il23a was associated with an increase in Il17a and Il22 expression in the whole mLN but not the small intestine of LRCR-colonized Rag1−/− mice (Figure 5F and S4B).
Figure 5. Lymphoid tissue-resident commensal bacteria colonize antibiotic-treated mice and elicit innate immune responses that enhance colonization.
Bacterial CFUs were determined in antibiotic (ABX)-treated control and ABX-resistant Achromobacter (LRCR)-colonized C57BL/6 PP and mLN homogenates (A and B) or Rag1−/− mLN homogenates (C). Data representative of at least 2 independent experiments. (D) LRCR 16S rDNA was measured in sorted CD11c+ cells from ABX-treated control or LRCR-colonized Rag1−/− mLN. (E) Expression of Il1b, Il10 and Il23a was measured in sorted CD11c+ cells from ABX-treated control or LRCR-colonized Rag1−/− mLN. Data pooled from 2 independent experiments for a total of 6 mice per group. (F) mLNs of CNV, ABX-treated and LRCR-colonized were analyzed for expression of Il17a and ll22. Data pooled from 2 independent experiments for a total of 6–8 mice per group. One-way ANOVA, Il17a – ****p < 0.0001; Il22 – **p < 0.01. (G and H) mLN and PP homogenates from LRCR-colonized C57BL6, Il17a−/− and Il22−/− mice were cultured to determine CFUs. (I) mLN homogenates from LRCR-colonized Rag1−/− and Rag1−/− Il22−/− mice were cultured to determine CFUs. Data pooled from 2 independent experiments for a total of 5–6 mice per group. (J) mLN homogenates from control or recombinant mouse IL-22 (rIL-22)-treated Rag1−/− mice were cultured to determine CFUs. Data representative of 2 independent experiments using recombinant mouse IL-22 or IL-22-Fc. (K and L) Whole PP and mLN were stained with Alcaligenes-specific 16S FISH probes ALBO (red) and BPA (green), and wheat germ agglutinin (blue). Data representative of 2 independent experiments with 2–5 mice per group. Number of puncta per mm3 of tissue was quantified. Data are represented as mean ± SEM. Statistics shown in panels E, F, J, K and L were performed using unpaired, two-tailed, student’s t test (E, J, K and L) and one-way ANOVA with uncorrected Fisher’s LSD test (F). *, p < 0.05; **, p < 0.01; ***, p < 0.001. ND, not detectable. See also Figure S4.
Multiple reports suggest that IL-17A and IL-22 limit intestinal colonization of the commensal bacterium SFB (Qiu et al., 2013; Shih et al., 2014; Upadhyay et al., 2012). Therefore, we hypothesized that induction of IL-17A and IL-22 by LRCs would be important in limiting its own colonization in intestinal-associated lymphoid tissues. To test this, ABX-treated C57BL/6, Il17a−/− and Il22−/− mice were inoculated with LRCR and lymphoid tissue colonization was assessed 10 days later. Based on the requirement for IL-22 in restricting Alcaligenes dissemination (Sonnenberg et al., 2012), we also predicted that colonization of Il22−/− mice with LRCR would result in systemic dissemination of LRCR. However, systemic dissemination of LRCR was not observed in the absence of either IL-17A or IL-22 (Figure S4C). Furthermore, analysis of intestinal-associated lymphoid tissues revealed an impaired ability of LRCR to colonize the mLN and PPs of Il22−/− but not C57BL/6 or Il17a−/− mice (Figure 5G and 5H). These data suggest that IL-22 may also facilitate colonization of lymphoid tissues by commensal bacteria. To test whether innate cell-derived IL-22 could impact lymphoid tissue colonization by commensal bacteria, we inoculated ABX-treated Rag1−/− and Rag1−/−Il22−/− mice with LRCR and measured lymphoid tissue colonization. Similar to our findings using lymphocyte-sufficient Il22−/− mice, inoculation of Rag1−/−Il22−/− mice with LRCR did not result in systemic dissemination of LRCR (Figure S4D), but resulted in impaired LRCR colonization in the mLN (Figure 5I). Collectively, these results suggest that induction of ILC3-derived IL-22 is important to facilitate LRC colonization of lymphoid tissues.
The IL-22 receptor is restricted to non-hematopoietic cells and IL-22-IL-22R signaling regulates antimicrobial responses in the intestinal lumen and the composition of the microbiota (Sonnenberg et al., 2011). Therefore, rather than a direct effect on DCs, we hypothesized that in the absence of IL-22 there may be an expansion of commensal bacteria that outcompete and limit LRC entry and colonization. To investigate this hypothesis, we examined luminal colonization in C57BL/6, Il22−/−, Il17a−/−, Rag1−/− and Rag1−/−Il22−/− mice that were treated with a limited ABX cocktail followed by LRCR inoculation. At 10 days post-inoculation, we detected an absence of fecal LRCR in Il22−/− and Rag1−/−Il22−/− but not in C57BL/6, Rag1−/− and Il17a−/− mice (Figure S4E–F). Il22+/+ littermates raised under the same conditions as Il22−/− mice also had impaired ABX-mediated fecal LRC colonization, and administration of additional antibiotics including neomycin, metronidazole and vancomycin, restored fecal LRC colonization in Il22−/− mice and Il22+/+ littermates (Figure S4G). Furthermore, cohousing Il22−/− mice with C57BL/6 mice impaired fecal and lymphoid tissue LRC colonization in C57BL/6 mice (Figure S4H). Altogether, our data indicate that IL-22 promotes LRC colonization by limiting competition with other commensal microbes.
To further interrogate the role of IL-22 in regulating LRC colonization, we pursued a gain-of-function approach. Consistent with a role for IL-22 in promoting lymphoid tissue colonization by LRCs, exogenous administration of recombinant mouse IL-22 to LRCR-colonized Rag1−/− mice significantly enhanced LRCR levels in the mLN (Figure 5J). The LRCR colonization model requires the use of antibiotics that can eliminates other commensal bacteria. To test the requirement for IL-22 in LRC colonization in the presence of a complex microbiota, we measured levels of the endogenous LRC, Alcaligenes by FISH in conventional Il22−/− mice and littermate controls. Compared to Il22+/+ littermates, conventional Il22−/− mice have reduced Alcaligenes puncta in the PP and mLN (Figure 5K and 5L). These data demonstrate that IL-22 plays a critical role in enhancing the colonization of intestinal-associated lymphoid tissues by LRCs in the presence of a complex microbiota.
Lymphoid tissue-resident commensal bacteria colonization provides protection from chemical-induced inflammation and intestinal injury in an IL-10-dependent manner
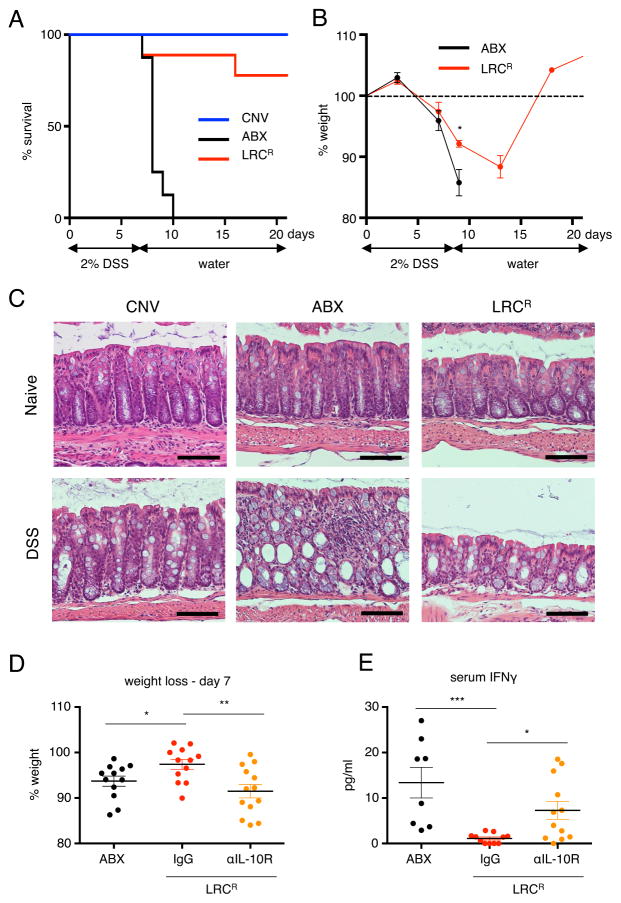
To test whether there was a benefit to mammalian hosts following LRC colonization, we employed a model of intestinal damage where administration of the chemical, dextran sodium sulfate (DSS), induces significant damage to the colonic epithelium. Compared to CNV mice, mice lacking commensal bacteria or treated orally with a broad-spectrum cocktail of antibiotics are highly sensitive to intestinal damage induced by DSS and succumb to disease (Ayres et al., 2012; Kitajima et al., 2001; Maslowski et al., 2009). To test the functional significance of LRC colonization following DSS treatment, we administered 2% DSS to CNV, ABX-treated alone and LRCR-colonized Rag1−/− mice for 6–7 days, then removed DSS, and re-administered normal drinking water. Compared to CNV mice, which survived throughout the duration of the experiment (21 days), ABX-treated mice exhibited significant systemic and intestinal morbidity and succumbed to disease by day 10 post DSS administration (Figure 6A, Figure S5A). In contrast, LRCR-colonized mice were largely protected from DSS-induced mortality, and demonstrated significantly reduced weight loss and elevated red blood cell counts as compared to ABX-treated controls (Figure 6A–B, S5A–B). Further, analyses on day 5 post DSS administration revealed that although CNV mice did not exhibit significant intestinal damage or inflammation in the colon at this low dose of DSS, the colons of ABX-treated mice displayed severe loss of crypt architecture and extensive inflammation (Figure 6C). In contrast, LRCR-colonized mice had visible but reduced intestinal tissue damage and inflammation (Figure 6C). LRC-conferred protection from DSS-induced mortality was also observed in gnotobiotic C57BL/6 (Figure S5C) and Rag1−/− (Figure S5D) mice monocolonized with Bordetella spp., suggesting that host protection can occur in the absence of other commensal bacteria. Collectively, our data demonstrate that colonization of lymphoid tissues by selective commensal bacteria is host beneficial in the context of intestinal damage and inflammation.
Figure 6. Lymphoid tissue-resident commensal bacteria promote IL-10R-dependent protection from DSS-induced intestinal tissue damage and mortality.
CNV, ABX-treated and ABX-resistant Achromobacter (LRCR)-colonized-colonized Rag1−/− mice were administered 2% DSS in their drinking water for 6 to 7 days and then placed on regular drinking water. Mice were monitored for survival (A) and weight loss (B) for up to 21 days. Data in panel A are pooled from 2 independent experiments for a total of 8–9 mice per group. (C) Mice were sacrificed on day 6 and analyzed for colon tissue pathology and inflammatory infiltrate by H&E. Data in panel C are representative of 2 independent experiments. Scale bar – 100 μm. (D and E) ABX control, IgG- or anti-IL-10R-treated LRCR-colonized Rag1−/− mice were administered 2% DSS for 8 days. Percentage of starting weight on day 7 (D) and serum IFNγ on day 8 are quantified (E). One-way ANOVA, D and E - **p < 0.01. Data in panel D are pooled from 3 independent experiments for a total of 12–13 mice per group. Data in panel E are pooled from 3 independent experiments for a total of 8–12 mice per group. Data are represented as mean ± SEM. Statistics shown in B were performed using unpaired, two-tailed, student’s t test. Statistics shown in panels D and E were performed using one-way ANOVA with uncorrected Fisher’s LSD test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. See also Figures S5 and S6.
To investigate whether LRC-induced IL-22 is providing host protection in LRCR-colonized mice, we administered anti-IL-22 neutralizing antibody to LRCR-colonized Rag1−/− mice during exposure to DSS. Anti-IL-22-treated mice exhibited comparable weight loss to IgG-treated mice (Figure S6B), suggesting that IL-22 is not involved in LRC-mediated host protection from DSS. As the anti-inflammatory cytokine IL-10 was also induced upon LRC colonization both in the steady state and during DSS exposure (Figure 2 and Figure S6A), we interrogated whether innate cell-derived IL-10 is providing host protection in LRCR-colonized mice. To test this, we administered anti-IL-10R neutralizing antibody to LRCR-colonized Rag1−/− mice during exposure to DSS. Blockade of IL-10-IL-10R interactions resulted in increased weight loss and colonic inflammation as compared to IgG-treated controls (Figure 6D, S6C–D). ABX-treated Rag1−/− mice on DSS display elevated levels of serum IFNγ, which was significantly diminished in LRCR-colonized Rag1−/− mice (Figure 6E). Conversely, IL-10R neutralization in LRCR-colonized mice partially restored serum IFNγ levels observed in ABX-treated mice (Figure 6E). Treatment of ABX-control Rag1−/− animals with anti-IL-10R during DSS exposure did not result in further weight loss or colon pathology compared to isotype-treated animals (Figure S6E-F). Collectively, these data suggest that LRC colonization protects mice from intestinal and systemic inflammatory responses through induction of innate cell-derived IL-10.
Discussion
Our results define a unique host-commensal bacteria dialogue whereby direct interactions between LRCs and the mammalian immune system orchestrate tissue-specific immune responses that are mutually beneficial for the host and microbe. While most studies of host-microbiota interactions to date have focused on populations of commensal bacteria residing in the intestinal lumen or attached to the intestinal epithelium, here we interrogate an under-appreciated class of commensal bacteria that has the potential to reside closely associated with immune cells in the interior of intestine-associated lymphoid tissues. This colonization pattern is paradoxical to our current understanding of interactions between the immune system and the majority of other commensal bacteria, which suggests that anatomical segregation must occur to limit the development of pathologic immune responses (Brenchley and Douek, 2012; Hooper et al., 2012; Hooper and Macpherson, 2010; Manichanh et al., 2012). Our data highlighting this unique colonization pattern and selective interactions with the healthy mammalian immune system represent a key advance in our understanding of host-commensal bacteria relationships.
Interrogating functional interactions between the host immune system and several species of LRCs revealed that LRCs but not luminal members of the microbiota could colonize and persist within murine DCs and modulate selective DC cytokine responses in a viability-dependent manner, as has previously been shown for IL-1β (Sander et al., 2011). Constitutive colonization of DCs by LRCs likely maintains expression of viability-dependent cytokines, including IL-1β, IL-10 and IL-23. This is in direct contrast to luminal-resident or epithelial-attached commensal bacteria in which viable bacteria belonging to these groups are not typically found beyond the intestinal epithelium (Hooper et al., 2012; Hooper and Macpherson, 2010; Obata et al., 2010). Therefore, these commensal bacteria predominantly influence immune cells through indirect interactions, such as via production of metabolites or modulation of intestinal epithelial cell responses (Arpaia et al., 2013; Furusawa et al., 2013; Ivanov et al., 2009; Smith et al., 2013). Furthermore, our data suggest that induction of tissue-specific immune responses by commensal bacteria is linked to their anatomical localization in the GI tract and associated lymphoid tissues. Consistent with this, SFB, which adheres to the surface of the terminal ileum in mice, induces potent Th17 cell responses in the underlying SI LP (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). Similarly, Bacteroides fragilis predominantly colonizes the colonic crypts in mice and promotes robust regulatory T cell responses in the underlying colon lamina propria (Lee et al., 2013; Round and Mazmanian, 2010). In our current study, we demonstrate that LRCs colonize and activate DCs to selectively promote Th17 and ILC3 responses within intestinal-associated lymphoid tissues.
Multiple reports suggest that induction of the ILC3-IL-22 pathway limits host colonization by pathogenic attaching and effacing bacterial pathogens such as Citrobacter rodentium or the commensal bacterium SFB (Qiu et al., 2013; Shih et al., 2014; Sonnenberg et al., 2011; Upadhyay et al., 2012). In contrast, our data demonstrate that IL-22 facilitates colonization of lymphoid tissues by LRCs. We demonstrate that Il22−/− mice harbor intestinal microbes that limit LRC colonization in the lumen likely through competition for space or nutrients. This is consistent with IL-22-induced antimicrobial functions on gram-positive commensal bacteria (Hooper and Macpherson, 2010; Sonnenberg et al., 2011). These data collectively suggest that IL-22 indirectly promotes LRC colonization in multiple contexts by restricting colonization of competing intestinal microbes. We previously reported that ILC3-derived IL-22 is a critical pathway that maintains anatomical containment of LRCs in intestinal-associated lymphoid tissues in healthy mice and that transient loss of this pathway results in dissemination of Alcaligenes to systemic lymphoid tissues (Sonnenberg et al., 2012). In the current study, we did not observe systemic dissemination of a distinct but related LRC, but rather impaired lymphoid tissue entry and colonization in mice deficient in IL-22. Therefore, our findings highlight a multifaceted role for IL-22 in maintaining anatomical localization of LRCs, by both promoting lymphoid tissue colonization and limiting systemic dissemination.
LRC colonization is sufficient to protect mice from lethal intestinal damage, supporting the hypothesis that host recognition of commensal bacteria provides beneficial microbial stimulation to limit intestinal damage. It was previously demonstrated that mice lacking the pattern recognition receptors TLR2, TLR4 or the downstream adaptor molecule MYD88 are also susceptible to lethal DSS-induced intestinal damage (Rakoff-Nahoum et al., 2004), suggesting that direct recognition of microbial signals is critical for maintenance of the intestinal barrier. Despite our understanding of microbial recognition pathways in host protection from intestinal injury, the downstream signals induced are not fully understood. Our current data suggest that MYD88-dependent IL-10 induction by LRCs is one pathway that provides tissue protective functions in the context of intestinal injury and systemic inflammation. These data are consistent with the anti-inflammatory and tissue protective roles of IL-10 in the intestine (Kuhn et al., 1993; Ouyang et al., 2011) and that IL-10 responses in the intestine are dramatically driven by the microbiota (Atarashi et al., 2011; Chiu and Ching, 2014; Geuking et al., 2011; Ueda et al., 2010). Our data support a model whereby, following LRC colonization, IL-10 produced by DCs in intestinal-associated lymphoid tissues has local effects in limiting Th17 cell responses in the steady state, as well as distal effects on the intestinal epithelium and systemic circulation in the context of intestinal damage.
Collectively, these findings demonstrate a previously unrecognized and mutually beneficial dialogue between the host and LRCs. Colonization of lymphoid tissues by commensal bacteria modulates the host immune system in a tissue-specific manner and confers protective effects in the context of intestinal damage. From an evolutionary perspective, there may be multiple beneficial functions for permitting colonization of mammalian lymphoid tissues by selective commensal bacteria. However, it is likely that a tightly regulated balance between beneficial, opportunistic and pathologic bacteria colonization must be maintained in the lymphoid tissues in order to prevent chronic inflammation. Further interrogation of the role and regulation of LRC colonization in health and disease will be critical to understand the complex microbial contexts where LRCs will influence tissue immunity and inflammation, and may reveal novel therapeutic targets for the treatment of multiple chronic inflammatory diseases.
Experimental Procedures
Mice
C57BL/6 and Rag1−/− mice were purchased from Jackson Laboratories and used at 6–12 weeks of age. Conventional mice used as controls in gnotobiotic mice experiments were purchased from Jackson Laboratories and co-housed with mice bred in our conventional animal facility for at least 7 days before use. C57BL/6 and Rag1−/− mice used as controls for knockout mice were either bred in the same animal facility as the knockout mice, purchased from Jackson Laboratories and co-housed with mice bred in our SPF animal facility for at least 7 days or littermates as indicated to normalize for microbiota differences. All conventional mice were maintained in specific pathogen-free facilities at Weill Cornell Medical College. Germ-free and gnotobiotic C57BL/6 and Rag1−/− mice were maintained within sterile vinyl isolators at Weill Cornell Medical College Gnotobiotic Mouse Facility and monitored for germ-free or gnotobiotic status by weekly aerobic and anaerobic culturing. Germ-free and gnotobiotic Il10+/+ and Il10−/− mice were maintained at the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill). Additional microbiology testing was performed on feces from mice under experimentation and at the endpoint of the experiment to confirm germ free or monocolonization status. DSS experiments using gnotobiotic mice were performed in a biosafety cabinet, and the gnotobiotic status of mice was confirmed by microbiology testing. Germ-free C57BL/6 mice were monocolonized with the mouse LRC isolate Bordetella spp., or SFB by oral gavage or cohousing with soiled bedding from previously monocolonized mice for at least 10 days. To establish LRC colonization in ABX-treated mice, conventional mice were treated in the drinking water with a limited ABX cocktail of ampicillin (1 mg/ml, Sigma) and gentamicin (1 mg/ml, Gemini Bio-Products) for C57BL/6, Il22−/− and Il17a−/− mice or ampicillin (1 mg/ml), gentamicin (1 mg/ml) and neomycin (0.25 mg/ml, Sigma) for Rag1−/− and Rag1−/− Il22−/− for 3 days and then orally gavaged with the LRC isolate (LRCR). Neomycin (0.25 mg/ml), metronidazole (0.5 mg/ml, Sigma) and vancomycin (0.25 mg/ml, Chem-Impex International) were added to the limited ABX cocktail as indicated. Antibiotic cocktails were supplemented with 1 packet of artificial sweetener (Sweet’N Low) per 250 ml. For cytokine treatments in vivo, rmIL-22 or PBS control (kindly provided by Pfizer) was injected i.p. at 25 μg/mouse every 2 days for 1 week and control- or IL-22-Fc (kindly provided by Pfizer) was injected i.p. at 50 μg/mouse every 3 days for 1 week. First injections were administered 1 day prior to LRCR colonization. All mice were used at least 10 days post LRCR colonization. All animals used are on a C57BL/6 background with the exception of Il10−/− and Il10+/+ germ free mice, which are on a 129S6/SvEv background. All experiments were performed according to the guidelines of the Weill Cornell Medical College Institutional Animal Care and Use Committee.
Microbiology
The LRC strain, Bordetella spp., used to colonized BMDCs or monocolonize germ-free mice was originally isolated from the spleen of an anti-CD90.2 mAb treated conventional Rag1−/− mice using a previously defined protocol (Sonnenberg et al., 2012). E. coli (human isolate) and Ochrobactrum spp. used to colonize BMDCs and the ABX-resistant (ampicillin, gentamicin, neomycin) LRC (LRCR), Achromobacter used to colonize antibiotic-treated mice were human clinical isolates kindly provided by Kaede V. Sullivan (Children’s Hospital of Philadelphia). Bacterial identities were determined by genomic sequencing (Wellcome Trust Sanger Institute) and confirmed by genus-specific 16S rDNA primers. E. coli R9 (ATCC 35328), Enterobacter cloacae (ATCC 13047) and gentamicin-sensitive Achromobacter spp. (ATCC 31444) used to colonize BMDCs were purchased from American Type Culture Collection. Feces from SFB-monocolonized mice were obtained from Dr. Yoshinori Umesaki and Dr. Tatsuichiro Shima (Yakult). All bacterial strains used to colonize GF mice and for in vitro DC assays were grown in LB broth and incubated at 250 RPM, 37°C for 16–20 hours. For enumeration of tissue and fecal CFUs, the spleen, liver, feces, mesenteric lymph nodes (mLN) and Peyer’s patches (PP) were isolated, homogenized in sterile PBS and plated on Brain Heart Infusion agar supplemented with 5% defibrinated horse blood. When culturing LRCR from tissues and feces, BHI blood agar plates were supplemented with 25 μg/ml ampicillin.
Bone marrow-derived and primary dendritic cell assays
Bone marrow-derived DCs were generated by culturing bone marrow cells in the presence of 20 ng/mL GM-CSF for 8–10 days. Culture media was replaced with fresh media every 3 days. Frequencies of CD11c+ cells were ≥ 95%. To prepare primary splenic and mLN DCs, C57BL/6 mice were first injected subcutaneously in the right flank with 5 x 106 cells GM-CSF-expressing B16 melanoma (kindly provided by Jedd D. Wolchok, MSKCC). 10 days later, CD11c+ cells were purified from Liberase TL (Roche)-digested spleen and mLN using CD11c positive selection beads (Miltenyi Biotec). Cell purities in the spleen were approximately 90% CD11c+ CD11b+ of non-T and B cells. For intracellular bacterial survival assays, BMDCs were seeded on 6-well plates at 5 x 106 cells/well in antibiotic-free media and co-cultured with bacteria at an MOI of 50 for 2 hours, harvested and washed 3 times with sterile PBS. BMDCs were then plated in media containing gentamicin (100 μg/mL) at 200,000 cells/well on 48-well plates and lysed with sterile water every 48 hours to enumerate CFUs. For ELISA and qPCR, BMDCs were co-cultured with bacteria at an MOI of 50 for 2 hours and then gentamicin-treated to kill extracellular bacteria. Culture supernatants and cell lysates were harvested for ELISA and qPCR, respectively, 24 hours later. Heat killing was performed by incubating bacterial suspensions at 70°C for 60 mins. For transmission electron microscopy, BMDCs colonized with Bordetella spp. for 5 days were harvested, processed and imaged using the Jeol-1010 transmission electron microscope. For microarray analysis, BMDCs were co-cultured with bacteria for 2 hours, washed extensively with PBS, incubated in media containing gentamicin (100 μg/mL) and lysed in TRIzol on day 4. All cell incubations were performed at 37°C and 5% CO2.
Immunofluorescent detection of intracellular bacteria
BMDC-bacteria co-cultures were harvested on days 0, 2 and 5 and transferred to glass slides via cytospin. BMDCs were fixed with 4% PFA for 30 minutes at room temperature and stained with the Live/Dead BacLight Bacterial Viability Kit according to the manufacturer’s instructions (Invitrogen). Cells were mounted with VectaShield mounting medium containing DAPI (Vector Laboratories) and imaged on the Nikon Eclipse Ti Fluorescence Inverted Microscope. PFA fixation did not affect the ability of the bacterial viability kit to distinguish between live and dead bacteria.
Gene expression profiling and quantitative real-time PCR
RNA was isolated using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. cDNA was generated using Superscript reverse transcriptase II (Invitrogen). Quantitative real-time PCR was performed using SYBR green chemistry (Invitrogen) and QuantiTect Primer Assays (Qiagen) on the ABI 7500 real-time PCR system (Applied Biosystems). Samples were normalized to β-actin and displayed as fold change over PBS-treated BMDCs or antibiotic-treated mice. For microarray analysis, BMDCs colonized with the LRC, Bordetella spp., for 4 days were lysed directly in TRIzol. RNA was isolated, amplified, reverse-transcribed to cDNA and hybridized to an Affymetrix GeneChip (Mouse Gene 1.0ST). Relative expression data was normalized by Z score transformation.
Histological sections
Tissues from the small and large intestines were fixed with 4% PFA, embedded in paraffin, and 5 μm sections were cut and stained with H&E.
DSS-induced intestinal damage
Mice were administered 2% DSS (MW 36,000–50,000, MP Biochemicals) in their drinking water ad libitum for the number of days indicated and then placed on regular drinking water. Disease severity was cumulatively scored based on rectal bleeding (out of 2), fecal consistency (out of 2), general appearance (out of 4), weight loss (out of 4) and rectal temperature (out of 4). For antibody treatments during DSS, mice were administered i.p. 500 μg/mouse of rat IgG, anti-IL22-01 (mouse neutralizing antibody developed by Pfizer) or anti-IL-10R (clone 1B1.3A, Bio X Cell) 1 day prior to DSS treatment, then on days 2, 5 and 7. Mice were sacrificed on day 8.
Statistical analyses
Results represent mean ± SEM and statistical analyses were performed by unpaired student’s t-test, one- or two-way ANOVA with or without multiple comparisons tests as indicated in figure legends.
Supplementary Material
Highlights.
Lymphoid-resident commensal bacteria (LRCs) colonize intestinal lymphoid tissues
LRCs promote tissue-specific Th17 cell and ILC3 responses in intestinal tissues
IL-22 enhances LRC colonization of intestinal lymphoid tissues
LRCs provide host tissue protection during DSS in an IL-10R-dependent manner eTOC blurb
Acknowledgments
We thank members of the Sonnenberg laboratory for discussions and critical reading of the manuscript. This research is supported by the National Institutes of Health (DP5OD012116, R56AI114724 and R01AI123368 to G.F.S.), and the NIAID Mucosal Immunology Studies Team (MIST) Scholar Award in Mucosal Immunity (to G.F.S.), the Crohn’s and Colitis Foundation of America (to M.R.H.), the Cancer Research Institute Student Training and Research in Tumor Immunology grant (to T.C.F), the Wellcome Trust (098051) and Medical Research Council UK (PF451) for library preparation, sequencing and genomic analysis, the Ministry of Education, Culture, Sports and Technology of Japan (Grants-in-Aid for Scientific Research B [J.K.] and for Scientific Research S [H.K.]), the Ministry of Health and Welfare of Japan (J.K. and H.K.), Japan Science and Technology Agency (JST), Core Research for Evolutional Science and Technology (CREST) (H.K.), P30-DK034987 (R.B.S.) and 5-P40-OD010995 (R.B.S.). We thank Sylvester S. Roundtree Jr. (Children’s Hospital of Philadelphia) for providing clinical bacterial isolates and National Gnotobiotic Rodent Resource Center for providing germ animals.
Footnotes
Author contributions
T.C.F., M.R.H., N.J.B., J.G., K.W., D.A. and G.F.S. designed and performed the experiments. D.K. and R.B.S provided assistance and expertise in gnotobiotic mice. N.K. and T.L. performed core genome sequencing and phylogeny analyses. N.S., J.K., H.K. performed FISH analyses. C.G.K.Z. performed analyses of microarray data. K.V.S. provided expertise and human clinical bacterial strains. T.S. and Y.U. provided expertise and feces from SFB-monocolonized mice. T.C.F. and G.F.S wrote the manuscript.
Data accessibility
Sequenced microbial genome data are available at the European Nucleotide Archive under study number ERP012121. Array data are available at GEO under accession number GSE76731.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nature medicine. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annual review of immunology. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CC, Ching YH. Monocolonization of germ-free mice with Bacteroides fragilis protects against dextran sulfate sodium-induced acute colitis. 2014;2014:675786. doi: 10.1155/2014/675786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Advances in immunology. 2010;107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunological reviews. 2014;260:35–49. doi: 10.1111/imr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annual review of immunology. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Morimoto M, Sagara E, Shimizu C, Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Experimental animals / Japanese Association for Laboratory Animal Science. 2001;50:387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Goppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Kunisawa J, Kiyono H. Alcaligenes is Commensal Bacteria Habituating in the Gut-Associated Lymphoid Tissue for the Regulation of Intestinal IgA Responses. Frontiers in immunology. 2012;3:65. doi: 10.3389/fimmu.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Tonkonogy SL, Sartor RB. Antigen-presenting cell production of IL-10 inhibits T-helper 1 and 17 cell responses and suppresses colitis in mice. Gastroenterology. 2011;141:653–662. 662.e651–654. doi: 10.1053/j.gastro.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nature immunology. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- Obata T, Goto Y, Kunisawa J, Sato S, Sakamoto M, Setoyama H, Matsuki T, Nonaka K, Shibata N, Gohda M, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annual review of immunology. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Shih VF, Cox J, Kljavin NM, Dengler HS, Reichelt M, Kumar P, Rangell L, Kolls JK, Diehl L, Ouyang W, Ghilardi N. Homeostatic IL-23 receptor signaling limits Th17 response through IL-22-mediated containment of commensal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13942–13947. doi: 10.1073/pnas.1323852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Kayama H, Jeon SG, Kusu T, Isaka Y, Rakugi H, Yamamoto M, Takeda K. Commensal microbiota induce LPS hyporesponsiveness in colonic macrophages via the production of IL-10. International immunology. 2010;22:953–962. doi: 10.1093/intimm/dxq449. [DOI] [PubMed] [Google Scholar]
- Upadhyay V, Poroyko V, Kim TJ, Devkota S, Fu S, Liu D, Tumanov AV, Koroleva EP, Deng L, Nagler C, et al. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nature immunology. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.