Abstract
Protein O-fucosyltransferase 2 (POFUT2) is an essential enzyme that fucosylates serine/threonine residues of folded thrombospondin type 1 repeats (TSRs). To date, the mechanism by which this enzyme recognizes very dissimilar TSRs remained unclear. By engineering of a fusion protein, we report the crystal structure of Caenorhabditis elegans POFUT2 (CePOFUT2) in complex with GDP and human TSR1 that suggests an inverting mechanism for fucose transfer assisted by a catalytic base, and shows that nearly half of the TSR1 is embraced by CePOFUT2. A small number of direct interactions and a large network of water molecules maintain the complex. Site-directed mutagenesis demonstrates that POFUT2 fucosylates threonine preferentially over serine and relies on folded TSRs containing the minimal consensus sequence CXX(S/T)C. Crystallographic and mutagenesis data together with atomic-level simulations uncover an unprecedented binding mechanism by which POFUT2 promiscuously recognizes the structural fingerprint of poorly homologous TSRs through a dynamic network of water-mediated interactions.
Protein glycosylation is the most abundant and complex post-translational modification in nature. The reaction in eukaryotes is performed by a limited number of glycosyltransferases (GTs) that specifically recognize acceptor substrates and catalyze transfer from different activated sugar donors1,2. Whereas many of these GTs have the capacity to glycosylate unstructured peptides, only four GTs have been proven to require disulfide bridge-containing folded domains to achieve glycosylation3,4. Of these GTs, protein O-fucosyltransferases 1 and 2 (POFUT1 and 2) have been more extensively studied. These inverting enzymes are classified as GT65 and GT68, respectively, according to the CAZy database5. POFUT1 and POFUT2 are located in the endoplasmic reticulum (ER), and fucosylate epidermal growth factor-like (EGF) repeats and thrombospondin type 1 repeats (TSRs), respectively, present in the extracellular domains of a large number of proteins (such as Notch, thrombospondin 1 and ADAMTS13)6. O-Fucosylation is an essential biological modification that is not only important as an ER quality control mechanism due to its influence in the correct folding and stability of EGF repeats and TSRs but also for protein-protein interaction7,8. In particular, it is well documented that glycosylation of Notch EGF repeats is important for Notch-ligand interactions9,10. The molecular aspects of this interaction have been very recently uncovered showing that O-fucose of EGF12 acts as a critical amino acid surrogate interacting with Notch-ligand Delta-like 47.
Crystal structures of the C. elegans POFUT1 and human POFUT2 (HsPOFUT2) have been solved in complex with nucleotides, revealing that both enzymes adopt the typical GT-B fold11,12. Despite large similarities at the structural level, their proposed catalytic mechanism differed. While POFUT1 was suggested to follow an SN1-like mechanism with the GDP β-phosphate acting as the catalytic base, POFUT2 was proposed to follow an inverting SN2-like mechanism, in which a glutamic acid acts as the catalytic base11,12. Site-directed mutagenesis on POFUT2 suggests that the specificity of POFUT2 on TSR domains is based on the three-dimensional structure of these domains.
Both EGF repeats and TSRs are small domains containing three disulfide bridges, and consensus sequences for fucosylation of each have been identified: C2X4(S/T)C3 (where C2 and C3 are the second and third cysteines) for POFUT1 modification of EGF repeats, and C1X2-3(S/T)C2X2G for POFUT2 modification of TSRs6. However, the molecular details of how these fucosyltransferases recognize these repeats and catalyze reactions are not currently known. The lack of ternary complexes has impeded answering the long-standing question of what strategies these broad-substrates enzymes employ to recognize and fucosylate such a vast number of protein substrates. In fact, up to ~75% of human TSRs, which are characterized by highly variable sequence identities and a very low number of conserved residues (identities oscillate between ~19% and ~37%; Supplementary Results, Supplementary Fig. 1a, b), contain the above consensus sequence and are fucosylated by POFUT2. Here, by engineering a fusion protein, we have captured a ternary complex formed between POFUT2, GDP and TSR1 by X-ray crystallography, uncovering the molecular basis of POFUT2 recognition on multiple protein substrates.
RESULTS
Strategy for producing a crystallizable construct
A multiple alignment of CePOFUT2 with higher eukaryotes POFUT2s clearly indicated the high level of sequence identity among species, with values ranging from 40% to 44% (Supplementary Fig. 2). POFUT2s from several species were expressed in Pichia pastoris, but only CePOFUT2 was successfully secreted, though the enzyme was partly degraded into two forms (Supplementary Fig. 3). We carried out trypsin digestion of the wild type form and the degraded products followed by MALDI-TOF-MS analysis, which helped us elucidate the potential cleavage site. Single and double mutants were designed and purified, but only the double mutant R298K-R299K (for simplicity, this double mutant will be henceforth named CePOFUT2) showed no degradation products (Supplementary Fig. 3). The stable enzyme was active at a very similar rate compared to the HsPOFUT2 and showed a decrease in activity in the presence of manganese, similarly to HsPOFUT2 (Supplementary Fig. 4). Different complexes formed between CePOFUT2, GDP and HsTSR1 (the first TSR from human thrombospondin 1) were isolated (Supplementary Fig. 5). However, none of these complexes rendered crystals containing the ternary complex. Of note, most of the proteins containing TSRs have multiple tandem repeats; this pointed us to consider a complex of the enzyme with TSRs in tandem such as the HsTSR1-2-3, which also did not result in crystals. Another strategy was to produce a complex of the enzyme with a fucosylated form of HsTSR1, though in this particular case the complex was not formed (Supplementary Fig. 5). This was further supported by isothermal titration calorimetry (ITC) analysis in which no energy was released upon titration of the fucosylated HsTSR1 under the same conditions measured for GDP-fucose, GDP or HsTSR1 (Online methods and Supplementary Fig. 6 and Supplementary Table 1).
Considering that HsTSR1 was able to moderately bind to CePOFUT1 either in the absence or presence of saturated GDP (Kd of HsTSR1 was 0.95 and 1.03 μM, respectively; Supplementary Table 1) and we could purify a complex by gel filtration (Supplementary Fig. 5), we envisaged a different strategy based on docking studies. We modeled the structure of CePOFUT2 based on the human POFUT2 crystal structure11 and then HsTSR1 was docked into the modeled enzyme (see details in Online methods). The complex showed that the N-terminus of HsTSR1 was ~30 Å away from the CePOFUT2 C-terminus (Supplementary Fig. 7a; Note that there is a large resemblance between our modeled complex and that described previously11). Consequently, a fusion protein was designed in which CePOFUT2 and HsTSR1, both located at the N- and C-terminal, respectively, are coupled by insertion of a flexible linker consisting of 22 Gly and Ser residues. The rationale was to increase the local concentration of CePOFUT2-HsTSR1 complex to increase the likelihood of getting crystals containing a functional ternary complex.
Architecture of the ternary complex
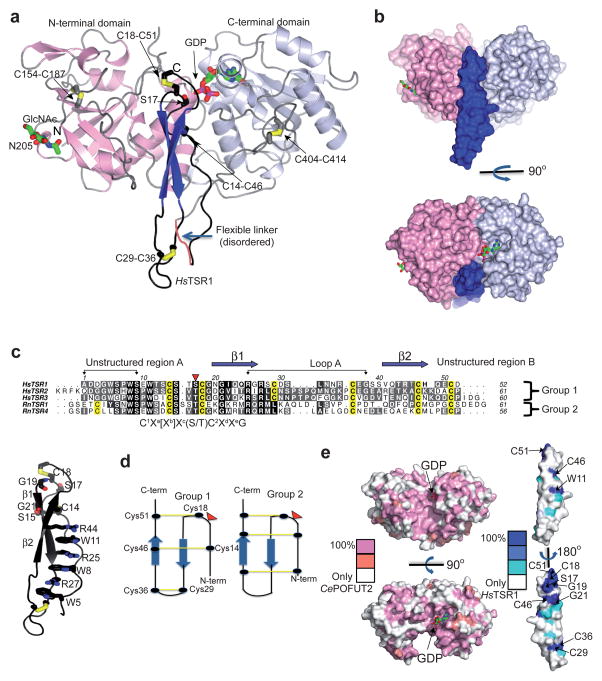
A synthetic DNA construct encoding the fusion protein was expressed in Pichia pastoris and the purified complex was able to transfer fucose from GDP-fucose to itself, but not to a mutant lacking the O-fucosylation site (Online methods and Supplementary Fig. 7b). Then, we crystallized the fusion protein in the presence of GDP. The monoclinic crystals diffracted at 1.98 Å (Online methods and Supplementary Table 2), enabling the structure to be solved and the electron density maps to be unambiguously interpreted (Fig. 1 and Supplementary Fig. 8; see details in Online methods). The asymmetric unit (AU) contained 2 independent ternary complexes with the expected 1:1:1 stoichiometry (1 molecule of CePOFUT2, HsTSR1 and GDP; Fig. 1a and Supplementary Fig. 9), which was also supported by the PISA software for quaternary structure prediction, gel filtration chromatography and atomic force microscopy (AFM) (Supplementary Fig. 5 and 10; see also Online methods). CePOFUT2 structure adopts the typical GT-B fold formed by two Rossmann-like domains, which face each other as previously reported for HsPOFUT2 and CePOFUT111,12. CePOFUT2 contains the two conserved disulfide bridges and one N-glycosylation site (N205 according to CePOFUT2 numbering; Fig. 1a and Supplementary Fig. 2). GDP is located in a shallow cavity of the C-terminus, whereas half of HsTSR1 is embraced by a pocket formed between both CePOFUT2 domains (Fig. 1a, b). Each side of the interface buries an area of 789 Å2, which agrees with the typical range of 600–800 Å2 shown for other heterocomplexes13.
Figure 1. Structure of CePOFUT2 in complex with GDP and HsTSR1.
a, Cartoon representation of the complex. The N- and C-termini of CePOFUT2 are colored in pink and bluewhite, respectively. Secondary structures of HsTSR1 are colored in blue whereas loops and unstructured regions are in black. The disulfide bridges are indicated in yellow. The GlcNAc moiety covalently bound to N205 and GDP are shown in green carbon atoms while N205 is shown as pink carbon atoms. The acceptor S17 of HsTSR1 is shown as black carbon atoms. The flexible linker is shown in orange. b, Surface representation of the complex in two different views. c, Multiple sequence alignment of human TSR1, TSR2 and TSR3 of thrombospondin 1 and Rattus novergicus F-spondin 1 and 4 (upper panel). The fucosylated S/T residues and the conserved cysteines are indicated in red and yellow, respectively. The numbering for each TSR does not correspond to the numbering to their location in thrombospondin and F-spondin, respectively. Residues delimited by arrows are exposed to the bulk solvent. The consensus sequence, C1Xa[Xb]Xc(S/T)C2XdXeG, is shown below the multiple sequence alignment. The lower panel shows a cartoon representation of HsTSR1 colored in black. The residues belonging to the CWR-layered structure and some other conserved residues (shown in bold) between human TSR1, TSR2 and TSR3 are shown as sticks with black carbon atoms. d, Diagram showing the different arrangement of disulfide bridges found in TSRs of group 1 and 2. The inverted triangles in red point out the fucosylation sites that in turn have the same location in the 3D fold of both groups. e, Surface representation of the CePOFUT2 (left) and HsTSR1 (right), colored by sequence conservation. GDP is shown as sticks with green carbon atoms.
POFUT2 is not only capable of fucosylating TSRs of group 1 but also group 2 containing the consensus sequence shown above11,14. These two types of TSRs share a very similar 3D fold and a common core structure (CWR-layered structure, where CWR stands out for cysteine, tryptophan and arginine residues) but differ in their disulfide bonding patterns (Fig. 1c, d)14. For example, only the second disulfide bridge is conserved (C18-C51, discussed below) (Fig. 1c, d). POFUT2 not only recognizes and reacts with TSRs showing slightly different structures but also accepts TSRs with very low sequence identity. In fact, a sequence conservation analysis of 63 human TSRs shows a very limited number of conserved residues among all these repeats (Fig. 1c, e, and Supplementary Fig. 1). This emphasizes the versatility of this enzyme in recognizing multiple TSRs. On the contrary, a sequence conservation analysis between different POFUT2s shows ~100% identity at both the binding site for the sugar nucleotide and the TSRs (Fig. 1e and Supplementary Fig. 2).
As above-mentioned, POFUT2 fucosylates proteins containing TSRs in tandem repeats and not as single entities (Supplementary Fig. 11), raising the question of whether this fucosyltransferase acts processively on these repeats. To address this question, the enzyme was incubated with a fixed concentration of fully folded HsTSR1-2-3 and variable concentrations of GDP-fucose (Online methods and Supplementary Fig. 12). The results clearly showed that the TSRs are randomly fucosylated, suggesting that this enzyme is non-processive in vitro (Supplementary Fig. 12). Our prior results suggested that POFUT2 modifies tandem TSRs sequentially from amino to carboxyl termini in vivo8. Our in vitro data presented here suggest that this is not an inherent property of POFUT2 but is likely due to sequential folding of newly synthesized TSRs in the endoplasmic reticulum. Our prior work showed that POFUT2 recognizes a newly folded TSR and adds a fucose, stabilizing the folded TSR and accelerating the net rate of folding, consistent with the quality control function of POFUT28.
E52 of CePOFUT2 is the catalytic base
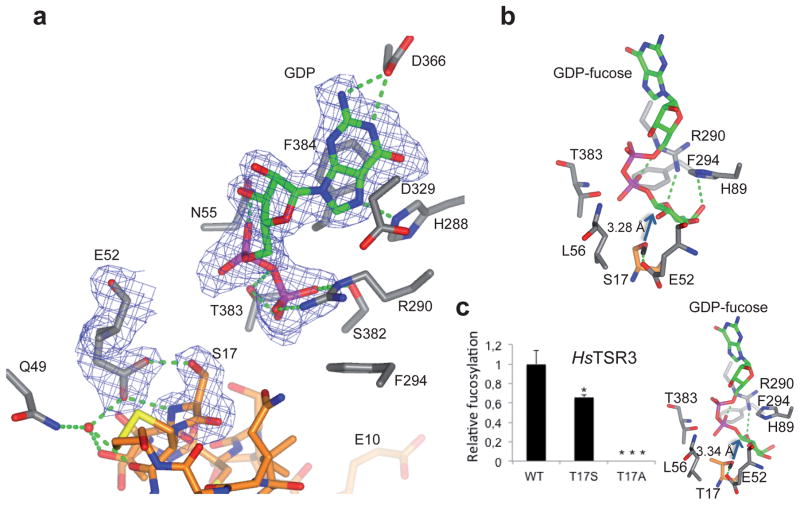
A close-up view at the intersection between the donor and acceptor substrate binding sites reveals that GDP is very close to the acceptor S17 of HsTSR1 (Fig. 2a), and enlightens mechanistic features of the glycosylation reaction. Specificity for the GDP is defined by several hydrogen bonds with the conserved residues N55, H288, R290, D329, D366, S382 and T383, and a stacking interaction between the guanine moiety and F384 (Fig. 2a and Supplementary Fig. 2). Some of these conserved residues were mutated in HsPOFUT2 and showed the essential role of R294 in catalysis (R290 in the CePOFUT2)11. This residue might be involved in facilitating the cleavage of the glycosidic bond and the stability of the product GDP, as was previously reported for the equivalent R240 of CePOFUT112. Similar roles have been usually attributed to metals in GTs that adopt a GT-A fold15. Even though POFUT2 adopts a GT-B fold, Mn+2 has been reported to play a critical role in catalysis for the human enzyme11. However, our human POFUT2 showed a ~50% decrease in activity in the absence of this metal (Supplementary Fig. 4). Furthermore, the role of the metal in CePOFUT2 catalysis was less important, supporting the non-essential role of Mn+2 in catalysis (Supplementary Fig. 4). The data are reinforced by our crystallized ternary complex that shows no metal in the structure, and the general observation that GTs with GT-B fold do not usually bind metals15. In our case, R294 replaces Mn+2 function by establishing electrostatic and hydrogen bond interactions with the β-phosphate (Fig. 2a).
Figure 2. Catalytic mechanism of POFUT2 and its preferences on threonine over serine residues.
a, Close-up view of the complex active site. The residues of CePOFUT2 and the HsTSR1 are depicted as grey and orange carbon atoms, respectively. GDP is shown as green carbon atoms. Hydrogen bond interactions are shown as dotted green lines. Electron density maps are FO–FC and 2FO–FC syntheses (blue) contoured at 2.2 and 1.0 σ for GDP and S17/E52, respectively. b, Close-up view of CePOFUT2 in complex with GDP-fucose and HsTSR1. c, Relative fucosylation of HsTSR3 compared to the mutants T17S and T17A (Left panel). Close-up view of CePOFUT2 in complex with GDP-fucose and the mutant S17T of HsTSR1 (Right panel). To note that the hydroxyl group of S17 and T17 is 3.28 and 3.34 Å to the anomeric carbon of GDP-fucose, respectively. Both complexes with GDP-fucose were obtained by MD simulations (see Online methods). All replicates were in triplicate. Error bars are standard deviation, and p-values were calculated using ANOVA.* p<0.10, ** p<0.05, ***p<0.01
The acceptor serine, S17, is engaged in a hydrogen bond with E52, reinforcing the role of this residue as the catalytic base. This has been supported by previous site-directed mutagenesis of E54 (numbering of the human enzyme; Supplementary Fig. 2), revealing that this residue is essential for catalysis11. The 0.5 μs molecular dynamics (MD) simulation performed on the enzyme in complex with GDP-fucose and HsTSR1 (see Online methods) shows that the hydroxyl group of S17 is very close to the anomeric carbon of GDP-fucose (O···C1= 3.28 Å) and in an optimal orientation for nucleophilic attack assisted by E52 (Fig. 2b). All these features suggest a classical inversion mechanism facilitated by an enzymatic general base catalyst (in our case a glutamate residue)1,11.
A comparison of the two ternary complexes present in the AU also revealed the presence of a loop (residues 88–94 of CePOFUT2; Supplementary Fig. 2) burying the pyrophosphate moiety of GDP in one of the complexes (Fig. 1b and Supplementary Fig. 13a). The loop is disordered in one complex of the AU (Fig. 1b) and ordered in the other (Supplementary Fig. 13b), revealing its dynamic character. In our MD simulation with GDP-fucose (Fig. 2b), the sugar is hydrogen bonded to H89 located in this loop, suggesting the implication of this motif in the entrance of GDP-fucose and released of GDP during the catalytic cycle. This agrees with previous observations of the importance of this loop in catalysis11.
Preference of threonine over serine residues
POFUT2 has been shown to fucosylate both serine and threonine residues, although the preference for one over the other is not known. To address this point, we mutated the HsTSR3 T17 to serine (T17S) and alanine (T17A) (Fig. 1c, Online methods and Supplementary Tables 3 and 4). We analyzed the secreted levels of HsTSR3 compared to those of the mutants by quantifying HsTSR3 in cell lysates and media from transiently transfected HEK293T cells (see Online methods). Both mutations, T17S and T17A decreased secretion significantly (Supplementary Fig. 14), consistent with the relationship between O-fucosylation and secretion previously observed for ADAMTS13 and ADAMTSL18,16,17. We evaluated then the activity of human POFUT2 on HsTSR3 and its mutants using a cell-based fucosylation assay, observing that the enzyme fucosylates T17 ~2-fold more efficiently than S17 (Fig. 2c and Supplementary Fig. 15). A possible explanation for this behavior might be the higher affinity of the enzyme toward T17 due to the hydrophobic interactions between its methyl group and highly conserved residues such as L56 and T383 (Fig. 2c and Supplementary Fig. 2; see below for further discussion). The better secretion of TSRs containing Thr-O-fucose and the preference of the enzyme to bind threonine might explain why nature has selected a major population of Thr-containing TSRs throughout evolution (~64% and ~36% of the human TSRs contain acceptor threonine and serine residues, respectively; Supplementary Fig. 1a, b). Similar preferences have been reported for the distant family of retaining polypeptide GalNAc-transferases18.
Specificity of POFUT2 on TSR domains
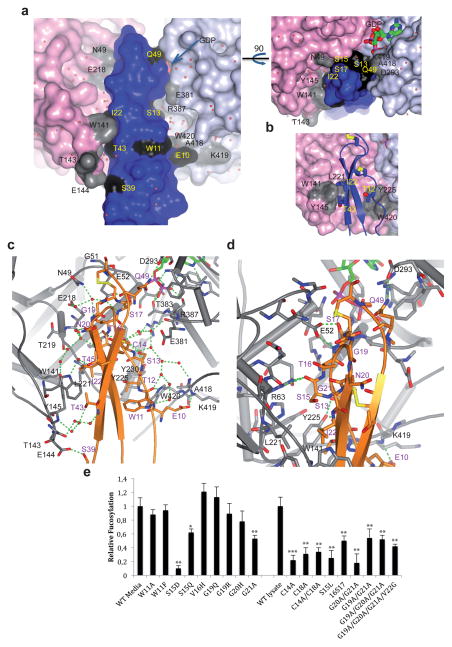
The interface between CePOFUT2 and HsTSR1 shows relevant features for the specificity of POFUT2 on TSR repeats. CePOFUT2 binding domain contains large cavities that are filled by a large network of water molecules along the interface (Fig. 3a). The complex is maintained by a limited number of direct hydrogen bonds and stacking interactions between CePOFUT2 and HsTSR1 that are complemented by water-mediated contacts (Fig. 3b–d, and Supplementary Table 5). Of the 10 residues in HsTSR1 directly involved in the binding, only W11, S15 and S17 are conserved. The side chain of S17 establishes a hydrogen bond with E52 (Fig. 2a), while W11, S15 and S17 backbones are engaged in hydrogen bond interactions with W420, R63 and E52, respectively (Fig. 3c, d, and Supplementary Table 5). In contrast, the interaction of HsTSR1 non-conserved residues with CePOFUT2 is exclusively through HsTSR1 side chains (Supplementary Table 5). In particular, E10, T12, S13, I22, S39, T43 and Q49 side chains interact with K419, Y225, Y225, W141/Y145/L221, E144, W141 and D293, respectively (Fig. 3b–d, and Supplementary Table 5). Note that the central part of HsTSR1 at both sides is tethered by two hydrophobic pockets formed by conserved residues in POFUT2 such as W141/Y145/L221 and Y225/W420 (Fig. 3b). The results also imply that each particular TSR might adopt a small number of unique direct interactions through their non-conserved residues. Unlike HsTSR1, CePOFUT2 engages most of its conserved residues in direct interactions (Fig. 1e, Supplementary Fig. 2 and Supplementary Table 5).
Figure 3. Interactions in the interface of the complex.
a, Surface representation of the complex interface in two different views. Colors are the same as in Fig. 1b. Residues colored in grey and black are engaged in direct or water-mediated interactions. Water molecules are indicated as red spheres. b, Close-up view of the interface showing the only stacking interactions in the complex. c, Close-up view of the direct and water-mediated interactions present in the complex. CePOFUT2 and HsTSR1 are depicted as grey and orange, respectively. Residues carbon atoms for each protein are also shown with the same color above. d, Close-up view of the few direct interactions present in the complex. Colors are the same as above. Hydrogen bond interactions are shown as dotted green lines. e, Relative fucosylation of HsTSR3 in comparison with different mutants located in the interface. The activity for some mutants is compared with HsTSR3 either from lysates or media (see Supplementary Fig. 15). All replicates were in triplicate. Error bars are standard deviation, and p-values were calculated using ANOVA.* p<0.10, ** p<0.05, ***p<0.01
Prompted by these results, we examined the significance of the X-positions in the consensus sequence, C1Xa[Xb]Xc(S/T)C2XdXeG (Fig. 1c, Supplementary Table 4). The additional residue, Xb, has only been found in a single TSR, TSR1 of rat F-spondin (Fig. 1c), and this TSR was only poorly glycosylated19. We generated 20 HsTSR3 mutants to examine effects on recognition by POFUT2 (Online methods and Supplementary Table 3 and 4). The mutations included substitutions in structural positions (W11, C14, C18), the Xa–e positions, and the conserved glycine in the consensus sequence (Fig. 1c). Mutations were generated in the Xa–e positions to evaluate whether certain types of amino acids not yet found in a mapped O-fucosylation site affect the efficiency of fucosylation (Supplementary Table 4). Mutants were evaluated for effect on secretion (Supplementary Fig. 14) and fucosylation efficiency (Fig. 3e and Supplementary Fig. 15) in cell-based assays.
There is a relationship between TSR folding, fucosylation, and secretion. Mutations predicted to cause defects in folding (e.g. C14A, C18A, C14A/C18A) caused severe decreases (>50%) in secretion (Online methods and Supplementary Fig. 14) and fucosylation (Fig. 3e), consistent with the concept that a properly folded TSR is required for recognition by POFUT2. For these mutants, fucosylation could only be measured in cell lysates. Several other mutations had similar effects (S15L, 16S17 (insertion of an additional S in the Xb position), G19A/G21A, G19A/G20A/G21A, G19A/G20A/G21A/V22G). Some fucosylation could be detected on many of these mutants, suggesting that the TSR was folded sufficiently for recognition by POFUT2.
Two mutations in the Xa position, S15D and S15Q, enhanced secretion (Supplementary Fig. 14), but caused a significant reduction in fucosylation, suggesting that human POFUT2 is quite selective for amino acids in the Xa position (Fig. 3e and Supplementary Fig. 15). S15 side chain is not engaged in direct interactions with CePOFUT2, though its side chain is facing L221 (Fig. 3d). Thus, the significant reduction in activity by aspartic acid or glutamine can be rationalized by the proximity of a negatively charged or polar amino acid to a nonpolar residue, which might lead to a decrease of the binding of TSR on POFUT2. In contrast, Xc, Xd and Xe positions were highly permissive for bulky amino acids (V16H, G19Q, G19R, G20H) for both secretion and fucosylation (Fig. 3e, Supplementary Fig. 14 and 15). Xc (T16 in HsTSR1, V16 in HsTSR3) can be explained because in the crystal structure T16 side chain is facing one of the water molecules network that are also surrounded by N49 and E218 (Fig. 3c). In the crystal structure, G19, N20 and G21 that correspond to Xd, Xe and the glycine in the consensus sequence, respectively, are residues exposed to the solvent or surrounded by the network of water molecules, explaining why single mutations on these residues have a little effect upon catalysis (Fig. 3c, d). Interestingly, the conserved glycine (G21) was not essential for secretion or fucosylation (Fig. 3e, Supplementary Fig. 14 and 15). Our data also indicate that the tryptophan immediately upstream of the consensus sequence is not required for fucosylation (Fig. 3e), suggesting the requirement of tryptophans may vary with different TSRs. In the crystal structure, W11 side chain is tethered by R25 and R44 side chains and also facing the bulk solvent, explaining why this residue is mainly important for the stability of the TSRs (Fig. 3c). On this basis, we propose a simplified consensus sequence for POFUT2 modification, CXaXb(S/T)C within a TSR, where certain amino acids are excluded from Xa (e.g. aspartic acid, leucine).
Water-mediated interactions
The limited number of direct interactions in the complex is likely insufficient to maintain the complex and cannot explain the plasticity required for POFUT2 to achieve recognition on such a diverse family of TSRs. As mentioned above, the protein-protein interface is mostly embedded by solvent, which provides ~50% of the interactions in the complex through an intricate network of water molecules (Fig. 3a, c, and Supplementary Table 6).
Most of the hydrogen-bonding interactions between the HsTSR1 and water molecules occur through the backbone of non-conserved residues. This is an important feature likely required for the plasticity to recognize such dissimilar TSRs. In contrast, CePOFUT2 residues interact with the bridging water molecules through mainly conserved side chains and to a lesser extent with the backbone of non-conserved residues, assuring the catalytic efficiency across different orthologues (Fig. 3c and Supplementary Table 5).
The existence of both direct and water-mediated stabilizing interactions agree with the favorable enthalpy value measured for this system (Online methods and Supplementary Table 1); the formation of the complex is in turn entropically unfavorable due to the persistence of ordered inter-protein solvent molecules that are not released to the water-bulk, and to the loss of conformational degrees of freedom upon binding (Supplementary Table 1 and Supplementary Fig. 16). The crucial role that water molecules play in stabilizing protein-protein contacts has been recently found in an antifreeze protein20.
Macromolecular crowding is known to cause osmotically-induced protein dehydration21 and to affect water-mediated protein interfaces22. To assess the relevance of the interstitial water molecules found in our crystallographic structure, we compared the effect of adding 15% and 20% PEG 400 on the Kd of the protein complex and found an increase of ~8 and ~60 fold, respectively (Online methods and Supplementary Table 1). This result suggests that PEG-induced water exclusion from the protein complex interface might reduce significantly the affinity between the two proteins, supporting the important role of water in the interaction between both POFUT2 and TSR. The secondary structure elements of the complex in the absence or presence of 20% PEG 400 did not change substantially, suggesting that the integrity of the complex was maintained (Supplementary Fig. 17).
The role of the interstitial water in the complex interface was examined through 0.5 μs MD simulations performed on HsTSR1 in complex with the enzyme and in the absence of the flexible linker (see Online methods). Exploratory MD simulations in the absence of explicit solvent but maintaining the crystallographic waters disrupted the binding of HsTSR1 and CePOFUT2, indicating the crucial role of discrete water molecules in the molecular recognition process. Conversely, fully solvated complexes between CePOFUT2 and HsTSR1 (wild-type and S17T/A mutants, both in the presence of GDP or GDP-fucose), maintained their associated structure during the total simulation time (0.5 μs). Remarkably, the first solvation shell around the bound TSR is preserved upon binding to CePOFUT2 in the MD simulations, as shown by the similar number of water molecules within 3.4 Å of the whole peptide and the residues located at the complex interface (Supplementary Table 6). The solvation patterns of HsTSR1 and S17T/A mutants are very similar in both the free and bound states. A detailed analysis of hydration around the acceptor residues revealed that S17 is, as expected, more solvated than T17 and A17; the water molecules in the first solvation of the CePOFUT2-GDP-fucose-HsTSR1 complexes was calculated to be 9 ± 2, 6 ± 1 and 5 ± 2 for Ser, Thr and Ala, respectively. The poorer solvation of T17 in the complex might increase the reactivity of its more hindered hydroxyl group compared to that of S17. This might explain, in combination with the higher affinity of CePOFUT2 toward threonine mentioned above (Fig. 2c), the higher rate of the fucosylation reaction measured for Thr-containing TSRs.
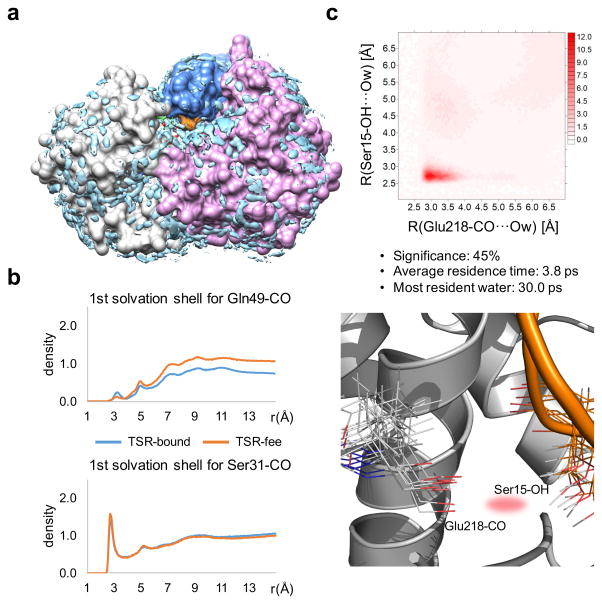
We then calculated the spatial distribution functions for water around HsTSR123. Whereas the isolated HsTSR1 presents marginal structured water molecules, a significant density of structured water can be observed for its complex with CePOFUT2 through all the simulation time (Fig. 4a). The radial distribution functions (rdf) for different atoms of HsTSR1 in both the free and bound states were also derived from the simulations. In general, for residues that are exposed to the solvent, such as the carboxylate group of D30 and the hydroxyl group of S31, these functions display a typical hydrophilic interaction with a well-defined first hydration shell showing a density peak at 2.8 Å and a second hydration shell around 4.7 Å24 (Fig. 4b). In contrast, those atoms involved in highly-populated hydrogen bonds, such as the carbonyl group of Q49, show a negligible water density in the first hydration shell. The existence of structured water molecules between CePOFUT2 and HsTSR1 was analyzed through normalized two-dimensional rdf functions (2D-rdf); these functions calculate the probability of finding a water molecule close to two selected atoms, in comparison to the one obtained in the bulk25, and quantify the magnitude of the localized water density (see Fig. 4c and Supplementary Fig. 18). The 2D-rdf functions were calculated for selected pairs of heteroatoms involved in contacts with crystallographic waters. The calculated pairwise values of shared water density were high (>8) in all cases, demonstrating the presence of persistent water pockets and the active role of solvent in binding. The highest shared water density (~32) was obtained between the carbonyl group of E218 and the hydroxyl group of S15. The average distance between these two atoms was 4.6 Å, ranging from 2.7 to 5.9 Å. This indicates that although these bridging water molecules are highly structured, they are also very dynamic, continuously changing their positions and exchanging with the bulk. The average residence time for a water molecule implicated in these pockets is 3.8 ps, and the most persistent bridging molecule has a residence time of 30 ps. This notion of dynamic structured water is also reflected by the time evolution of the 2D-rdf. As shown in Supplementary Fig. 19, the density peak between two selected atoms gradually changes in intensity and position, leading to a new water pocket involving a different atom nearby.
Figure 4. Hydration structure and dynamics of CePOFUT2-GDP-fucose-HsTSR1 supports water-mediated binding.
a, Water oxygen density over 0.5 μs calculated for the ternary complex through MD simulations. The two domains of CePOFUT2-GDP are shown in grey and pink, respectively, HsTSR1 in blue and GDP-fucose as green sticks. Reactive S17 is highlighted in orange. Regions with water density greater than 1.5 times the density of the bulk are represented as blue isosurfaces. b, Examples of 1D-rdf functions calculated for buried (upper panel) and solvent-exposed (lower panel) atoms of HsTSR1 in the isolated (in orange) and bound (in blue) states. c, (Upper panel) 2D-rdf function calculated for two selected atoms of the protein-protein interface hydrogen bonded to a crystallographic water, together with its significance (population in the MD ensemble) and average residence time. The atoms are labeled according to AMBER force field nomenclature. (Lower panel) Ensembles for the ternary complex obtained through MD simulations for the residues involved in the water pocket. One conformation of the rest of the residues is shown for clarity.
All these features stress the crucial role that solvent plays in the molecular recognition process of TSR. The hydration water could make the binding surface highly adaptable and thus somewhat promiscuous26. The lack of specificity in the complex is likely due to the large number of water-mediated interactions. The dynamic character of the water molecules of the first hydration shell plays a pivotal role in the recognition event, allowing water molecules to adopt different positions for singular substrates, acting as an extension of the protein surface.
DISCUSSION
The results described herein provide the molecular basis for the promiscuous behavior of POFUT2 acting on folded TSRs with different primary sequences, which might apply to other enzymes that modify a wide variety of peptide sequences. Our data support a general recognition mechanism in which water participates actively in the protein association. The presence of dynamically structured water allows the binding of different TSRs to POFUT2 in a sequence-independent manner that overrides the need for multiple direct protein-protein interactions. This shows a striking example of how nature has produced multispecific enzymes that overcome sequence recognition by harnessing nonspecific protein-protein interactions mediated by water molecules.
ONLINE METHODS
Cloning, expression and purification of HsTSR1 and HsTSR1-2-3
The DNA sequence encoding amino acid residues 378–550 of human thrombospondin 1, defined as HsTSR1-2-3, was made synthetically and codon optimized by GenScript to be expressed in E. coli. The DNA, containing at the 5′ end a recognition sequence for EcoRI and a PreScission protease cleavage site, and at the 3′ end a sequence for SalI, was cloned into the pUC57 vector (GenScript). Following digestion with EcoRI and SalI the construct was subcloned into the protein expression vector pMALC2x (NEB), resulting in the expression plasmid pMALC2xHsTSR1-2-3 (A378-G550). The plasmid pMALC2xHsTSR1-2-3 was used as a template for introducing a stop codon by site-directed mutagenesis after the triplet encoding D429 in order to only express HsTSR1. The resulting plasmid was named as pMALC2xHsTSR1. Site-directed mutagenesis was carried out following the ‘QuikChange’ kit protocol (Stratagene), using the Phusion High-Fidelity DNA polymerase (Thermo Scientific). To facilitate the purification of HsTSR1-2-3, we introduced a histidine tag preceding the PreScission protease cleavage site. HsTSR1-2-3 was amplified from pMALC2xHsTSR1-2-3 using the forward primer,
5′-CTGGAAGTTCTGTTCCAGGGGCCCGGTAGCGGTAGCGCAGACGATGGTTGG-3′
containing a sequence encoding a PreScission protease cleavage site (italic), and the reverse primer
5′-CGGAATTCGTCGACTCAGCCATCAATCGGGCAATC-3′
containing a SalI site (italic) and a stop codon (underlined). HsTSR1-2-3 was amplified from the first PCR product and using the forward primer,
5′-CGGAATTCGAATTCAGCGGCGGCCATCACCATCACCATCACCATCACGGCGGCAGCCTGGAAGTTCTGTTCCAGGGGCCC-3′
containing a EcoRI site (underlined), a sequence encoding a histidine tag (bold) and a PreScission protease cleavage site (italic), and the reverse primer used above.
Subsequently the PCR product was digested with EcoRI and SalI and cloned into pMAC2x, resulting in the expression plasmid pMALC2xHistHsTSR1-2-3. All plasmids were verified by sequencing (Sistemas Genómicos; www.sistemasgenomicos.com).
Both pMALC2xHsTSR1 and pMALC2xHistHsTSR1-2-3 were transformed into Rosettagami 2 (DE3). Cells were grown and disrupted by sonication as described before27.
The supernatant containing the maltose binding protein (MBP) bound to HsTSR1 was loaded into 1 × 5 ml MBP Trap HP column (GE Healthcare) previously equilibrated with buffer A (25 mM Tris/HCl, 150 mM NaCl pH 7.5). The fusion protein was eluted with buffer A + 10 mM maltose, dialyzed with buffer A, and subsequently was cleaved overnight with PreScission protease (PP; GE Healthcare) at 4ºC. HsTSR1 was further purified by size exclusion chromatography using a Superdex 75 XK26/60 column (SIGMA) previously equilibrated with buffer A. The eluted protein was dialyzed against buffer B (25 mM Tris/HCl pH 7.5), concentrated and used for biophysical experiments.
The supernatant containing the MBP-HsTSR1-2-3 was loaded into 1 × 5 ml His Trap HP column (GE Healthcare) previously equilibrated in buffer C (25 mM Tris/HCl, 150 mM NaCl, 20 mM imidazole pH 7.5). The fusion protein was eluted with a gradient of imidazole from 20 to 200 mM. Then, buffer exchange of the fusion protein into buffer A was carried out using a HiPrep 26/10 desalting column, and the fusion protein was subsequently cleaved overnight with PP at 4ºC. The PP and the maltose binding protein were removed by a 1 × 5 ml GSTrap (GE Healthcare) and MBP Trap HP columns, respectively. The unbound HsTSR1-2-3 was further purified by size exclusion chromatography as described above. The eluted protein was dialyzed against buffer B, concentrated and used for biophysical experiments.
Cloning, expression and purification of CePOFUT2
The DNA sequence encoding amino acid residues 40–424 of the Caenorhabditis elegans POFUT2 (CePOFUT2), defined as cepofut2, was made synthetically and codon optimized by GenSript for expression in Pichia pastoris. The DNA, containing at the 5′ end a recognition sequence for XhoI and a KEX2 cleavage signal, and at the 3′ end a sequence for SacII, was cloned into the pUC57 vector (GenScript). Following digestion with XhoI and SacII the construct was subcloned into the protein expression vector pPICZαA, resulting in the expression plasmid pPICZαAcepofut2. Site-directed mutagenesis was carried out following the ‘QuikChange’ kit protocol (Stratagene), using the KOD HotStart DNA polymerase (Novagene). The plasmid pPICZαAcepofut2 was used as a template for introducing the following single and double amino acid changes by site-directed mutagenesis: R299D, R298K-R299K and R299K-A300V. The plasmid pPICZαAcepofut2-R298K-R299K that encodes the double mutant R298K-R299K will be named as CePOFUT2 along the manuscript.
The DNA sequence encoding the fusion protein formed by amino acid residues 40–424 of the CePOFUT2 (this construct contains two triplets encoding for the double mutant R298K-R299K), the flexible linker (formed by a combination of 22 glycine and serine residues) and HsTSR1 (Fig. 1c) was made synthetically and codon optimized by GenScript for expression in Pichia pastoris. The DNA, containing at the 5′ end a recognition sequence for XhoI and a KEX2 cleavage signal, and at the 3′ end a sequence for SacII, was cloned into the pUC57 vector (GenScript). Following digestion with XhoI and SacII the construct was subcloned into the protein expression vector pPICZαA (Invitrogen), resulting in the expression plasmid pPICZαAcepofut2-linker-HsTSR1. The plasmid pPICZαAcepofut2-linker-HsTSR1 was used as a template for introducing the mutant S17A in the HsTSR1 by site-directed mutagenesis. All plasmids were verified by sequencing (Sistemas Genómicos).
All plasmids were isolated from E. coli strain DH5α, linearized with SacI and used to transform the Pichia pastoris strain X-33 by electroporation. Transformants were selected and cells were grown as described before12. Supernatant containing CePOFUT2 was dialyzed against buffer D (25 mM Tris/HCl pH 8.5) and loaded into 1 × 5 ml HiTrap Blue column. The protein was eluted with a NaCl gradient from 0 to 1 M. Supernatant containing the fusion protein was dialyzed against buffer D as well and loaded into 1 × 5 ml HiTrap QFF column. The protein was also eluted with a NaCl gradient from 0 to 1 M. Buffer exchange of both CePOFUT2 and the fusion protein into buffer E (25 mM MES pH 6.7) was carried out using a desalting column as described above. Then, both proteins were treated with Endo Hf (NEB) overnight at 18ºC. Samples were loaded into 1 × 5 ml MBPTrap HP column where both CePOFUT2 and the fusion protein were collected in the flow through. Finally, both proteins were further purified by size exclusion chromatography as described above. The eluted proteins were dialyzed against buffer B, concentrated and used for biophysical experiments.
For the formation of the different complexes, a 5:1 ratio of either HsTSR1 or fucosylated HsTSR1 versus CePOFUT2, and 3:1 ratio of HsTSR1-2-3 versus CePOFUT2 were purified by size exclusion chromatography. In these experiments, the column was previously equilibrated with buffer F (25 mM Tris/HCl, 10 mM NaCl, 50 μM GDP pH 7.5). Buffer exchange of the eluted complexes into buffer F without NaCl was carried out, and then complexes of HsTSR1/CePOFUT2 and HsTSR1-2-3/CePOFUT2 were concentrated and used for biophysical experiments.
Crystallization
Crystals of the fusion protein in complex with GDP were obtained by mixing 0.5 μl of protein solution (a mix formed by 20 mg/ml of the fusion protein and 5 mM GDP in buffer C) with 0.5 μl of precipitant solution (16–20% PEG 3000, 100 mM sodium citrate pH 5.5) against 60 μl of precipitant solution. The crystals were obtained by sitting drop vapour diffusion at 18ºC. The crystals were cryoprotected in the precipitant solution plus 20% etilenglicol and frozen in a nitrogen gas stream cooled to 100 K.
Structure determination and refinement
The data was collected in the beamline I02 of DLS at a wavelength and temperature of 0.97 Å and a temperature of 100 K, respectively. The data was processed and scaled using the XDS package28 and CCP429,30 software. Relevant statistics are given in Supplementary Table 2. The crystal structure was solved by molecular replacement with Phaser29,30 and using the PDB entry 4AP5 as the template that corresponds to the human POFUT2. Initial phases were further improved by cycles of manual model building in Coot31 and refinement with REFMAC532. After CePOFUT2 was built and refined, ARP/wARP29,30 was used to build the HsTSR1. The final model was validated with PROCHECK, model statistics are given in Supplementary Table 2. The asymmetric units of the primitive monoclinic crystals contain 2 molecules of the ternary complex in a stoichiometry of 1:1:1 (Supplementary Fig. 9). The Ramachandran plot shows that 95.19%, 3.76% and 1.06% of the amino acids are in most favored, allowed and disallowed regions, respectively.
Spectroscopic characterization by Far-UV circular dichroism (CD)
CD spectra were acquired at 25ºC in a Chirascan spectropolarimeter (Applied-Photophysics). Far-UV CD spectra were recorded from 190 to 250 nm using a 1-mm path-length cuvette. The experiments consisted of the fusion protein at 15 μM in 50 mM HEPES pH 7.5 in the presence and absence of 20% PEG 400.
Radioactive assay for POFUT2 activity
POFUT2 assays were performed as previously described33. Briefly, HsTSR3 from thrombospondin 1 (100 μM final, purified from BL21 cells) and GDP-[3H]fucose (10 μM final, 19.8 Ci/mmol, American Radiolabeled Chemicals) was incubated with purified CePOFUT2 or HsPOFUT2 (11.2 ng) in 50 mM HEPES, 10 mM MnCl2 pH 6.8 for 20 min at 37°C. The reaction was stopped by adding 1000 reaction volumes of 0.1 M EDTA. TSRs were then purified from the mixture using a C18 cartridge (100 mg, Agilent Technologies) and radioactivity was measured on a Beckman scintillation unit.
Fucosylation of HsTSR1
HsTSR1 was fucosylated in the presence of CePOFUT2 and GDP-fucose. The reaction was carried out in 25 mM Tris/HCl, 150 mM NaCl pH 7.5 at 18ºC overnight. HsTSR1 was in 5-fold excess compared to the enzyme whereas GDP-fucose was added at 700 μM. The fucosylated HsTSR1 sample was then loaded into 1 × 5 ml HiTrap QFF column (GE Healthcare), previously equilibrated with 25 mM BIS-TRIS pH 6.5. Fucosylated HsTSR1 was collected in the flow through, concentrated, quantified and used in both ITC and size exclusion chromatography experiments.
Isothermal titration microcalorimetry (ITC)
ITC was used to characterize the interaction of CePOFUT2 with GDP, GDP-fucose, HsTSR1 and fucosylated HsTSR1. All experiments were carried out in an Auto-iTC200 (Microcal, GE Healthcare) at 25ºC with CePOFUT2 at 50 μM and concentrations of ligands at 300 μM, in both 50 mM HEPES and 50 mM PIPES pH 7.0. To determine how PEG 400 might affect the Kd of HsTSR1, we used the same conditions as above but using 15% and 20% PEG 400 in buffer HEPES pH 7.0.
The buffer independent binding enthalpy ΔH0 was estimated from linear regression using different ionization enthalpies ΔHion for each buffer (ΔHion (HEPES) = 4.88 kcal/mol and ΔHion (PIPES) = 2.74 kcal/mol)27. Data integration, correction and analysis were carried out in Origin 7 (Microcal). The data was fitted to a one-site equilibrium-binding model.
Atomic Force Microscopy (AFM)
AFM has enabled the direct visualization of single CePOFUT2 molecules to characterize the different conformational states upon ligand binding. Methods and instrumentation were as reported34. 5× GDP, GDP-fucose, HsTSR1 and MnCl2 were added to CePOFUT2. AFM images were analyzed with the WSxM software35 as described36. Though CePOFUT2 presents a slightly net negative charge at neutral pH, the enzyme was adsorbed electrostatically on the negative mica surface due to the presence of Mn2+ that inverts its polarity. Adsorption of enzymes on mica was evaluated and found they preserve the enzymatic activity, even later desorbed37. CePOFUT2 appears mainly as monomer, but the images allowed the identification of several distinguishable conformational states and compaction degree. CePOFUT2 heights vary from 3 to 7 nm due to the charged ligands, Mn2+, GDP and/or GDP-fucose, attract the AFM tip making a “flatten” effect on the molecules; while the addition of HsTSR1 makes the features their overall volume increase between 7 and 9 nm (Supplementary Fig. 10). Z-height has sub-nm resolution, while in the X–Y plane the scanned features suffer the AFM tip broadening effect that does not affect the comparative analysis of the width related to the size due to proportionality. When incubating only with Mn2+, the molecules exhibited clearly the two domains in a separate disposition, composing an open or extended structure (Supplementary Fig. 10). Though, negative GDP-fucose binds strongly to the substrate hindering visualization of enzyme molecules, the analysis was also possible. Binding to GDP or GDP-fucose introduces a conformational change, and the structure increases the volume of one of the modules, making the two domains to approach (Supplementary Fig. 10). Simultaneous binding of GDP and HsTSR1 leads to a global reorganization of the structure, generating a feature of a higher volume, where the modules cannot be distinguished, composing a closed conformation (Supplementary Fig. 10).
Fucosylation of the fusion protein CePOFUT2-HsTSR1
The fusion protein CePOFUT2-HsTSR1 (WT or S17A mutant, 0.11 mg/ml) was incubated in the presence of radioactive 2.5 μM GDP-[1-3H]fucose (19.8 Ci/mmol, American Radiolabeled Chemicals). The reaction was carried out in 10 μl of 50 mM HEPES pH 6.8, 10 mM MnCl2, and 0.5% Nonidet P-40 at 37ºC for 20 min and stopped by adding 900 μl of 100 mM EDTA pH 8.0. The sample was loaded onto a C18 cartridge (100 mg, Agilent Technologies). After the cartridge was washed with 5 ml of H2O, the bound fusion protein was eluted with 1 ml of 80% methanol. Incorporation of [3H]fucose into the fusion proteins was determined by scintillation counting of the eluate. Reactions without the fusion proteins were used as background control. Data are from three independent assays. The values indicate mean ± S.E.M.
Fucosylation of HsTSR1-2-3 to test processivity of CePOFUT2
HsTSR1-2-3 (10 μM) was fucosylated in the presence of 1.5 μg of CePOFUT2 and GDP-fucose at the concentration of 200, 20, or 2 μM. The reaction was carried out in 30 μl of 50 mM HEPES pH 6.8, 10 mM MnCl2 at 37ºC overnight. An aliquot of the products were analyzed by LC-MS using an Agilent 6340 ion-trap mass spectrometer with a nano-HPLC CHIP-Cube interface as previously described38. Extracted ion chromatograms for the most abundant charge state of the unmodified form, or mono-, di-, or tri-fucosylated forms of HsTSR1-2-3 were generated. The remainder of the products were purified by reverse phase high performance liquid chromatography (Agilent Technologies, 1200 Series) equipped with a C18 column (4.6 mm × 150 mm, VYDAC) with a linear gradient of solvent B (80% acetonitrile, 0.1% trifluoroacetic acid (TFA) in water) from 10% to 90% in solvent A (0.1% TFA in water) for 30 min, monitoring absorbance at 214 nm. The lyophilized samples were reduced, alkylated, digested with trypsin, and analyzed by nano-liquid chromatography/tandem mass spectrometry (nano-LC-MS/MS) using an Agilent 6340 ion-trap mass spectrometer with a nano-HPLC CHIP-Cube interface auto sampler as reported previously38.
Analysis of secretion and fucosylation of HsTSR3 mutants using cell-based assays
Mutations in the variable X positions of the O-fucose consensus sequence (Supplementary Table 4) were generated by site-directed mutagenesis using the pSecTag-HsTSR3 (with C-terminal Myc-His6 tag) and the primers listed in Supplementary Table 3. Plasmids encoding wild type and mutant forms of HsTSR3 were co-expressed with hIgG controls in HEK293T cells by transient transfection. Briefly, 6 μL of polyethylenimine (PEI) was added to 0.8 μg pSecTag-HsTSR3 and 0.2 μg pRK5-hIgG in 100 μL 150 μM sodium chloride. Transfection mixtures were vortexed, incubated 20 minutes at room temperature, then added dropwise onto 80% confluent HEK293T cells in 1 mL DMEM in 32 mm dishes. Four hours post-transfection cells were washed with PBS, and then incubated in 1 mL OptiMEM containing 200 μM peracetylated alkynylfucose (Invitrogen) for 72 hours as previously described39. Media was collected for analysis and cells lysed in RIPA buffer (0.1% sodium dodecyl sulfate, 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% sodium doxycholate, 1% Nonidet P-40). Samples were then analyzed for protein production and secretion by Western blot with anti-Myc antibodies. Fucosylation was analyzed following cycloaddition “click” reaction of fucosylated proteins with azido-biotin as previously described39. Briefly, media or lysates containing alkynylfucose-tagged glycoproteins were incubated with 1 mM copper sulfate and 2 mM sodium ascorbate in PBS with 0.1 mM Azido-Biotin and 0.1 mM tris-(benzyltriazolylmethyl)amine for one hour at room temperature. Clicked samples were centrifuged for 5 minute at 14,000 rpm prior to analysis by immunoblot using IRDye 800 labeled streptavidin.
For Western blotting, proteins were separated by 15% SDS-PAGE then transferred to 0.45 micron nitrocellulose membrane (BioRad). Blots were blocked 1 hour at room temperature in 5% milk in Phosphate-buffered saline, 0.1% Tween (PBST) then probed with 1:2000 anti-Myc antibody in PBST. Samples containing unclicked protein were incubated with Alexa Fluor 680 Goat Anti-Mouse IgG (Invitrogen) and IRDye800 Anti-Human IgG (Invitrogen), both 1:2000 in PBST, for one hour in the dark at room temperature. Clicked samples were incubated with 1:2000 Alexa Fluor 680 Goat Anti-Moues (Invitrogen) and 1:20000 streptavidin IRDye800 (Rockland Immunochemicals) in PBST for one hour at room temperature in the dark. Western blots were visualized using an Odyssey Imager (LI-COR). Intensities of bands were quantified from captured images using Odyssey Software. Relative mutant secretion was determined by calculating the ratio of Myc signal to that of the IgG control in the media, with wild type protein normalized to 1. Relative fucosylation was calculated from captured images by dividing the intensity of streptavidin by the intensity of Myc signal, and expressing as a percentage of wild type ratio. Standard error of the mean was calculated for each sample mean comparing the mutant streptavidin to Myc ratio to wild type ratio.
Molecular dynamics simulations
Parameters for the substrates were generated with the antechamber module of Amber1440 using a combination of GLYCAM0641 parameters for the fucose unit and the general Amber force field (GAFF) for GDP, with partial charges set to fit the electrostatic potential generated with HF/6-31G(d) by RESP42. The charges are calculated according to the Merz-Singh-Kollman scheme using Gaussian 0943. Each protein was immersed in a truncated octahedral box with a 10 Å buffer of TIP3P44 water molecules and neutralized by adding explicit counter ions (Na+Cl−). All subsequent simulations were performed using the ff14SB force field45. A two-stage geometry optimization approach was performed. The first stage minimizes only the positions of solvent molecules and ions, and the second stage is an unrestrained minimization of all the atoms in the simulation cell. The systems were then gently heated by incrementing the temperature from 0 to 300 K under a constant pressure of 1 atm and periodic boundary conditions. Harmonic restraints of 30 kcal/mol were applied to the solute, and the Andersen temperature coupling scheme46 was used to control and equalize the temperature. The time step was kept at 1 fs during the heating stages. Water molecules are treated with the SHAKE algorithm such that the angle between the hydrogen atoms is kept fixed. Long-range electrostatic effects are modeled using the particle-mesh-Ewald method47. An 8 Å cutoff was applied to Lennard-Jones and electrostatic interactions. Each system was equilibrated for 2 ns with a 2 fs timestep at a constant volume and temperature of 300 K. Production trajectories were then run for additional 0.5 μs under the same simulation conditions. Water density properties were derived from the production trajectories using a cubic grid consisting of 160 × 160 × 160 bins with 0.5 Å spacing, through the grid command available in the cpptraj module of AmberTools1440. Minimizations in explicit solvent for the modeled complex (Supplementary Fig. 11) was performed running 2000 steepest-descent steps followed by 2000 conjugate gradient optimization steps using the AMBER force field. MD simulations were performed on CePOFUT2 in complex with HsTSR1. Two additional calculations were run with mutants of HsTSR1 (S17T, S17A). The simulations were duplicated to study the influence of GDP or GDP-fucose. In all cases, the MD calculations were run without the linker between the protein and the corresponding TSR. In addition, MD simulations on HsTSR1 ligand in the free state were run.
Volume maps of the water density were obtained using UCSF Chimera48. The radial distribution functions (RDF) of water (O atom) for selected protein atoms were measured using the radial command in cpptraj. 2D-RDF functions and the residence times were calculated using in-house programs developed in collaboration with Prof. S. B. Engelsen (University of Copenhagen)25.
PDB deposition
Coordinates and structure factors have been deposited in the Worldwide Protein Data Bank (wwPDB) with the pdb code 5foe.
Supplementary Material
Acknowledgments
We thank synchrotron radiation sources DLS (Oxford) and in particular beamline I02 (experiment number MX10121-2). We thank ARAID, MEC (BFU2010-19504, CTQ2013-44367-C2-2-P, CTQ2012-36365), NIH (GM061126 and CA123071) and the DGA (B89) for financial support, and BIFI (Memento cluster) for supercomputer support. The research leading to these results has also received funding from the FP7 (2007–2013) under BioStruct-X (grant agreement N°283570 and BIOSTRUCTX_5186). We also acknowledge Prof. S. B. Engelsen (University of Copenhagen) for providing the software to calculate 2D-rdf functions and residence times for water molecules.
Footnotes
Supplementary information includes Online methods, 19 Figures and 6 Tables.
Author contribution: R.H-G. designed the crystallisation construct, solved the crystal structure and refined the final model. J.V-G., E.L-N., C.H-R. and R.H-G. cloned the different constructs, purified the enzymes and crystallised the complex. R.H.-G. and J.V-G. performed the ITC experiments. G.J-O. and F.C. performed the molecular dynamics experiments. C.L-M., D.P, H.T. and R.S.H. cloned the different constructs for expression in mammalian cells, performed the site-directed mutagenesis, the analysis of the enzymatic studies (including the mutants in this work), the study of the non-processivity of CePOFUT2 and the studies in mammalian cells (both the secretion and the activity experiments). M.C.P. and A.L. performed the AFM studies. Y.R. and R.H-G. performed the multiple alignment of the TSRs and POFUT2s. R.H-G. wrote the article with the main contribution of H.T., R.S.H., F.C., A.L. and G.J-O. All authors read and approved the final manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Accession codes: Coordinates and structure factors have been deposited in the Worldwide Protein Data Bank (wwPDB) with the pdb codes 5foe (see Supplementary Table 2).
References
- 1.Hurtado-Guerrero R, Davies GJ. Recent structural and mechanistic insights into post-translational enzymatic glycosylation. Curr Opin Chem Biol. 2012;16:479–87. doi: 10.1016/j.cbpa.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–62. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luther KB, Haltiwanger RS. Role of unusual O-glycans in intercellular signaling. Int J Biochem Cell Biol. 2009;41:1011–24. doi: 10.1016/j.biocel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakaidani Y, et al. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat Commun. 2011;2:583. doi: 10.1038/ncomms1591. [DOI] [PubMed] [Google Scholar]
- 5.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasudevan D, Haltiwanger RS. Novel roles for O-linked glycans in protein folding. Glycoconj J. 2014;31:417–26. doi: 10.1007/s10719-014-9556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luca VC, et al. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347:847–53. doi: 10.1126/science.1261093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasudevan D, Takeuchi H, Johar SS, Majerus E, Haltiwanger RS. Peters plus syndrome mutations disrupt a noncanonical ER quality-control mechanism. Curr Biol. 2015;25:286–95. doi: 10.1016/j.cub.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi H, Haltiwanger RS. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun. 2014;453:235–42. doi: 10.1016/j.bbrc.2014.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor P, et al. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc Natl Acad Sci U S A. 2014;111:7290–5. doi: 10.1073/pnas.1319683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CI, et al. Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012;31:3183–97. doi: 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lira-Navarrete E, et al. Structural insights into the mechanism of protein O-fucosylation. PLoS One. 2011;6:e25365. doi: 10.1371/journal.pone.0025365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan C, Wu F, Jernigan RL, Dobbs D, Honavar V. Characterization of protein-protein interfaces. Protein J. 2008;27:59–70. doi: 10.1007/s10930-007-9108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan K, et al. Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol. 2002;159:373–82. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–55. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 16.Ricketts LM, Dlugosz M, Luther KB, Haltiwanger RS, Majerus EM. O-fucosylation is required for ADAMTS13 secretion. J Biol Chem. 2007;282:17014–23. doi: 10.1074/jbc.M700317200. [DOI] [PubMed] [Google Scholar]
- 17.Wang LW, et al. O-fucosylation of thrombospondin type 1 repeats in ADAMTS-like-1/punctin-1 regulates secretion: implications for the ADAMTS superfamily. J Biol Chem. 2007;282:17024–31. doi: 10.1074/jbc.M701065200. [DOI] [PubMed] [Google Scholar]
- 18.Lira-Navarrete E, et al. Substrate-guided front-face reaction revealed by combined structural snapshots and metadynamics for the polypeptide N-acetylgalactosaminyltransferase 2. Angew Chem Int Ed Engl. 2014;53:8206–10. doi: 10.1002/anie.201402781. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez de Peredo A, et al. C-mannosylation and o-fucosylation of thrombospondin type 1 repeats. Mol Cell Proteomics. 2002;1:11–8. doi: 10.1074/mcp.m100011-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Sun T, Lin FH, Campbell RL, Allingham JS, Davies PL. An antifreeze protein folds with an interior network of more than 400 semi-clathrate waters. Science. 2014;343:795–8. doi: 10.1126/science.1247407. [DOI] [PubMed] [Google Scholar]
- 21.Parsegian VA, Rand RP, Rau DC. Macromolecules and water: probing with osmotic stress. Methods Enzymol. 1995;259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- 22.Rajapaksha A, Stanley CB, Todd BA. Effects of macromolecular crowding on the structure of a protein complex: a small-angle scattering study of superoxide dismutase. Biophys J. 2015;108:967–74. doi: 10.1016/j.bpj.2014.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naidoo KJ, Kuttel M. Water structure about the dimer and hexamer repeat units of amylose from molecular dynamics computer simulations. Journal of Computational Chemistry. 2001;22:445–456. [Google Scholar]
- 24.Soper AK. The radial distribution functions of water and ice from 220 to 673 K and at pressures up to 400 MPa. Chemical Physics. 2000;258:121–137. [Google Scholar]
- 25.Andersson C, Engelsen SB. The mean hydration of carbohydrates as studied by normalized two-dimensional radial pair distributions. J Mol Graph Model. 1999;17:101–5. 131–3. doi: 10.1016/s1093-3263(99)00022-4. [DOI] [PubMed] [Google Scholar]
- 26.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 27.Sahun-Roncero M, et al. The mechanism of allosteric coupling in choline kinase alpha1 revealed by the action of a rationally designed inhibitor. Angew Chem Int Ed Engl. 2013;52:4582–6. doi: 10.1002/anie.201209660. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The CCP4 suite: programs for protein crystallography. Acta crystallographica. Section D, Biological crystallography. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y, Nita-Lazar A, Haltiwanger RS. Two distinct pathways for O-fucosylation of epidermal growth factor-like or thrombospondin type 1 repeats. J Biol Chem. 2006;281:9385–92. doi: 10.1074/jbc.M511974200. [DOI] [PubMed] [Google Scholar]
- 34.Lira-Navarrete E, et al. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat Commun. 2015;6:6937. doi: 10.1038/ncomms7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horcas I, et al. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum. 2007;78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 36.Lostao A, Peleato ML, Gomez-Moreno C, Fillat MF. Oligomerization properties of FurA from the cyanobacterium Anabaena sp. PCC 7120: direct visualization by in situ atomic force microscopy under different redox conditions. Biochim Biophys Acta. 2010;1804:1723–9. doi: 10.1016/j.bbapap.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Marcuello C, Arilla-Luna S, Medina M, Lostao A. Detection of a quaternary organization into dimer of trimers of Corynebacterium ammoniagenes FAD synthetase at the single-molecule level and at the in cell level. Biochim Biophys Acta. 2013;1834:665–76. doi: 10.1016/j.bbapap.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Leonhard-Melief C, Haltiwanger RS. O-fucosylation of thrombospondin type 1 repeats. Methods Enzymol. 2010;480:401–16. doi: 10.1016/S0076-6879(10)80018-7. [DOI] [PubMed] [Google Scholar]
- 39.Al-Shareffi E, et al. 6-alkynyl fucose is a bioorthogonal analog for O-fucosylation of epidermal growth factor-like repeats and thrombospondin type-1 repeats by protein O-fucosyltransferases 1 and 2. Glycobiology. 2013;23:188–98. doi: 10.1093/glycob/cws140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Case DA, et al. AMBER. Vol. 14. University of California; San Francisco: 2014. [Google Scholar]
- 41.Kirschner KN, et al. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. Journal of Computational Chemistry. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. The Journal of Physical Chemistry. 1993;97:10269–10280. [Google Scholar]
- 43.Frisch MJ, et al. Gaussian 09, Revision D.01. 2009. [Google Scholar]
- 44.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926. [Google Scholar]
- 45.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Structure, Function, and Bioinformatics. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrea TA, Swope WC, Andersen HC. The role of long ranged forces in determining the structure and properties of liquid water. Journal of Chemical Physics. 1983;79:4576–4584. [Google Scholar]
- 47.Darden T, York D, Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. The Journal of Chemical Physics. 1993;98:10089–10092. [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.