Abstract
Data in the biological, chemical, and clinical domains are accumulating at ever-increasing rates and have the potential to accelerate and inform drug development in new ways. Challenges and opportunities now lie in developing analytic tools to transform these often complex and heterogeneous data into testable hypotheses and actionable insights. This is the aim of computational pharmacology, which uses in silico techniques to better understand and predict how drugs affect biological systems, which can in turn improve clinical use, avoid unwanted side effects, and guide selection and development of better treatments. One exciting application of computational pharmacology is drug repurposing- finding new uses for existing drugs. Already yielding many promising candidates, this strategy has the potential to improve the efficiency of the drug development process and reach patient populations with previously unmet needs such as those with rare diseases. While current techniques in computational pharmacology and drug repurposing often focus on just a single data modality such as gene expression or drug-target interactions, we rationalize that methods such as matrix factorization that can integrate data within and across diverse data types have the potential to improve predictive performance and provide a fuller picture of a drug's pharmacological action.
Modern pharmaceutical research faces serious challenges1-4 with decreasing productivity in drug development and a persistent gap between the many therapeutic needs and the types of available treatments. The number of drugs approved per dollar spent on research and development is declining2, 4, with recent studies estimating 15 years and over $1 billion to bring a new drug to market5. This is partially due to high attrition rates; only 10% of compounds that make it to Phase II clinical trials are eventually approved6, with the majority of failures either resulting from safety concerns or poor efficacy7, 8. Amidst the declining productivity, there is also a pressing need to provide treatments for rare diseases. According to the National Organization for Rare Disorders9, there are roughly 7,000 rare diseases that, taken together, affect about 10% of the first-world population, and yet only a few percent of these diseases have any pharmacological treatments available10. With current research and development costs, developing de novo therapies for each of these rare diseases is infeasible. All of this taken together points to a need for innovative approaches, both for identifying new therapeutic opportunities, as well as improving our knowledge surrounding drug action and side effects of investigational compounds.
Against this backdrop, advances in genomics and computational methods present new opportunities in research and drug development. Data such as gene expression, drug-target interactions, protein networks, electronic health records, clinical trial reports, and drug adverse event reports are rapidly accumulating and becoming increasingly accessible and standardized11, 12. However, these data are often complex, high-dimensional, and noisy, presenting new challenges and opportunities to develop computational methods that can assimilate these data in order to accelerate drug discovery and generate novel insights surrounding drug mechanisms, side effects, and interactions.
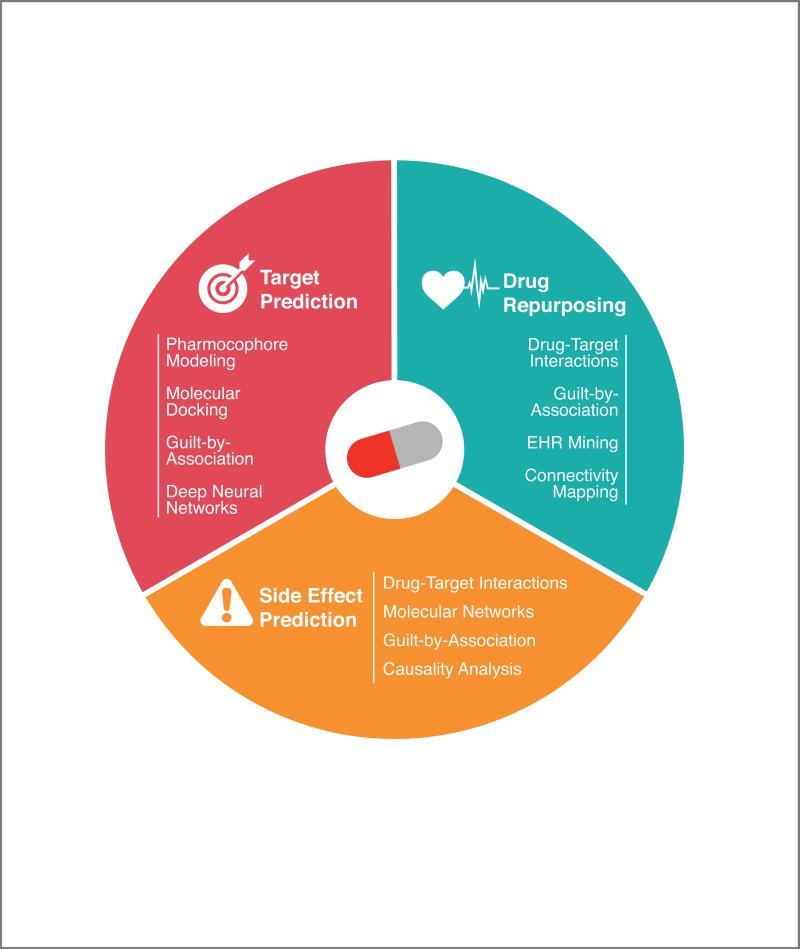
Computational pharmacology is the growing set of techniques aiming to address precisely the challenges above. In this review, we will cover three specific aims within the realm of computational pharmacology (see Figure 1). The first is the prediction of drug-target interactions, which are fundamental to the way that drugs work and often provide an important foundation for other aims in computational pharmacology. Next, we will discuss methods to predict or explain potential side effects or adverse drug reactions. This is important, as an improved understanding of off-target effects would result in fewer therapeutic failures due to unintended physiological responses. Third, we will discuss methods for drug repurposing, i.e. finding new uses for existing drugs.
Figure 1. A visual map of this article.
We can discover new associations between drugs and molecular targets, side effects, or diseases, using a variety of techniques. Some of the existing strategies reviewed in this article are listed in the three segments.
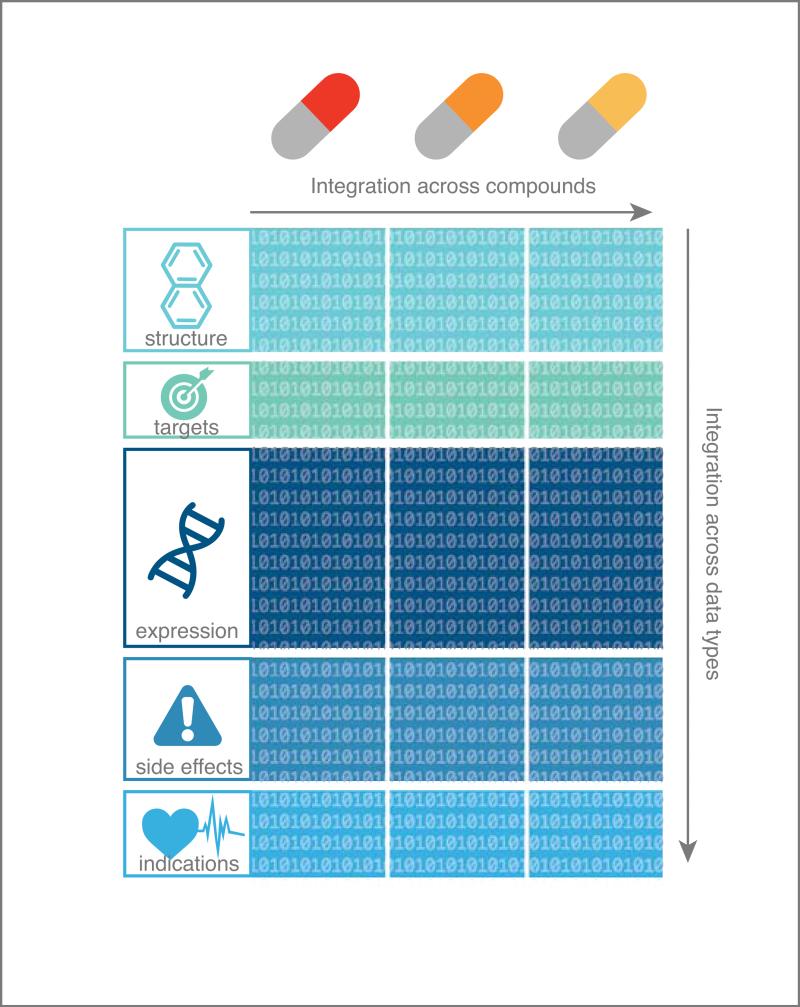
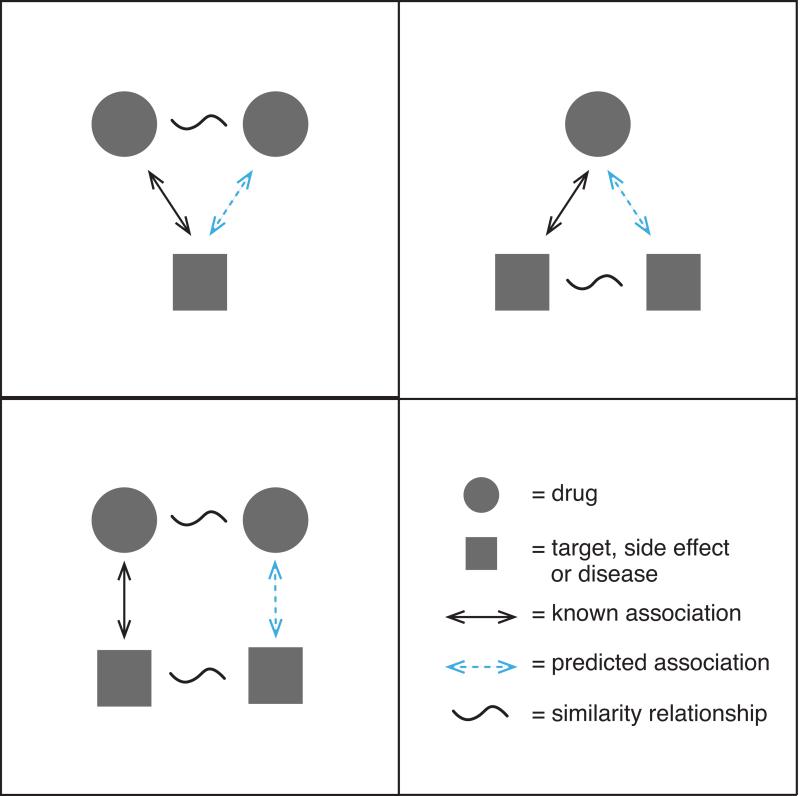
In this review, we discuss integrative or “multi-scale” methods in computational pharmacology that integrate across multiple data resources or data for many compounds (see Figure 2). Pharmacological space comprises a variety of data types, each one having its peculiarities and challenges. Integration of non-orthogonal dimensions of data can help to reduce noise and improve the predictive ability of high-dimensional data sets13-18. Systematic data integration across a single dimension can also enable new types of data inquiry; e.g., “What can information about one drug teach us about another drug?” Examples include similarity-based approaches (also sometimes called guilt-by-association) that evaluate if “similar” drugs could share common targets19-23, or have similar side effects24, or treat the same disease25-28 (see Figure 3). There are many different ways to define similarity and to make use of this idea, and herein we illustrate several examples.
Figure 2. Data can be integrated across compounds and/or across data types.
Note that this is a simplified illustration in the sense that both targets and gene expression responses to a compound can vary depending on the biological conditions in which they are assayed, e.g. different cell lines, dosages, etc.135
Figure 3. Various guilt-by-association strategies in computational pharmacology.
The top-left panel could be expressed by the statement “similar drugs may have common targets [or side effects or diseases]”; the top-right panel could be expressed as “similar targets may interact with the same drug” while the bottom-left panel expresses “similar drugs may interact with similar targets.”
We start by discussing how different aspects of pharmacological space can be measured and quantified, including a description of some important databases and resources. We then give an overview of three applications of computational pharmacology: predicting drug-target interactions, predicting and explaining side effects, and drug repurposing. We close with a discussion of data integration in computational pharmacology and some comments on future directions in the field.
QUANTIFYING AND REPRESENTING DRUG SPACE
The properties of a drug or drug-like compound and its interactions with the human body can be described or quantified in a variety of ways. We can quantify the physicochemical properties of a drug such as chemical structure, melting point, or hydrophobicity. We can quantify interactions between compounds and biological targets using measures of binding and kinetic activities. We can quantify downstream biological perturbations by measuring changes in cellular state or gene expression. We can also represent drugs using categorical metadata, such as diseases and conditions for which use of a drug is indicated, side effects, or known physiological interactions with other drugs. Such quantities and metadata lend themselves to numerical representations, which can then be analyzed to find patterns and relationships between compounds and generate new hypotheses.
Chemical structure
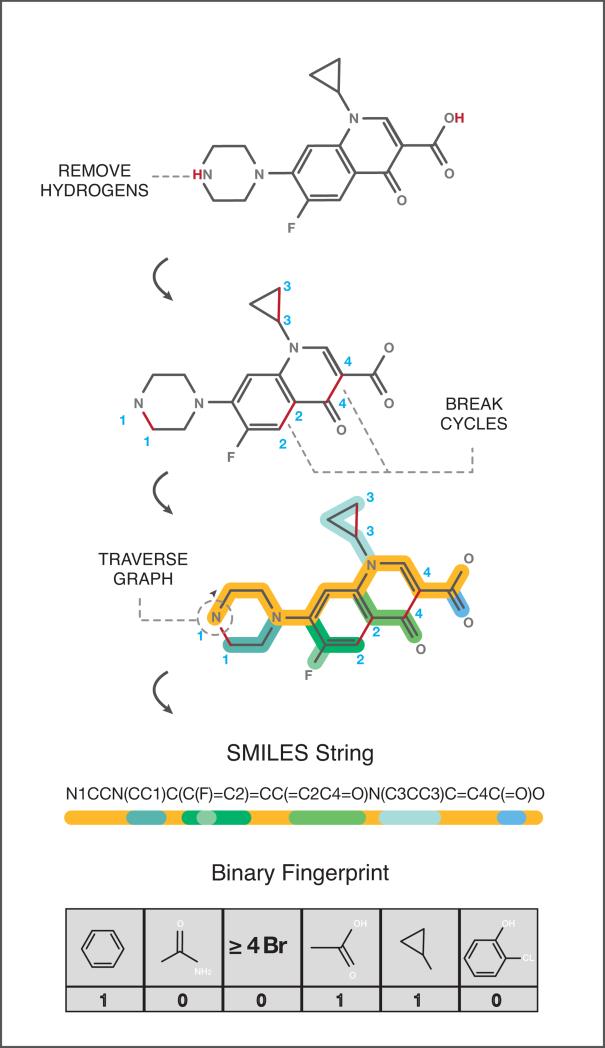
There are several different approaches to represent the chemical structure of small molecule compounds. The three-dimensional geometry of atoms and their electronic structure can be used in simulation-based analyses such as molecular docking. Alternatively, the structure can be codified into a character string or line notation such as SMILES29, which is obtained by printing the atomic symbols during a depth-first tree traversal of the chemical graph (See Figure 4A) or the more recently introduced InChI30 string (pronounced in-chee), which encodes various layers of information such as atoms, bonds, electronic charge, and tautomers. While SMILES is generally considered more human-readable, InChI can capture more information and in contrast to SMILES, is unique, making database mapping easier31. While these character string representations can be analyzed algorithmically, they are variable-length and non-numeric, which can be difficult to work with. To address this, fixed-length binary fingerprints32, 33 have been developed (see Figure 4A), where each bit might correspond to the presence or absence of a particular atom, moiety, aromatic ring, etc. Distance between two chemical structures can then be quantified easily, e.g. using the Tanimoto coefficient (Tc), which is the Jaccard similarity (|A∧B|/|A∨B|) of the two fingerprints. PubChem34 and ChEMBL35 are both widely used databases of chemical compounds containing chemical structure as well as many other properties, with information on over 60 million and 1 million compounds, respectively.
Figure 4. Quantifying and representing drug space.
4A: Representing chemical structure. A two-dimensional representation of chemical structure can be processed into line notation such as the SMILES string, or into a binary fingerprint, as shown in the figure. To construct the SMILES string, first hydrogens are removed and any cycles are broken by removing one edge from each cycle. The SMILES string is then generated by printing the node symbols during a depth-first tree traversal of the chemical graph, and using parentheses to denote branches of the tree. In the example, the gold-colored path represents the main backbone for traversal. A binary fingerprint can be generated by pre-defining chemical features such as the ones shown, and then using a ‘1’ or ‘0’ to indicate the presence or absence of each feature in the chemical structure.
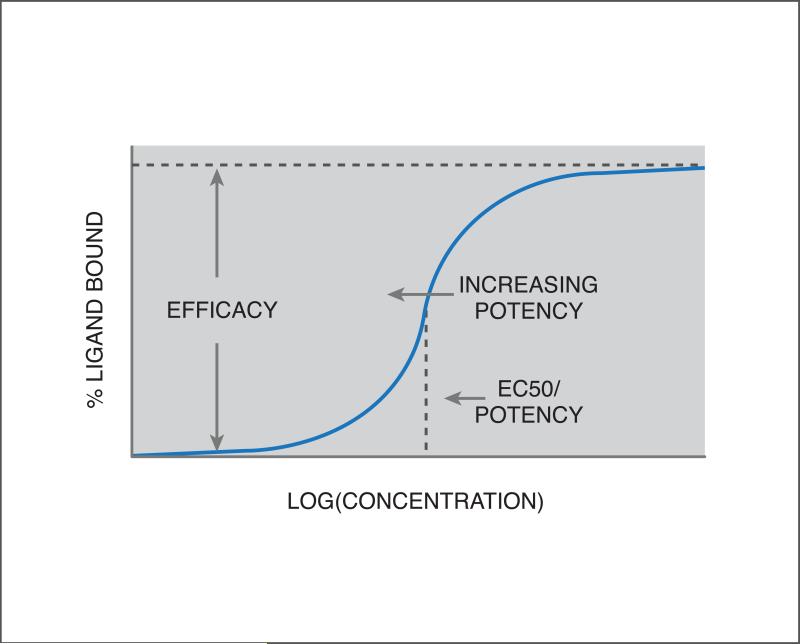
4B: Quantifying drug-target interactions. A dose-response curve is shown, plotting the percent of ligand (drug) bound with a candidate target, as a function of the logarithm of ligand concentration. Since the slope is positive, the inflection point is called the EC50 value (see text). This is a measure of potency, with a lower EC50 corresponding to a more potent effect of the drug on the target. The height of the curve at the inflection point is a measure of the strength of the effect, i.e. efficacy.
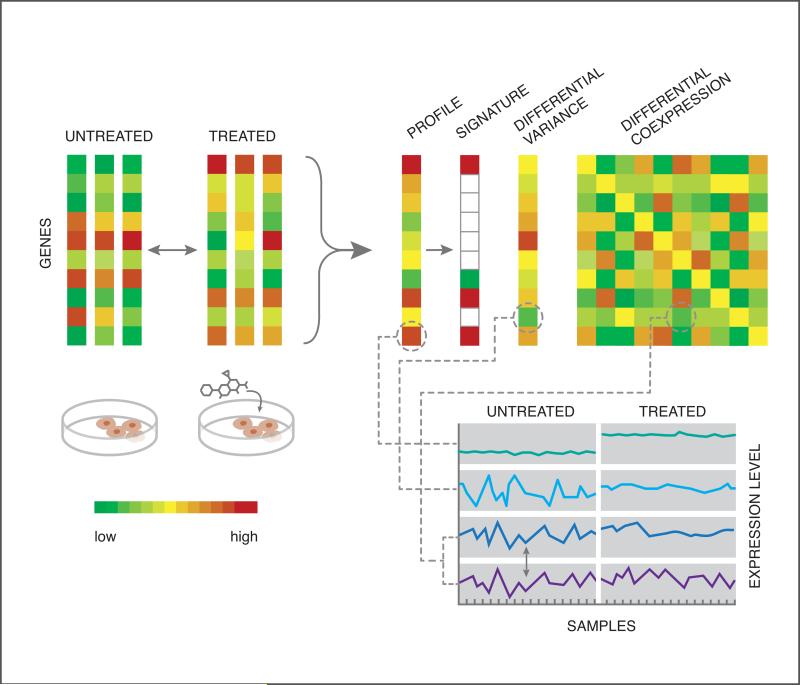
4C: Quantifying drug-perturbed gene expression. Gene expression can be used to characterize the effect of a drug on a group of cells by comparing expression between treated and untreated samples. The data can be processed into a differential expression profile, or processed further into a signature of up- and down-regulated genes. One could also summarize the perturbation using differential variance or differential coexpression.
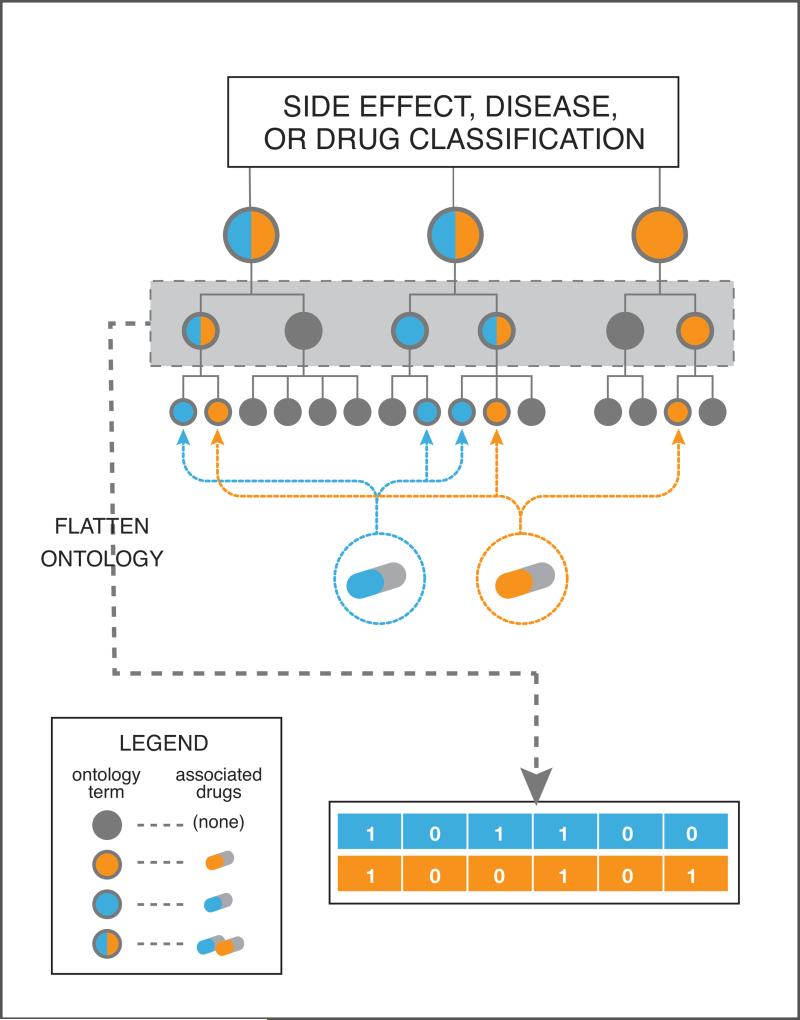
4D: Representing categorical associations such as side effects, diseases, or therapeutic classes. Categorical metadata can often be mapped to a structured ontology (see text for examples), where the highest level of the tree corresponds to the broadest categorization, and deeper levels divide these into more and more detailed distinctions. A numerical representation can be generated by selecting a level of detail in the ontology tree and indicating presence or absence of a drug's association with each category using a ‘1’ or ‘0’.
Drug-target interactions
A drug-target interaction (DTI) can be measured using a variety of experimental techniques such as direct binding or competition binding assays22, and can be summarized in a dose-response curve, plotting some readout corresponding to the amount of protein-ligand complexes formed relative to the logarithm of ligand (drug) concentration. If there is significant interaction, this curve is generally sigmoidal, with the inflection point and height of the curve characterizing the compound's potency and efficacy against the target, respectively (see Figure 4B). This inflection point is either called the EC50 or IC50 value, for the half-maximal [inhibitory/effective] concentration, depending on whether the curve is increasing or decreasing with concentration. While the EC50/IC50 values can vary depending on the experimental setup (e.g. target concentration), these can sometimes be related36 to the binding affinity, denoted by Ki, which is an unchanging property of the intrinsic strength of the interaction.
Various levels of DTI information are available in public databases. Binary-level information, i.e. simply indicating the presence or absence of an interaction, is available in DrugBank37 for several thousand drugs, representing over four thousand unique targets. This could naturally be constructed into a binary target interaction profile vector for each drug, with length equal to the number of targets. Alternatively, more detailed, experimentally-determined binding data for hundreds of thousands of drugs and drug-like compounds are captured in databases such as ChEMBL35, PubChem Bioassay34, and BindingDB38.
Drug perturbations of gene expression
Genome-wide mRNA expression levels can be used as a proxy to measure chemical perturbations of cellular state by comparing expression in cellular samples with and without exposure to a chemical compound. Each perturbation can be represented as an expression profile, where each gene is assigned a number corresponding to the degree of up- or down-regulation relative to control (e.g. the difference of mean expression values); or this can be further processed by discretizing the values into a signature, defined here to mean the sets of significantly up- and down-regulated genes. Though less commonly used, one could alternatively consider differential variance39, or drug-induced changes in the gene-gene covariance, also called differential coexpression40 (see Figure 4C). Several publicly available resources are worth mentioning here. The Connectivity Map41 (Cmap) and its recent update utilizing the L1000 technology as part of the LINCS42 project have generated publicly available expression measurements from thousands of in vitro drug perturbations to multiple human cell lines; while GEO43 serves as a public gene expression repository with over one million samples to date, covering a wide variety of experiments including both drug and disease perturbations. Also, as part of a crowdsourcing project44 organized by the LINCS data integration and coordination center (DCIC), over 900 drug-perturbation experiments have been extracted from GEO and processed into signatures that are freely available for download. Different metrics can be used to evaluate the similarity between two expression profiles and/or signatures45, 46, including correlation, cosine distance, and Gene Set Enrichment Analysis (GSEA)41.
Cell and animal phenotypes
Moving beyond the molecular level, one can also measure or observe a compound's phenotypic effects on a cellular sample or in an animal model, e.g. cytotoxicity in cancer cells47-49 or sleeping patterns in zebrafish50. In fact, until roughly thirty years ago this was the primary approach to drug discovery until it was largely replaced by rational (i.e. target-centric) drug discovery, and yet has remained an important source of new therapies, e.g. contributing the majority of first-in-class FDA approvals between 1998 and 200851. Phenotypic screens are advantageous in that they evaluate a drug's effects within the complexities of biological systems, enabling identification of hits whose mechanism may depend on novel and/or multiple targets, and which may translate more easily into the clinic52. Within the phenotypic screening paradigm, Zheng, et al.52 discusses trade-offs between the use of cellular versus animal models, e.g. cell-based screens usually have higher throughput while animal models enable probing of more complex phenotypes.
While phenotypic screens are usually performed one assay at a time with a particular disease or outcome in mind, data from multiple screens could potentially be aggregated to provide a phenotypic profile for each compound. For example, the Bioassay feature of PubChem53 contains over 740 million data points from both biochemical and phenotypic screens covering over 1 million small molecules, with many compounds having results from hundreds or even thousands of assays. ChEMBL also contains bioassay data, with over 12 million data points35. There are also some publicly available data resources containing (relatively) full drug-by-phenotype matrices. For example, NPCPD255 contains results of nearly 2,500 clinically approved compounds screened in 35 phenotypic assays designed to focus on cardiovascular disease, diabetes, and cancer. Additionally, the NIH Chemical Genomics Center has also compiled a dataset56 of roughly 2,500 approved compounds screened in about 200 phenotypic and target-based assays, focusing on various cancers, malaria, nuclear receptors, and signaling pathways.
Finally, a noteworthy set of cell-based phenotypic screens are cancer cell line sensitivity studies47-49, where cellular growth rates (also called cell viability) are measured before and after drug exposure, for a panel of cancer cell lines. For example, the Cancer Therapeutic Response Portal47 (CTRP) measured sensitivity of 242 genetically characterized cancer cell lines to 354 small molecule probes and drugs. Another example is the Genomics of Drug Sensitivity in Cancer49 (GDSC) database, which measured 138 anti-cancer drugs across 700 cell lines. The Cancer Cell Line Encyclopedia57 (CCLE) provides complementary information to these data, providing detailed genetic characterization of 1,000 cancer cell lines, which e.g. might be used to assess cell line similarity and predict drug-perturbed growth rates in additional cell lines58.
Drug classifications
Various drug classifications, e.g. based on therapeutic usage or pharmacological action, provide an additional layer of information to the drug space. These classification systems are generally organized into some sort of hierarchical, structured ontology, where higher levels refer to more general categorizations and lower levels to more specific terms and associations, and oftentimes multiple, synonymous terms are stored together in the hierarchy, to support a wide variety of queries and mappings. Such information can be translated into binary feature vectors for each drug by simply flattening the ontology tree at a particular depth and only respecting distinctions up to that level (See Figure 4D). Similarity between these vectors could then be computed using Jaccard similarity.
There are many examples of drug ontologies. The Anatomical Therapeutic Class59 (ATC) coding system classifies the active ingredients of drugs into five levels, starting with the organ system[s] on which the compound acts (e.g. the nervous or respiratory system), and subsequently drilling down into more detail such as chemical or pharmacological categories. Drug ATC codes are available on DrugBank's website60. The National Drug File Reference Terminology61 (NDFRT) provides an alternative classification of drugs based on properties such as mechanism of action, physiologic effect, and therapeutic category, and is cross-referenced to other vocabularies including MESH62 and RxNorm63. A third example is the ChEBI64 ontology, which contains multiple sub-ontologies: one based on molecular structure, e.g. dividing organic and inorganic compounds; another based on chemical role, e.g. as an inhibitor, ligand, or surfactant; another based on biological role, e.g. antibiotic, antiviral agent, coenzyme, or hormone; and finally another for applications, e.g. pesticide or anti-rheumatic drug. Additional drug classifications or controlled vocabularies are provided by KEGG65, MeSH62, and MedDRAa.
A practical issue that can arise when working with multiple drug databases is that a single drug often carries many different names and identifiers. RxNorm63 addresses this problem by providing standardized compound names that are mapped to many other names and identifiers, enabling easier data integration.
Disease indications
Known therapeutic indications of a drug can be treated as additional metadata providing clues e.g. for predicting side effects or new indications. Drug-disease associations are available from a variety of sources, including DrugBank, Pharos66, and PharmGKB67. Pharos is a relatively new resource that connects drugs, targets, and diseases, where drug-disease associations include both those in clinical trials as well as approved indications, and disease terms are mapped to Disease Ontology ids (see below). PharmGKB is a database focusing on pharmacogenetics and pharmacogenomics (i.e. identifying drug/gene associations) but contains drug-disease relationships from FDA labels, such as those used in the work of Yang and Agarwal15. Information can also be directly mined from the FDA, e.g. using “the Orange Book”68 or FDALabel69, the latter enabling full-text searching of drug labels including prescription drugs, biologics, and over-the-counter medicines. Finally, clinical trials information can be considered a “noisy” indication of drug-disease relationships, with later-stage clinical trials representing increased confidence in the association, relative to early-stage trials70. At the time of writing, ClinicalTrials.gov71 contained information on nearly 200,000 trials.
Similar to the above-described drug classifications, disease terms and indications are also organized into various classifications and ontologies. Both the Disease Ontology72 and MedDRA® provide structured ontologies over disease terms, hence enabling numerical representations for each drug based on its known disease indications, in a similar way as just described in the previous section. Mappings of unstructured disease terms between datasets is made easier by controlled vocabularies such as Medical Subject Headings62 (MeSH) and others within the Unified Medical Language System73 (UMLS).
Side effects and adverse drug events
A final example of potentially useful drug-related information is given by side effects and adverse drug events (ADEs). Similar to disease indications, side effects terms and adverse events are represented in structured ontologies such as MedDRA®. Several important resources organize complementary aspects of side effect information. First, SIDER74 is a public side effect database with compiled information from FDA package inserts connecting 888 drugs to 1,450 side effect terms. Another resource is the OFFSIDES75 database, generated by analyzing over 400,000 adverse effects not listed on the FDA's official drug label, and identifying an average of 329 off-label ADEs per drug. Finally, the FDA Adverse Event Reporting System (FAERS) is a database of information on adverse event and medication error reports submitted to the FDA by manufacturers, health care professionals, and the general public.
Now that we have considered various ways to quantify and represent drug-related information, we will see how such information can be used in several different applications of computational pharmacology, starting with target prediction.
PREDICTING DRUG-TARGET INTERACTIONS
At the most basic level, drugs exert their effects on biological systems by binding with protein targets and affecting their downstream activity, and hence knowledge of these interactions provides a key toward understanding and predicting higher-level information such as side effects, therapeutic mechanisms and novel indications. However, there are still many gaps in our knowledge of which drugs bind to which targets. At the time of writing, DrugBank37 lists on average less than 2 targets per drug, whereas a recent article76 predicted that the true average number of targets per drug is a staggering 329. Even if this is a gross overestimation, it provides some indication that there are many more interactions than are currently known. Filling these gaps by experimentally testing all drugs against all possible protein targets is currently infeasible, and hence a variety of computational methods have been developed to predict likely interactions. De novo prediction, i.e. based only on structure, is useful for virtual screening of large compound libraries, while other methods make use of related interactions to generate new predictions for compounds that have already been shown to have pharmacological activity.
De novo structure-based prediction
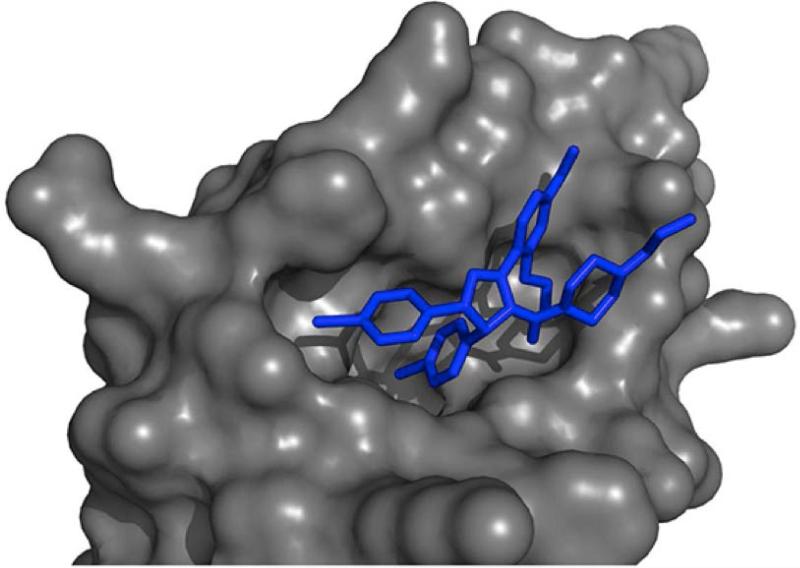
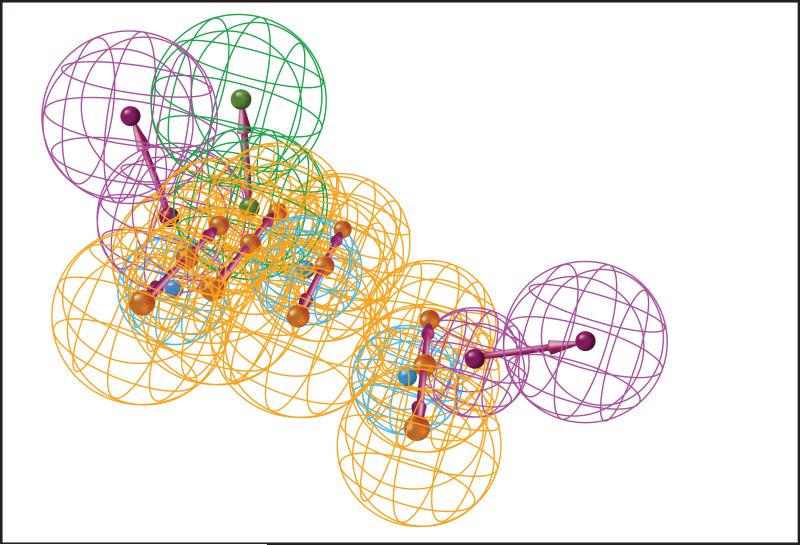
Molecular docking is a popular approach that uses three-dimensional modeling and computer simulation to dock a candidate drug into a protein binding pocket and then score the energetic favorability or likelihood of the pair's interaction77, 78. This approach is advantageous in that it can provide structural insights into the nature of the interaction (see Figure 5A), which might enable further optimization of the compound's structure to increase binding affinity for its target. However, molecular docking depends on the existence of a reliable three-dimensional model of the protein, and for certain target classes such as membrane-bound proteins, this often does not exist due to experimental limitations. Further, the approach is very computationally demanding, limiting its feasibility for large scale, many-to-many DTI prediction tasks.
Figure 5. Computational chemistry approaches for target prediction.
5A: Result of a molecular docking simulation. The globular surface of the protein is shown in grey, and its docked ligand is in blue.
5B: Example of a pharmacophore model. A pharmacophore model is used to represent the chemical features deemed to be important for interaction with a chosen target. The features are arranged in three-dimensions along with some tolerance radius in an attempt to account for dynamic conformational changes of both protein and ligand. A pharmacophore model can be constructed from structural analysis of the target's binding pocket, or could be based on previously known interactions with the target. Compounds can then be aligned and scored against a pharmacophore model in order to prioritize likely interactions. Colors indicate different chemical descriptors such as hydrogen bond donor, or hydrogen bond acceptor, or hydrophobic region.
While molecular docking is considered a target-based approach since each compound is evaluated against the selected target's structure, one can alternatively take a ligand-based approach, constructing a sort of abstract “pseudo-drug” representation called a pharmacophore model (see Figure 5B), containing the chemical features deemed to be important for interaction with the chosen target79. Compounds can then be aligned and scored against the model through a process that is much less computationally demanding than molecular docking. Pharmacophore models can be constructed from analysis of the target's binding pocket, or (moving beyond the de novo prediction setting) could alternatively be derived using a set of positive and negative examples of compounds interacting with the target. Compared with molecular docking, this approach is more computationally efficient, and some studies indicate that it generally has better accuracy80, 81. Pharmacophore models are often used to screen large compound libraries (e.g. millions of compounds) in order to prioritize potential lead compounds for experimental follow-up82, sometimes improving hit rates by an order of magnitude. However, the hit rate will naturally depend on the quality of the pharmacophore model, which can be sensitive to the specific compounds or algorithm used and hence prone to high false-positive and false-negative rates.79
Learning from related interactions
If there are already established examples of compounds that interact with the same or a similar target, this information can be included as an additional layer useful for predicting new interactions. This is accomplished by employing (either implicitly or explicitly) a guilt-by-association (GBA) principle, i.e. that similar drugs may share common targets, or likewise, similar proteins may be targeted by the same drug. This line of thinking is supported by recent work which found that among the roughly 20,000 human proteins, there are only about 1,000 unique shapes of binding pockets83, implying that proteins have many shared binding pockets and in turn, shared binding partners. Providing additional support for the guilt-by-association approach, Paolini, et al.84 integrated drug-target interaction data from multiple sources to construct a bipartite DTI network and found that proteins from the same class tend to share common drug interaction partners.
Various approaches exist that incorporate knowledge of related interactions. One was already mentioned in the previous section: DTI-based pharmacophore modeling. Another common approach85, 86 is to frame the problem as binary classification and employ supervised machine learning models where the inputs are physicochemical features of the drug and/or protein in question, and the output (either known or predicted), is the presence or absence of an interaction. For example, Nidhi, et al.85 used a Naïve Bayes framework to predict targets based only on chemical structure, achieving 77% recall of known interactions among the top three predicted targets for each drug. As an alternative to binary classification, one can also formulate DTI prediction as a regression problem where the aim is to estimate binding affinities. Examples of this include the work of Bock and Gough87, in which support vector regression was used to identify high-affinity ligands for orphan GPCRs; and the more recent work of Cao et al.88 in which random forest regression on both drug and target features achieved AUC's of up to 0.96.
Deep neural networks have also recently been explored to predict drug-target interactions from chemical structure along with known interactions90, 91. For example, Ramsundar, et al.90 integrated millions of data points representing both positive and negative examples of DTIs for over 200 unique targets. They used a “multi-task” framework, in which prediction for each target was considered a separate task requiring its own [linear] classifier, but where all classifiers used the same feature representation, which was optimized using the neural network. The deep-learning based approach achieved a maximum cross-validated AUC (area under the receiver operating curve) of 0.87 and demonstrated that the multi-task aspect of their approach consistently provided slight improvements (roughly 0.01 increase in AUC) over an equivalent single-task analysis with the same amount of data. Note that the task-specific linear classifiers as well as the previously mentioned machine learning models are in some sense analogous to a pharmacophore model, in that all of these models “decide” which structural features are most important for the interaction.
All of the above-described methods only implicitly invoke the similarity principle, e.g. by fitting coefficients to drug and/or protein features, so that drugs with similar features would have similar predictions. However a number of machine learning methods have been developed which explicitly employ a similarity-based framework by working directly with similarity matrices between drugs and/or targets. A very simple example is a nearest-neighbor method20, where, e.g. one could predict whether an interaction would occur between drug D and target T based on whether the drug “nearest” to D interacts with T, or alternatively, whether the target nearest to T interacts with D. In this same vein, Bleakley, et al.19 propose a slightly more sophisticated approach they call bipartite local models, training a different support vector machine (SVM) classifier for each drug and each target, where user-specified drug- and target-similarity matrices are input to the SVM algorithm, and known interactions serve as labels. Ding, et al.20 provide a cogent and insightful review of similarity-based machine learning approaches, along with some experiments benchmarking the ability of eight different algorithms to recover known DTIs. While their results did not reveal a clear winner, AUCs reached as high as 0.98 for ion channels, but varied significantly per target class (likely due at least in part to varying amounts of available data per class), and are hence difficult to compare against the AUCs from the deep-learning approach described above.
While compound structural similarity is perhaps the most natural and well-supported metric used for DTI prediction, other notions of similarity have also found success. For example, Campillos, et al.21 developed a metric for side effect similarity over a set of 746 marketed compounds, finding ~1,000 side effect driven drug-drug relationships and confirming 9 out of 20 subsequent DTI predictions in cell-based assays. Interestingly, about one quarter of their identified drug pairs were both chemically dissimilar and also had different therapeutic indications, demonstrating that side effect information provides a somewhat orthogonal view of compound relationships that is still informative of target activity. Keiser, et al.92 present an alternative framework based on their similarity ensemble approach (SEA)22, where each target is represented by its known binding ligands (including endogenous ligands), and then similarity between the candidate drug and the ligand set is evaluating using a statistical framework developed by the authors22. 23 of 30 tested predictions were experimentally confirmed, including the activity of the drug DMT on serotonergic receptors, indicating a different mechanism of action for DMT than was currently understood. A final example using an alternative notion of similarity is the network-based inference (NBI) method23, which simply uses known DTIs to predict new ones; i.e. the drug similarity metric in this case (though not explicit in the NBI framework) is based on the target interaction profiles.
One important consideration when employing any such technique based on related interactions is the paucity of high-confidence negative examples; i.e. it is difficult to know whether a particular drug-target interaction is, in fact, not possible, or if an interaction may occur in a different biological context. Recent work89 aimed to address this problem by developing an in silico method to identify high-confidence negative examples and further demonstrating that such examples boost predictive performance.
DTI prediction is a fairly well studied problem, with many different techniques that together use a variety of data including chemical structure, protein structure, side effect associations, ligand sets, and other drug-target interactions. While computational chemistry can be used to generate de novo predictions and hence explore new areas of pharmacological space, similarity-based techniques offer the advantage that they can improve in accuracy as more data become available. Many of these methods have demonstrated a high degree of accuracy and have proven to be useful both in virtual screening settings to prioritize compounds for HTS, as well as for identifying new targets for known drugs. We will see in the next sections how these techniques can also provide a foundation to predict side effects and discover new therapeutic indications.
PREDICTING AND EXPLAINING SIDE EFFECTS AND ADVERSE EVENTS
Drug safety is a critical factor in the success of commercial drug development. Improved ability to model and predict drug side effects and adverse events is crucial for improving the efficiency of drug discovery, as early identification of undesirable toxicity can prevent further investment of resources in a non-viable drug entity. The current standard approach to safety screening is pre-clinical testing in animal disease models. However, such experiments are costly93, and leave a large degree of uncertainty as to whether the results will translate into humans94, 95 due to genetic and environmental differences.
Computational approaches can help address some of these challenges. In silico techniques have the potential to predict unwanted side effects at earlier stages in the drug development pipeline, e.g. based on predicted drug-target interactions76, 96 or in vitro drug-induced gene expression perturbations97. Further, Lum et al.98 suggest that translational uncertainty between animal models and humans could be lessened by taking a computational systems biology approach, modeling the conserved responses of molecular networks across species.
Identification of new side effect associations with approved compounds is also an important aim and falls under the heading of pharmacovigilance. Such associations might be missed in clinical trials, e.g. due to the rarity of occurrence, or a delay between start of medication and onset of symptoms99. Computational techniques are particularly relevant in this case, given the added ability to mine data surrounding the compound's post-market use and effects24, 99, 100.
Target-mediated connections
Some protein targets have been identified as causally implicated for undesirable effects101, 102, and this information can be used to link drugs with such effects. For example, Lounkine, et al.96 used the SEA method22 described in the previous section to evaluate the activity of 656 marketed drugs on 73 side effect-associated proteins. They developed a method to identify predicted off-targets that explained side effects better than any of a drug's established targets. From this came a prediction that abdominal pain from the synthetic oestrogen chlorotrianisene is mediated by its newly discovered and validated interaction with the enzyme cyclooxygenase-1. Zhou, et al.76 took a similar approach, using their FINDSITEcomb method103 to predict DTIs for all drugs in DrugBank compared against a majority of proteins in the human proteome. Combining these predicted DTIs with known drug-side effect associations enabled association of targets with side effects, even if the targets had no experimentally verified drug interactions. Finally, the authors introduced a killing index, which estimates the likelihood that a compound has serious side effects such as death, stroke or heart failure. They found that 44% of small molecules from DrugBank were predicted to have a killing index > 0, whereas this was true for only 16% of FDA approved drugs, providing some validation to their analysis and suggesting that this killing index might be useful, e.g. to filter out investigational compounds in early stages of drug development.
Molecular network modeling
While the approaches just described are based on established connections between targets and side effects, molecular network modeling can be used to hypothesize new connections between targets and side effects and help to elucidate physiological mechanisms. This is exemplified by the work of two different groups aiming to explain a fatal hypertensive response among some people taking the CETP inhibitor torcetrapib which lead to the drug (intended for atherosclerosis) failing Phase III clinical trials104. An understanding of the molecular mechanisms inducing the fatal response would help to avoid repetitions of this scenario in the future and clarify whether other CETP inhibitors should continue to be pursued. Chang, et al.105 developed a framework using structure-based target prediction and a technique called metabolic modeling106 to implicate targets for the hypertensive response, hypothesizing that the side effect was due to renal regulation of blood pressure via metabolite reabsorption and secretion. Using structure-based target prediction, they identified a list of 41 metabolic proteins predicted to be off-targets of the drug. Then they used a renal metabolic network model constructed over 338 genes to simulate phenotypic consequences of inhibition of each of the targets, yielding 6 out of 41 “hits” predicted to alter renal function. Two of these hits had literature support connecting the targets to hypertension in humans, mice and/or rats, while the remaining four were novel hypotheses. Fan et al.107 also used network analysis to explore potential explanations for the torcetrapib-induced hypertension. They constructed a context-specific human signaling network filtered by a set of genes that were differentially expressed in adrenal carcinoma cells treated with torcetrapib, identifying several enriched signaling pathways with previous associations to hypertension.
Other approaches
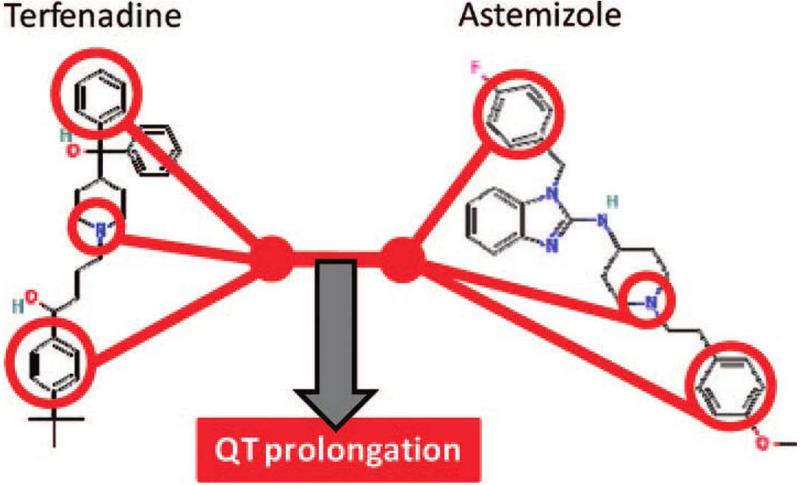
A variety of other approaches have been used to analyze or predict connections between drugs and adverse effects. Scheiber, et al.108 used known drug-ADE associations to connect specific chemical features of drugs to 4,210 ADE terms using an extension of Naïve Bayes modeling. An example of a resulting model is shown in Figure 6, depicting a well-known example of structural associations with QT interval prolongation, which causes cardiac arrhythmia. Along similar lines, Liu, et al.109 used causality analysis based on Bayesian network structure learning to connect both chemical and biological features of drugs to ADEs, in a way that could be causally interpreted. As a final example, Vilar et al.24 used a guilt-by-association approach on a large insurance claims database to estimate drug associations with four diverse ADEs: acute renal failure, acute liver failure, acute myocardial infarction, and upper gastrointestinal (GI) ulcer. The authors evaluated various compound similarity metrics such as chemical structure, targets, ATC code, and other ADEs, finding that the latter two metrics, both informed by phenotypic associations, achieved the top AUPRs (Area Under the Precision-Recall Curve) in three of the four ADEs tested.
Figure 6. Connecting chemical features to side effects.
Scheiber, et al.108 used known drug-ADE associations to connect specific chemical features of drugs to 4,210 ADE terms using an extension of Naïve Bayes modeling. An example of a resulting model is shown here, associating specific chemical features with QT interval prolongation, which causes cardiac arrhythmia.
Side effect prediction and analysis is an important yet challenging aim in computational pharmacology. Part of the challenge stems from the difficulty in defining side effects unambiguously. Additionally, relative to drug-target interactions, side effects are generally quite downstream in a cascade of biological events initiated by drug exposure, and hence drug-side effect relationships are more indirect and hence elusive.
DISCOVERING NEW CONNECTIONS BETWEEN DRUG AND DISEASE
One area of computational pharmacology that has gained increasing amounts of attention in recent years is drug repurposing (also called “repositioning”), which seeks to find new uses for known drugs as well as for early-stage assets or shelved compounds. Two key insights help explain why drugs could be used for more than one purpose: first, many drugs have multiple protein targets84, 110, and second, different diseases can share genetic factors, molecular pathways, and/or clinical manifestations111, 112, and hence a drug which acts on such overlapping factors may be beneficial to both conditions. Drug repurposing is not a new idea. Examples of successfully repurposed drugs include Minoxidil, developed for hypertension and now indicated for hair loss, Viagra, repurposed from angina to erectile dysfunction, and Thalidomide, originally for morning sickness and now used to treat symptoms of leprosy113, 114. However, while these examples were due to serendipitous observations, we will discuss computational methods that explore the drug repurposing space systematically.
Drug repurposing offers many benefits over de novo drug development. The time and cost toward approval of a new indication can be greatly reduced for a drug with an established safety record, with estimates of 3-10 years for a repurposed compound as compared with 10-17 years for a new molecular entity (NME)115. Approval rates are also much higher, e.g. 25% of repurposed candidates succeed from Phase II to approval, compared with 10% for a NME6. Furthermore, drug repurposing is a promising avenue to address unmet therapeutic needs for rare and neglected diseases116-122, and can also identify drugs that are more efficacious or cost-effective than existing ones. Finally, some of the in silico techniques described here provide the additional benefit of generating hypotheses about biological mechanisms of a drug or disease in the process of predicting new repositioning candidates, compared with traditional drug development strategies which sometimes treat biological systems as “black boxes”.
Note that, in the following paragraphs, we will describe how various sources of information, including DTIs and side effects, can provide clues for drug repurposing. This can be effective even when these clues are predicted from other information sources such as chemical structure15, and hence in this way we can view drug repurposing as a natural extension of methods described in the previous sections.
Target-centric approaches
One approach for identifying a new indication is to repurpose a drug based on the biological role that a target plays in disease. A rather straightforward example in this regard is the work of Chavali, et al.120, who used metabolic modeling to generate a list of 15 genes and 8 double-gene combinations predicted to be relevant targets for the neglected tropical disease, leishmaniasis major. The authors were able to associate these genes with 254 FDA-approved compounds based on drug-target interactions, and found validation for 14% (10 out of 71) of these compounds which overlapped with an independent HTS screen against leishmaniasis. Another example that employs this approach in a more complex manner is the work of Chen, et al.70, who integrated a large number of information sources including drug-target interations, disease-gene associations and protein-protein interactions networks into a heterogeneous network they call DrugNet, connecting drugs, targets, and diseases. The authors use a network propagation algorithm called ProphNet123 that, given an input query node, either a drug or disease, ranks the remaining nodes of the other type, i.e. drugs for a disease query, and vice versa. They achieved a leave-one-out cross-validation AUC of 0.96 in recapitulating known drug-disease associations.
From side effects to discoveries
While side effects usually carry negative connotations, sometimes these unintended consequences offer clues toward new therapeutic directions. For example, the testosterone reductase inibitor Finasteride was initially tried and ultimately approved to treat benign prostatic hyperplasia124. During the trials, however, an unintended treatment outcome was hair growth. Rather than dismissing this side effect as a negative, this observation ignited the idea to repurpose Finasteride for the hereditary condition Androgenetic Alopecia (colloquially called male pattern baldness). In another example, the anti-depressant drug bupriopion was noted to have an anti-smoking effect during the clinical trials for treating depression125. This finding lead to the development of a new smoking cessation drug marketed as Zyban126.
These serendipitous observations raise the question of whether the discovery of new indications can be accelerated by automated, systematic mining of side effect information. Indeed, Zhang, et al.16 found that side effect information was even more predictive of disease indications than chemical structure or protein target information. Yang and Agarwal15 merged drug-side effect data from SIDER127 with drug-disease information from PharmGKB128 to identify a set of side-effect-disease relationships, which were then used to build Naïve Bayes models for 145 disease indications using the side effects as features (see Figure 7), achieving AUCs above 0.8 for 92% of the models. Ye, et al.25 similarly hypothesized that drugs with similar side effects might share common indications. They constructed a network over drugs based on Jaccard similarity of their associations with 6,495 side effects. Disease indications were then predicted based on enrichments of FDA-approved indications among neighboring compounds, and while the authors did not compute ROC curves, they found that over 70% of the predictions were FDA-approved, and another 10% supported by pre-clinical/clinical trials or scientific literature. Interestingly, the results varied widely for different classes of drugs, with the best performance for treatments of diabetes and obesity as well as laxatives and antimycobacterials.
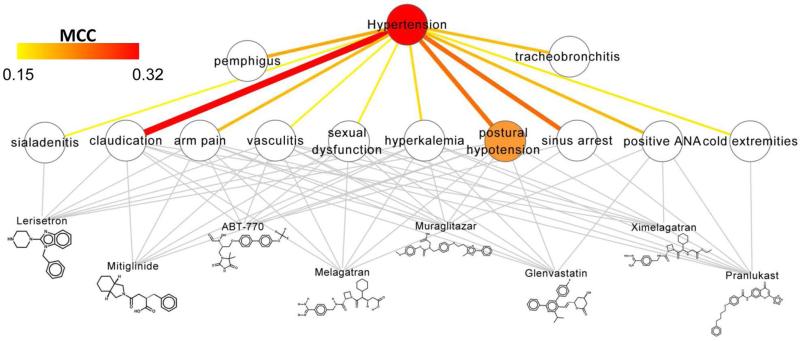
Figure 7. Connecting side effects to diseases.
Yang and Agarwal15 constructed 145 disease-specific models using drug side effects as predictive features to evaluate each drug's therapeutic potential for the disease. Shown is their predictive model for hypertension, where the association between hypertension and each side effect (quantified by the Matthews correlation coefficient, MCC) is depicted by both color and edge-thickness. Binary associations between drugs and side effects are shown in grey. Notice that many of the features such as postural hypotension and cold extremities seem reasonable in that they are commonly associated with low blood pressure.
Of course, side effect information is only available for drugs that have at least reached clinical trials if not approval, and hence approaches using side effect information alone would generally only apply in these cases. However, there are ways around this; for example, predicting side effects from chemical structure and then connecting these side effects to potential indications15.
Gene expression as a common language between drug and disease
Gene expression data provide a high dimensional readout of cellular state and biological perturbation resulting from drug treatment or the presence of disease. Gene expression profiling enables quantitative molecular comparisons between drug- and disease-perturbed states. One advantage of transcriptomic approaches is that this type of data can be generated for nearly any chemical compound or disease, regardless of the compound's approval status, and agnostic to drug or disease mechanisms. Further, while information on side effects and targets has many false negatives, expression profiling provides an unbiased, genome-scale view for each drug and disease perturbation.
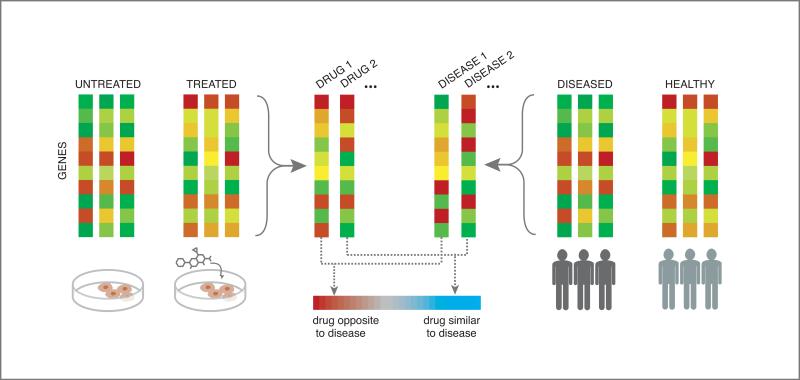
One key approach used in many expression-based drug repurposing studies113, 129-132 is alternatively called signature reversion, signature matching, or connectivity mapping41, which matches drugs and diseases with opposing or anti-correlated expression profiles, reasoning that if gene expression is perturbed in one direction in a diseased state, and in the reverse direction upon exposure of a drug, then perhaps that drug could “push” the disease-perturbed expression back toward a more normal state, and hence provide therapeutic benefit for the disease133 (see Figure 8). For example, Sirota, et al.129 systematically compared gene expression signatures derived from Cmap for 164 small molecule compounds against a set of expression signatures derived from GEO for 100 different diseases, generating over one thousand drug repurposing predictions, connecting at least one of the 164 compounds to each of 53 diseases. Two predictions from this work were selected for experimental validation in animal models, both yielding positive results. Specifically, topiramate, an anti-convulsant predicted to be therapeutic for both ulcerative colitis and Crohn's disease, was shown to ameliorate symptoms in a rat model of irritable bowel disorder134, and also cimetidine, an antihistamine approved for inhibition of gastric acid secretion was predicted to treat lung adenocarcinoma (LA), and showed dose-dependent reduction of LA tumor growth in mice129.
Figure 8. “Connectivity mapping” for drug repurposing.
The connectivity mapping approach hypothesizes that a drug and disease with opposing or anti-correlated expression profiles might be a therapeutic match, reasoning that if gene expression is perturbed in one direction in a diseased state, and in the reverse direction upon exposure of a drug, then perhaps that drug could “push” the disease-perturbed expression back toward a more normal state, and hence provide therapeutic benefit for the disease.
Signature matching can also be used to connect between drugs. Iorio, et al.97 used pairwise similarity been drug-perturbed gene expression profiles to construct a network over 1,302 drugs. Highly connected communities in this graph were significantly enriched for compounds with similar MoA (Mechanisms of Action) and also revealed new mechanisms and indications, for example predicting and subsequently verifying that the drug Fasudil enhances cellular autophagy, indicating potential for certain neurodegenerative disorders. Note that this is another similarity-based approach, since mechanisms are hypothesized to be shared between similar compounds, where similarity is now based on gene expression perturbations.
One drawback of these expression-based approaches is that some drugs and diseases do not induce strong expression perturbations, and hence the signal for such perturbations would be noisy and hence lead to higher false-positive or false-negative rates. Another consideration is that the signature reversion principle may fail, e.g. if the disease expression profile is a result instead of a cause of the diseased state, in which case reverting the profile with a drug may not be therapeutic.
Finally, there are some interesting opportunities for future work here. First, there is an opportunity to better explore and leverage the tissue- or cell type-specificity of drug transcriptional perturbations, as most existing approaches ignore this dimension of information, and in some cases such context has been shown to be very important135. Also, instead of simple, pairwise-comparisons of expression profiles, it might be fruitful to better understand or map out these drug- or disease-perturbed transcriptional landscapes, providing more meaningful context or metrics for subsequent comparisons. This is one example of data integration within a single modality. Some work has already ventured in this direction, e.g. analyzing bi-clusters136 of genes co-regulated by a subset of compounds, or applying a generalization of Bayesian principal component analysis to project drug-perturbed expression profiles into a lower-dimensional linear subspace137. One simple, yet relatively unexplored direction within this vein would be to incorporate covariance between genes (often called coexpression) into a similarity metric.
Drug- and disease-similarity
The guilt-by-association principle can also be applied to make new connections between drug and disease26-28. For example, Chiang and Butte26 hypothesized that if two diseases have medications in common, then other medications currently used for only one of the two diseases may also be therapeutic for the other. They compiled FDA-approved as well as off-label uses connecting 2,022 drugs to 726 diseases, and applying this simple guilt-by-association rule, generated about 57,000 novel drug-use suggestions. As validation, the authors found that their predicted drug-disease pairs were 12 times as likely to be found in recent clinical trials than those that were not suggested by their method. Another example is the work of Zhang, et al.27, who developed a matrix factorization framework to implement a more general version of a drug and disease guilt-by-association rule, where instead of connecting disease pairs based on sharing the exact same medication, they incorporate a variety of both disease similarity and drug similarity information. They achieve 10-fold cross-validation AUC of 0.87. This method offers the added benefit of a quantitative estimate of the relative contribution of each source of similarity information, here finding that side effect information had the largest contribution, followed by chemical structure and then known targets.
Mining and validating drug repurposing signals in electronic health records
Electronic health records (EHR) offer a promising new resource to be explored for generating and validating drug repurposing hypotheses. EHR provide massive, longitudinal data on thousands or even millions of patients, including lab results, diagnosis codes, prescriptions, and physician notes. As EHR databases are becoming more standardized and integrated across multiple hospital systems, they are gaining increasing attention from the informatics community as a resource to be mined, e.g. to assess quality of patient care, build early prediction models for disease, re-evaluate medication usage, and identify off-label usage138. By identifying matched cohorts within an EHR database that either have or have not been prescribed a particular medication, one could conceivably perform observational studies as proxy for randomized controlled clinical trials, mining for unexpected effects associated with the prescribed medication. In contrast to many observational study contexts, the vast scale of EHR databases would enable this to be done in parallel to test a large number of drug repurposing hypotheses and analyze effects over larger patient populations and longer time durations. While we are not yet aware of any such published analyses generating novel drug repurposing hypotheses from EHR analysis, Xu and collagues139 demonstrated the utility of EHR data for a similar use- to provide external validation to an existing drug repurposing hypothesis. They used the case study of metformin, a drug traditionally used to treat type 2 diabetes (T2D) but recently hypothesized to be associated with reduced cancer mortality. To test the hypothesis, the authors identified patients in two separate EHR databases diagnosed with both cancer and T2D, and applied Kaplan-Meier survival analysis to find that patients taking metformin indeed had improved survival. While we foresee that more drug repurposing studies will be published using EHR data, current work with EHR databases is often impeded by privacy concerns as well as data cleaning and modeling issues, including incomplete and irregularly sampled information, inaccurate diagnosis codes, and unstructured clinical notes138.
While computational drug repurposing is still waiting to see its first compound reach the market, experimental and quantitative evidence is accumulating in support of the feasibility of this approach. The field will likely continue to draw new attention, both from researchers; as new data such as internet search queries100 and electronic health records are incorporated into analytic pipelines; as well as from the general public, as evidence for this strategy continues to grow. One way to make this evidence more convincing and advance the field more systematically would be to adopt standardized validation datasets so that different methodologies could be compared on the same footing (e.g. see Cheng, et al.46). Potential datasets that could serve this purpose might come from clinical trials data as used by Martinez, et al.70, drug therapeutic classification as used by Napolitano, et al.17, or drug-disease associations, such as those used by Cheng, et al.46
Finally, enough examples of repurposed compounds have been generated that we can begin to ask questions like, “Are there certain features of a compound that make it more or less repurposeable”? Or similarly, “Are there certain classes of diseases or conditions for which repurposing hits are more likely”?
DATA INTEGRATION IN COMPUTATIONAL PHARMACOLOGY
While we have already described many integrative approaches as they apply to different pharmacological aims, in this section we bring data integration into the spotlight, first highlighting the benefits of such approaches, and then critically discussing several computational methodologies that lend themselves well to data integration.
The case for data integration
As pharmacological space comprises a variety of data types, each one having its own peculiarities and challenges, it is natural that the first attempts to mine these data often focus on just one or two of these information sources. However, in the same way that multiple camera angles can help clarify a sports play, it is intuitive that multiple data angles would generally improve predictive performance and help clarify the story surrounding a particular drug and its potential effects on the human body. A statistical rationale for the benefit of integrative approaches is that some component of the noise contained in each data modality will be independent, and hence, combining these modalities would lessen the obfuscating effects of such noise.
Quantitative evidence in support of integration across data types has been demonstrated for a variety of tasks in areas related to computational pharmacology13-18. For example, Napolitano, et al.17 demonstrated the benefits of data integration for predicting drug therapeutic class; incorporating gene expression, chemical structure, and target interaction profiles into a single drug-similarity matrix that was input to a multi-class SVM classifier. The authors compared ROC curves generated using the multi-source similarity matrix against curves generated using three different single-data source similarity matrices, achieving higher accuracy with the integrative approach, as shown in Figure 9. Vilar, et al.13 used principal component analysis to integrate five different types of features including chemical structure and target interaction profiles to predict drug-drug interactions, and showed that the integrated features were as good or better than any individual feature, and the advantage, as measured by AUC, was magnified in an independent test dataset. Another example is the work of Zitnic et al.14 who incorporated a variety of drug, gene and disease information sources using a simultaneous matrix factorization approach to build a data-driven disease classification system that, impressively, found literature support for all 14 predicted disease-disease associations not already present in the Disease Ontology. Furthermore, by systematically removing each data source one at a time and measuring the change in recall of disease-gene associations, the authors demonstrated that each individual data source contributed positively to model performance. All of these examples demonstrate the power of combining multiple data dimensions in the chemical, biological, and phenotypic spaces to build predictive models related to drug and disease.
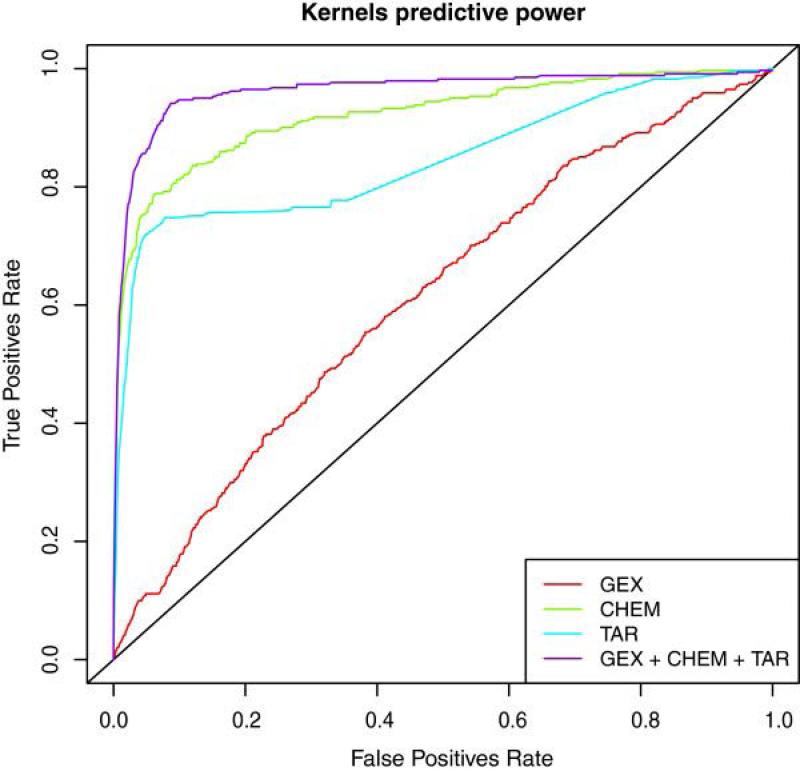
Figure 9. Improved performance of data integration.
Napolitano, et al.17 demonstrated the benefits of data integration for predicting drug therapeutic class; incorporating gene expression (GEX), chemical structure (CHEM), and target interaction profiles (TAR) into a single drug-similarity matrix that was input to a multi-class SVM classifier. They compared the ROC curve generated using the multi-source similarity matrix against curves generated using three different single-data source similarity matrices, with the former achieving higher accuracy, as shown.
Comparison of integrative methods
Here, we highlight three algorithmic frameworks that stand out among recent work in computational pharmacology as relatively well-suited for multi-scale data integration, namely: similarity-based methods, network modeling, and matrix factorization. Each makes certain modeling assumptions and has various practical benefits and drawbacks.
Similarity-based methods
Similarity-based methods13, 20, 21, 24-28 comprise a wide variety of approaches applicable to all three aims reviewed in this article. Similarity-based approaches lend themselves naturally to data integration in that a variety of information sources and metadata can be used to define similarity between compounds, targets, side effects, and diseases. Further, multiple similarity measures for the same type of entity can often be combined into a single similarity matrix, e.g. combining multiple drug-similarity matrices into an SVM classifier13, 17. However, one should use caution when combining similarity information, as different modalities can be somewhat orthogonal, as found in the work of Campillos, et al.21 where drugs were connected based on side effects but did not share targets or known indications (as described earlier). In this case, averaging different similarity measures might hide a signal that comes from only one dimension, and therefore, one might consider alternative ways to combine similarity information, such as taking the maximum similarity among all measures.
The premise that similar compounds have similar properties, though not always true140, is an intuitive notion and has substantial empirical support, particularly in the case of structural similarity revealing shared target interactions21, 83, 84. Further, based on the premise that DTIs are the fundamental effectors of downstream biological and physiological perturbations, it is reasonable to extend the similarity principle to such downstream effects, e.g. side effects and disease indications. One drawback of similarity-based approaches is the reliance on data existing “nearby” in pharmacological space, hence limiting applicability for discovery of truly novel classes of compounds, targets, etc.
Network methods
Networks provide an intuitive framework to integrate a wide variety of information sources, capturing both quantitative and qualitative relationships between entities, such as gene expression correlation, or the presence or absence of an interaction70. Network topology can be utilized in graph-based algorithms such as label propagation methods141 that iteratively propagate information to neighboring nodes; network-based inference23 methods that make new connections based only on local topology; and shortest-path algorithms to identify parsimonious explanations of network perturbations142.
Molecular networks such as gene regulatory networks and metabolic models have many applications in computational pharmacology143. Gene regulatory networks constructed from genome-wide transcriptional profiles and intrinsic genomic variation are able to estimate causal relationships between molecules and identify key drivers of disease144-146. This new field of network pharmacology is still in the early days147 but is already illuminating fruitful drug targets for treating diseased states148-150 and producing accurate estimates of off-target effects151, 152. Alternatively, metabolic models constructed from sets of metabolic reactions can be used to simulate enzyme kinetic activities and perform in-silico gene knockouts, which can e.g. help to identify and prioritize new drug targets120, 153.
One practical drawback of some network approaches is a tendency to be somewhat ad hoc in nature, having many tuning parameters154, e.g. thresholds to determine presence or absence of edges, or how exactly to extract subnetworks, or to what degree nodes should share information with their neighbors. Systematic exploration of the robustness of the analysis to various parameter settings should ideally be performed, however this can be time-consuming and may lead to ambiguous conclusions, and hence a researcher may simply resort to using default parameters. However, in some cases, selection155 of parameters can be guided by [relatively crude] heuristics such as a constraint that the network structure satisfies the scale-free property156.
Bayesian network modeling, used e.g. to model gene regulatory networks157, 158, presents some specific practical challenges. Bayesian network inference is computationally intensive and requires a large number of samples (at least hundreds or thousands) in order to derive accurate results159. In addition, there are sometimes multiple Bayesian network graph structures that can equally represent the same dataset. To illustrate, the two graphs, X -> Y and Y <- X are semantically equivalent in the language of Bayesian networks, essentially representing the idea that correlation does not imply causation. In such cases, however, additional data can sometimes be used to resolve directional ambiguities, e.g. using intrinsic genomic variation160 or time series data161.
While it is natural to model biological systems and, more abstractly, pharmacological space, as a set of entities with local interactions, an implicit modeling assumption that is often made in network-based approaches is that information travels along paths consisting of local relationships23, 70, 141, i.e. long-range interactions and pathway cross-talks leading to nonlinearities might be ignored. While these assumptions simplify modeling and analysis considerably and may provide reasonable results, this should be considered carefully before proceeding down this path.
Matrix factorization
Recent applications of matrix factorization-based methods14, 27, 162-164 demonstrate some important advantages of this type of approach, in particular, ease of multi-scale integration as well as data imputation within a mathematically rigorous formulation. Matrix factorization approximates a [usually large] matrix as a product of lower-rank matrices. This approximation can be interpreted as making the modeling assumption that there are a small number of “factors” (i.e. less than the number of data points) that are responsible for the main variations in the data. Another interpretation is that the data can be projected from a higher-dimensional space to a lower-dimensional linear space, by applying some linear transformation. When the factorized matrix represents self-similarity (e.g. a drug-drug matrix) the lower-dimensional space corresponds to a more succinct, and hopefully more natural, feature representation of the data. Alternatively, if the factorized matrix represents relationships between two entities, e.g. a drug-disease matrix, then this lower-dimensional space corresponds to one into which both drugs and diseases can be projected27, 162, and hence compared quantitatively.
The method developed by Zhang, et al.27 (described earlier) presents a unified framework for incorporating multiple drug- and disease-similarity measures along with known drug-disease associations in order to predict new therapeutic associations. All three relationships (drug-drug, disease-disease, and drug-disease) are represented by matrices, which are factorized. The key idea here that makes this a truly integrative approach, is that a single, common low-dimensional drug projection is sought to be maximally consistent with all of the drug-similarity matrices, and likewise, a common disease-projection matrix is sought to be consistent with all of the disease-similarity matrices. Then, these two projections are used in combination to factorize the drug-disease matrix. The entire process is optimized to maximally recapitulate known drug-disease associations. A similar approach was presented by Zitnic et al.14 (also described earlier) for disease classification, where e.g., five different gene-gene similarity matrices were all factorized using a common projection matrix. In addition to disease classification and drug repurposing, matrix factorization has also been used for DTI prediction163 and drug-ADR associations164.
Another advantage of matrix factorization is its utility for data imputation via “matrix completion,” where missing entries in a matrix are filled in based on the observed entries. Many techniques165-167 to solve this problem were developed as a result of the Netflix competition168, which posed movie recommendation as a matrix completion problem. As an example of a biological application, Chi, et al.169 used matrix completion to impute genotype information.
Conclusion
Computational pharmacology and drug repurposing are burgeoning areas of research that are enabling new ways to systematically explore the drug space and generate novel hypotheses surrounding drug action and indications. These techniques are helping to accelerate the drug discovery process and generate novel hypotheses from diverse data, helping to augment research beyond what might be possible based solely on human intuition or observation.
As new sources of drug-related information become available and molecular measurements (e.g., -omics) become more routine, we foresee several new and exciting directions that could be explored within the space of computational pharmacology. First, under-utilized data sources such as quantitative binding data38 and phenotypic screens170, as well as newer data sources such as EHR and internet search engine queries100 will likely provide further avenues of development in the near future. Second, we believe that matrix factorization is a very promising approach that should be explored in terms of its ability to integrate across diverse data and impute missing information. Tensor decomposition, the natural extension of matrix factorization to data structures that extend across more than two dimensions (e.g. gene expression across many drugs and cell types) might also be explored, e.g. to analyze LINCS L1000 gene expression data or to construct low-dimensional representations of patient state from EHR data171. Third, recent big-data approaches are generating improved disease classifications14 and subtype stratifications172, and this will likely lead to an improved ability to identify therapies targeted to more specific patient populations. Finally, some drug repurposing methodologies translate naturally into a personalized medicine setting; most notably, signature matching techniques, where the disease signature could easily be replaced by an individual's expression signature. Perhaps there are other techniques developed for drug repurposing that could be easily translated into this new setting. For example, it is likely that more types of -omics data will soon be available on a large scale, providing alternative, high-dimensional disease quantifications that could readily translate into personalized medicine applications in the coming years.
Table 1.
A selection of databases and resources useful for computational pharmacology and drug repurposing.
| Resource Type | Resource | Description | URL |
|---|---|---|---|
| General resource for compound information | PubChem | Data base of over 60 million compound structures, chemical features, bioactivity, etc. | https://pubchem.ncbi.nlm.nih.gov/ |
| General resource for compound information | ChEMBL | Data base of over 1 million compound structures, chemical features, bioactivity, etc. | https://www.ebi.ac.uk/chembl/ws |
| DTIs (binary) | DrugBank | Drug and drug target information for over 7000 drugs | http://www.drugbank.ca/ |
| DTIs (detailed) | BindingDB | Information on binding affinities and other quantities related to DTIs | https://www.bindingdb.org |
| Predicted DTIs | SEA | DTIs predicted using the SEA method | http://sea.bkslab.org/ |
| Predicted DTIs | DR. PRODIS | DTIs predicted using the Findsitecomb method, can also access killing index (see text) | http://cssb.biology.gatech.edu/repurpose |
| Drug-induced transcriptional perturbations | Cmap v2 | 1309 compounds exposed to 5 different cancer cell lines | https://www.broadinstitute.org/cmap/ |
| Drug-induced transcriptional perturbations, etc. | LINCS | L1000 data: over 1 million profiles generated by chemical and genetic perturbation of dozens of cancer and primary cell lines; other drug-related datasets also available. | http://www.lincscloud.org/, http://www.lincsproject.org/ |
| Disease-related genetic/genomic perturbations | TCGA | RNAseq, microarray, and/or sequence information on cancerous tissues, covering over 30 types of cancer | http://cancergenome.nih.gov/ |
| Disease-related transcriptional perturbations, etc. | GEO | An archive of microarray, next-generation sequencing, and other forms of high-throughput functional genomic data submitted by the scientific community, covering a wide variety of experimental conditions including disease characterizations. | http://www.ncbi.nlm.nih.gov/geo/ |
| Phenotypic drug screen | NPC (NCGC) | Results of roughly 2,500 approved compounds screened in ~200 phenotypic and target-based assays, focusing on various cancers, malaria, nuclear receptors, and signaling pathways | http://tripod.nih.gov/npc/ |
| Phenotypic drug screen | PD2 | Results of nearly 2,500 approved compounds screened in 35 phenotypic assays covering 5 phenotypic modules (angiogenesis, Wnt potentiation, insulin secretion, GLP-1 secretion, and KRAS) | https://ncats.nih.gov/expertise/preclinical/pd2 |
| Drug-disease associations | Pharos | Resource connecting drugs, targets, and diseases | https://pharos.nih.gov/idg/index |
| Drug-disease associations | ClinicalTrials | A registry and results database of publicly and privately supported clinical studies of human participants conducted a round the world | https://clinicaltrials.gov/ |
| Drug-side effect associations | SIDER | Information from public documents and package inserts on marketed compounds and their recorded ADEs, including side effect frequency | http://sideeffects.embl.de/ |
| Drug-side effect associations | Offsides | Side effects and ADEs not listed on FDA's official drug label | http://tatonettilab.org/resources/tatonetti-stm.html |
| Drug-side effect associations | FAERS | Adverse event and medication error reports submitted to FDA | http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ |
| Signature-matching repositioning pipeline | DvD (Drug vs. Disease) | An R/Cytoscape pipeline providing dynamic access to public gene expression repositories and enabling drug/disease comparisons | www.ebi.ac.uk/saezrodriguez/DVD/ |
Acknowledgements
The authors would like to acknowledge the efforts of Carmen Lopez for her design expertise in helping to create Figures 1, 2, 4, 5, and 8.
Funding source:
This work was funded by the following grants from National Institutes of Health (NIH): Illuminating the Druggable Genome (IDG), Knowledge Management Center sponsored by NIH Common Fund, National Cancer Institute (NCI, U54-CA189201-02) and National Center for Advancing Translational Sciences (NCATS, UL1TR000067) Clinical and Translational Science Award (CTSA).
Footnotes
MedDRA®, the Medical Dictionary for Regulatory Activities terminology, is the international medical terminology developed under the auspices of the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH).
Conflicts of Interest Disclosures
Corresponding author Joel T. Dudley owns equity in NuMedii Inc., Ayasdi Inc., and LAM Therapeutics.
Contributor Information
Rachel A Hodos, New York University and Icahn School of Medicine at Mt. Sinai, New York, NY.
Brian A Kidd, Icahn School of Medicine at Mt. Sinai, New York, NY.
Shameer Khader, Icahn School of Medicine at Mt. Sinai, New York, NY.
Ben P Readhead, Icahn School of Medicine at Mt. Sinai, New York, NY.
Joel T Dudley, Icahn School of Medicine at Mt. Sinai, New York, NY.
References
- 1.Horrobin DF. Realism in drug discovery—could Cassandra be right? Nature biotechnology. 2001;19:1099–1100. doi: 10.1038/nbt1201-1099. [DOI] [PubMed] [Google Scholar]
- 2.Pammolli F, Magazzini L, Riccaboni M. The productivity crisis in pharmaceutical R&D. Nat Rev Drug Discov. 2011;10:428–438. doi: 10.1038/nrd3405. [DOI] [PubMed] [Google Scholar]
- 3.Cressey D. Traditional drug-discovery model ripe for reform. Nature. 2011;471:17–18. doi: 10.1038/471017a. [DOI] [PubMed] [Google Scholar]
- 4.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nature reviews Drug discovery. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 5.Kaitin KI. Deconstructing the drug development process: the new face of innovation. Clin Pharmacol Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barratt MJ, Frail DE. Drug repositioning: Bringing new life to shelved assets and existing drugs. John Wiley & Sons; 2012. [Google Scholar]
- 7.Arrowsmith J, Miller P. Trial watch: phase II and phase III attrition rates 2011-2012. Nat Rev Drug Discov. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 8.Landis IKaJ. Can the pharmaceutical industry reduce attrition rates? Nature Reviews Drug Discovery. 2004;3:711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 9.NORD National Organization for Rare Disorders (NORD) Available at: https://www.rarediseases.org/rare-disease-information.
- 10.Denis A, Mergaert L, Fostier C, Cleemput I, Simoens S. A comparative study of European rare disease and orphan drug markets. Health Policy. 2010;97:173–179. doi: 10.1016/j.healthpol.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Bellazzi R, Diomidous M, Sarkar IN, Takabayashi K, Ziegler A, McCray AT. Data analysis and data mining: current issues in biomedical informatics. Methods of information in medicine. 2011;50:536. doi: 10.3414/ME11-06-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, Guyer M, Green ED. The National Institutes of Health's Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. Journal of the American Medical Informatics Association. 2014;21:957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilar S, Uriarte E, Santana L, Lorberbaum T, Hripcsak G, Friedman C, Tatonetti NP. Similarity-based modeling in large-scale prediction of drug-drug interactions. Nature protocols. 2014;9:2147–2163. doi: 10.1038/nprot.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Žitnik M, Janjić V, Larminie C, Zupan B, Pržulj N. Discovering disease-disease associations by fusing systems-level molecular data. Scientific reports. 2013:3. doi: 10.1038/srep03202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, Agarwal P. Systematic drug repositioning based on clinical side-effects. PLoS One. 2011;6:e28025. doi: 10.1371/journal.pone.0028025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Wang F, Hu J, Sorrentino R. AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2013. Exploring the relationship between drug side-effects and therapeutic indications. [PMC free article] [PubMed] [Google Scholar]
- 17.Napolitano F, Zhao Y, Moreira VM, Tagliaferri R, Kere J, D'Amato M, Greco D. Drug repositioning: a machine-learning approach through data integration. J. Cheminformatics. 2013;5:30. doi: 10.1186/1758-2946-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.English SB, Butte AJ. Evaluation and integration of 49 genome-wide experiments and the prediction of previously unknown obesity-related genes. Bioinformatics. 2007;23:2910–2917. doi: 10.1093/bioinformatics/btm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleakley K, Yamanishi Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics. 2009;25:2397–2403. doi: 10.1093/bioinformatics/btp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H, Takigawa I, Mamitsuka H, Zhu S. Similarity-based machine learning methods for predicting drug–target interactions: a brief review. Briefings in bioinformatics. 2014;15:734–747. doi: 10.1093/bib/bbt056. [DOI] [PubMed] [Google Scholar]
- 21.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 22.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 23.Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, Zhou W, Huang J, Tang Y. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS computational biology. 2012;8:e1002503. doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilar S, Ryan PB, Madigan D, Stang PE, Schuemie MJ, Friedman C, Tatonetti NP, Hripcsak G. Similarity-based modeling applied to signal detection in pharmacovigilance. CPT Pharmacometrics Syst Pharmacol. 2014;3:e137. doi: 10.1038/psp.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye H, Liu Q, Wei J. Construction of drug network based on side effects and its application for drug repositioning. PloS one. 2014;9:e87864. doi: 10.1371/journal.pone.0087864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang AP, Butte AJ. Systematic evaluation of drug-disease relationships to identify leads for novel drug uses. Clin Pharmacol Ther. 2009;86:507–510. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, Wang F, Hu J. AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2014. Towards drug repositioning: a unified computational framework for integrating multiple aspects of drug similarity and disease similarity. [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. Journal of chemical information and computer sciences. 1988;28:31–36. [Google Scholar]
- 30.McNaught A. The iupac international chemical identifier. Chemistry international. 2006:12–14. doi: 10.1186/s13321-015-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Boyle NM. Towards a Universal SMILES representation-A standard method to generate canonical SMILES based on the InChI. J. Cheminformatics. 2012;4:22. doi: 10.1186/1758-2946-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue L, Godden JW, Stahura FL, Bajorath J. Design and evaluation of a molecular fingerprint involving the transformation of property descriptor values into a binary classification scheme. Journal of chemical information and computer sciences. 2003;43:1151–1157. doi: 10.1021/ci030285+. [DOI] [PubMed] [Google Scholar]
- 33.Durant JL, Leland BA, Henry DR, Nourse JG. Reoptimization of MDL keys for use in drug discovery. Journal of chemical information and computer sciences. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 34.Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Annual reports in computational chemistry. 2008;4:217–241. [Google Scholar]
- 35.Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Krüger FA, Light Y, Mak L, McGlinchey S. The ChEMBL bioactivity database: an update. Nucleic acids research. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yung-Chi C, Prusoff WH. Relationship between the inhibition constant (K i) and the concentration of inhibitor which causes 50 per cent inhibition (I 50) of an enzymatic reaction. Biochemical pharmacology. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 37.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic acids research. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic acids research. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Dayan MM, MacCarthy T, Schlecht NF, Belbin TJ, Childs G, Smith RV, Prystowsky MB, Bergman A. Cancer as the Disintegration of Robustness: Population-Level Variance in Gene Expression Identifies Key Differences Between Tobacco-and HPV-Associated Oropharyngeal Carcinogenesis. Archives of pathology & laboratory medicine. 2015 doi: 10.5858/arpa.2014-0624-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu C-L, Juan H-F, Huang H-C. Functional Analysis and Characterization of Differential Coexpression Networks. Scientific reports. 2015:5. doi: 10.1038/srep13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 42.Duan Q, Flynn C, Niepel M, Hafner M, Muhlich JL, Fernandez NF, Rouillard AD, Tan CM, Chen EY, Golub TR. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic acids research. 2014:gku476. doi: 10.1093/nar/gku476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lab Ma. [November 25, 2015];BD2K-LINCS-DCIC Crowdsourcing Portal. Available at: http://www.maayanlab.net/crowdsourcing.
- 45.Cheng J, Xie Q, Kumar V, Hurle M, Freudenberg JM, Yang L, AGARWAL P. Pac Symp Biocomput. World Scientific; 2013. Evaluation of analytical methods for connectivity map data. [PubMed] [Google Scholar]
- 46.Cheng J, Yang L, Kumar V, Agarwal P. Systematic evaluation of connectivity map for disease indications. Genome Medicine. 2014;6:540. doi: 10.1186/s13073-014-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nature Reviews Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 49.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic acids research. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swinney D. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clinical Pharmacology & Therapeutics. 2013;93:299–301. doi: 10.1038/clpt.2012.236. [DOI] [PubMed] [Google Scholar]
- 52.Zheng W, Thorne N, McKew JC. Phenotypic screens as a renewed approach for drug discovery. Drug discovery today. 2013;18:1067–1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA. PubChem's BioAssay database. Nucleic acids research. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang J, Xiao J, Zhang J, Bryant SH. An overview of the PubChem BioAssay resource. Nucleic acids research. 2010;38:D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JA, Shinn P, Jaken S, Oliver S, Willard FS, Heidler S, Peery RB, Oler J, Chu S, Southall N. Novel Phenotypic Outcomes Identified for a Public Collection of Approved Drugs from a Publicly Accessible Panel of Assays. PloS one. 2015;10:e0130796. doi: 10.1371/journal.pone.0130796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang R, Southall N, Wang Y, Yasgar A, Shinn P, Jadhav A, Nguyen D-T, Austin CP. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Science translational medicine. 2011;3:80ps16–80ps16. doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costello JC, Heiser LM, Georgii E, Gönen M, Menden MP, Wang NJ, Bansal M, Hintsanen P, Khan SA, Mpindi J-P. A community effort to assess and improve drug sensitivity prediction algorithms. Nature biotechnology. 2014;32:1202–1212. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Organization WH. Guidelines for ATC classification and DDD assignment. World Health Organization; 1996. [Google Scholar]
- 60. [September 28, 2015];DrugBank ATC classification browser. Available at: http://www.drugbank.ca/atc.
- 61. [November 8, 2015];2015AA UMLS National Drug File - Reference Terminology Source Information. Available at: https://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/NDFRT/index.html.
- 62.Lipscomb CE. Medical subject headings (MeSH). Bulletin of the Medical Library Association. 2000;88:265. [PMC free article] [PubMed] [Google Scholar]
- 63.Liu S, Ma W, Moore R, Ganesan V, Nelson S. RxNorm: prescription for electronic drug information exchange. IT professional. 2005;7:17–23. [Google Scholar]
- 64.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Research. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Research. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. [September 28, 2015];Pharos. Available at: https://pharos.nih.gov/idg/index.
- 67.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. Pharmacogenomics Knowledge for Personalized Medicine. Clinical pharmacology and therapeutics. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Food U, Administration D. Orange book: approved drug products with therapeutic equivalence evaluations. US FDA; Silver Spring, MD: 2010. [Google Scholar]
- 69.Administration FaD [November 8, 2015];FDALabel: Full-Text Search of Drug Labeling. Available at: http://www.fda.gov/ScienceResearch/BioinformaticsTools/ucm289739.htm - Issues.
- 70.Martínez V, Navarro C, Cano C, Fajardo W, Blanco A. DrugNet: network-based drug-disease prioritization by integrating heterogeneous data. Artificial Intelligence in Medicine. 2015 doi: 10.1016/j.artmed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 71.Health NIo [October 6, 2015];ClinicalTrials.gov. Available at: ClinicalTrials.gov.
- 72.Schriml LM, Arze C, Nadendla S, Chang Y-WW, Mazaitis M, Felix V, Feng G, Kibbe WA. Disease Ontology: a backbone for disease semantic integration. Nucleic acids research. 2012;40:D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Molecular systems biology. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tatonetti NP, Patrick PY, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Science translational medicine. 2012;4:125ra131–125ra131. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou H, Gao M, Skolnick J. Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Scientific reports. 2015:5. doi: 10.1038/srep11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meng X-Y, Zhang H-X, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Current computer-aided drug design. 2011;7:146. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng T, Li Q, Zhou Z, Wang Y, Bryant SH. Structure-based virtual screening for drug discovery: a problem-centric review. The AAPS journal. 2012;14:133–141. doi: 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang S-Y. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug discovery today. 2010;15:444–450. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 80.Krüger DM, Evers A. Comparison of Structure-and Ligand-Based Virtual Screening Protocols Considering Hit List Complementarity and Enrichment Factors. ChemMedChem. 2010;5:148–158. doi: 10.1002/cmdc.200900314. [DOI] [PubMed] [Google Scholar]
- 81.Chen Z, Li H-l, Zhang Q-j, Bao X-g, Yu K-q, Luo X-m, Zhu W-l, Jiang H-l. Pharmacophore-based virtual screening versus docking-based virtual screening: a benchmark comparison against eight targets. Acta Pharmacologica Sinica. 2009;30:1694–1708. doi: 10.1038/aps.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alvarez J, Shoichet B. Virtual screening in drug discovery. CRC press; 2005. [Google Scholar]
- 83.Gao M, Skolnick J. A comprehensive survey of small-molecule binding pockets in proteins. 2013 doi: 10.1371/journal.pcbi.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 85.Nidhi, Glick M, Davies JW, Jenkins JL. Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogenomics databases. Journal of chemical information and modeling. 2006;46:1124–1133. doi: 10.1021/ci060003g. [DOI] [PubMed] [Google Scholar]
- 86.Jacob L, Vert J-P. Protein-ligand interaction prediction: an improved chemogenomics approach. Bioinformatics. 2008;24:2149–2156. doi: 10.1093/bioinformatics/btn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bock JR, Gough DA. Virtual screen for ligands of orphan G protein-coupled receptors. Journal of chemical information and modeling. 2005;45:1402–1414. doi: 10.1021/ci050006d. [DOI] [PubMed] [Google Scholar]
- 88.Cao D-S, Liang Y-Z, Deng Z, Hu Q-N, He M, Xu Q-S, Zhou G-H, Zhang L-X, Deng Z, Liu S. Genome-scale screening of drug-target associations relevant to Ki using a chemogenomics approach. PloS one. 2013;8:e57680. doi: 10.1371/journal.pone.0057680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu H, Sun J, Guan J, Zheng J, Zhou S. Improving compound–protein interaction prediction by building up highly credible negative samples. Bioinformatics. 2015;31:i221–i229. doi: 10.1093/bioinformatics/btv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramsundar B, Kearnes S, Riley P, Webster D, Konerding D, Pande V. Massively Multitask Networks for Drug Discovery. arXiv preprint arXiv:1502.02072. 2015 [Google Scholar]
- 91.Dahl GE, Jaitly N, Salakhutdinov R. Multi-task neural networks for QSAR predictions. arXiv preprint arXiv:1406.1231. 2014 [Google Scholar]
- 92.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bottini AA, Hartung T. Food for Thought on the Economics of Animal Testing. Altex. 2009;26:3–16. doi: 10.14573/altex.2009.1.3. [DOI] [PubMed] [Google Scholar]
- 94.Jucker M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nature medicine. 2010;16:1210–1214. doi: 10.1038/nm.2224. [DOI] [PubMed] [Google Scholar]
- 95.Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. American journal of translational research. 2014;6:114. [PMC free article] [PubMed] [Google Scholar]
- 96.Lounkine E, Keiser M, Whitebread S, Mikhailov D, Hamon J, Jenkins J, Lavan P, Weber E, Doak A, Côté S, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, Murino L, Tagliaferri R, Brunetti-Pierri N, Isacchi A, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proceedings of the National Academy of Sciences. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lum PY, Derry JM, Schadt EE. Integrative genomics and drug development. 2009 doi: 10.2217/14622416.10.2.203. [DOI] [PubMed] [Google Scholar]
- 99.Yom-Tov E, Gabrilovich E. Postmarket drug surveillance without trial costs: discovery of adverse drug reactions through large-scale analysis of web search queries. Journal of medical Internet research. 2013:15. doi: 10.2196/jmir.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.White RW, Tatonetti NP, Shah NH, Altman RB, Horvitz E. Web-scale pharmacovigilance: listening to signals from the crowd. Journal of the American Medical Informatics Association. 2013;20:404–408. doi: 10.1136/amiajnl-2012-001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bender A, Scheiber J, Glick M, Davies JW, Azzaoui K, Hamon J, Urban L, Whitebread S, Jenkins JL. Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. ChemMedChem. 2007;2:861–873. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- 102.Azzaoui K, Hamon J, Faller B, Whitebread S, Jacoby E, Bender A, Jenkins JL, Urban L. Modeling promiscuity based on in vitro safety pharmacology profiling data. ChemMedChem. 2007;2:874–880. doi: 10.1002/cmdc.200700036. [DOI] [PubMed] [Google Scholar]
- 103.Zhou H, Skolnick J. FINDSITEcomb: A threading/structure-based, proteomic-scale virtual ligand screening approach. Journal of chemical information and modeling. 2012;53:230–240. doi: 10.1021/ci300510n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif J-C, Waters DD. Effects of torcetrapib in patients at high risk for coronary events. New England journal of medicine. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 105.Chang RL, Xie L, Xie L, Bourne PE, Palsson BØ. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. 2010 doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim HU, Sohn SB, Lee SY. Metabolic network modeling and simulation for drug targeting and discovery. Biotechnol J. 2012;7:330–342. doi: 10.1002/biot.201100159. [DOI] [PubMed] [Google Scholar]
- 107.Fan S, Geng Q, Pan Z, Li X, Tie L, Pan Y, Li X. Clarifying off-target effects for torcetrapib using network pharmacology and reverse docking approach. BMC systems biology. 2012;6:152. doi: 10.1186/1752-0509-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scheiber J, Jenkins JL, Sukuru SCK, Bender A, Mikhailov D, Milik M, Azzaoui K, Whitebread S, Hamon J, Urban L. Mapping adverse drug reactions in chemical space. Journal of medicinal chemistry. 2009;52:3103–3107. doi: 10.1021/jm801546k. [DOI] [PubMed] [Google Scholar]
- 109.Liu M, Cai R, Hu Y, Matheny ME, Sun J, Hu J, Xu H. Determining molecular predictors of adverse drug reactions with causality analysis based on structure learning. Journal of the American Medical Informatics Association. 2014;21:245–251. doi: 10.1136/amiajnl-2013-002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koch U, Hamacher M, Nussbaumer P. Cheminformatics at the interface of medicinal chemistry and proteomics. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2014;1844:156–161. doi: 10.1016/j.bbapap.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 111.Glicksberg BS, Li L, Cheng WY, Shameer K, Hakenberg J, Castellanos R, Ma M, Shi L, Shah H, Dudley JT, et al. An integrative pipeline for multi-modal discovery of disease relationships. Pac Symp Biocomput. 2015;20:407–418. [PMC free article] [PubMed] [Google Scholar]
- 112.Piro RM. Network medicine: linking disorders. Human genetics. 2012;131:1811–1820. doi: 10.1007/s00439-012-1206-y. [DOI] [PubMed] [Google Scholar]
- 113.Dudley JT, Deshpande T, Butte AJ. Exploiting drug–disease relationships for computational drug repositioning. Briefings in bioinformatics. 2011:bbr013. doi: 10.1093/bib/bbr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews Drug discovery. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 115.Elvidge S. Getting the drug repositioning genie out of the bottle. Life Science Leader. 2010 [Google Scholar]
- 116.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. In silico repositioning of approved drugs for rare and neglected diseases. Drug discovery today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 117.Muthyala R. Orphan/rare drug discovery through drug repositioning. Drug Discovery Today: Therapeutic Strategies. 2011;8:71–76. [Google Scholar]
- 118.Sardana D, Zhu C, Zhang M, Gudivada RC, Yang L, Jegga AG. Drug repositioning for orphan diseases. Briefings in bioinformatics. 2011;12:346–356. doi: 10.1093/bib/bbr021. [DOI] [PubMed] [Google Scholar]
- 119.Molineris I, Ala U, Provero P, Di Cunto F. Drug repositioning for orphan genetic diseases through Conserved Anticoexpressed Gene Clusters (CAGCs). BMC bioinformatics. 2013;14:288. doi: 10.1186/1471-2105-14-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chavali AK, Blazier AS, Tlaxca JL, Jensen PA, Pearson RD, Papin JA. Metabolic network analysis predicts efficacy of FDA-approved drugs targeting the causative agent of a neglected tropical disease. BMC Syst Biol. 2012;6:27. doi: 10.1186/1752-0509-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu Z, Borlak J, Tong W. Deciphering miRNA transcription factor feed-forward loops to identify drug repurposing candidates for cystic fibrosis. Genome medicine. 2014;6:94. doi: 10.1186/s13073-014-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Andrews KT, Fisher G, Skinner-Adams TS. Drug repurposing and human parasitic protozoan diseases. International Journal for Parasitology: Drugs and Drug Resistance. 2014;4:95–111. doi: 10.1016/j.ijpddr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martínez V, Cano C, Blanco A. ProphNet: A generic prioritization method through propagation of information. BMC bioinformatics. 2014;15:S5. doi: 10.1186/1471-2105-15-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gormley GJ, Stoner E, Bruskewitz RC, Imperato-McGinley J, Walsh PC, McConnell JD, Andriole GL, Geller J, Bracken BR, Tenover JS, et al. The effect of finasteride in men with benign prostatic hyperplasia. The Finasteride Study Group. N Engl J Med. 1992;327:1185–1191. doi: 10.1056/NEJM199210223271701. [DOI] [PubMed] [Google Scholar]
- 125.Ferry L, Johnston JA. Efficacy and safety of bupropion SR for smoking cessation: data from clinical trials and five years of postmarketing experience. Int J Clin Pract. 2003;57:224–230. [PubMed] [Google Scholar]
- 126.Wu P, Wilson K, Dimoulas P, Mills EJ. Effectiveness of smoking cessation therapies: a systematic review and meta-analysis. BMC Public Health. 2006;6:300. doi: 10.1186/1471-2458-6-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science translational medicine. 2011;3:96ra77–96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McArt DG, Zhang S-D. Identification of candidate small-molecule therapeutics to cancer by gene-signature perturbation in connectivity mapping. PloS one. 2011;6:e16382. doi: 10.1371/journal.pone.0016382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ramachandran S, Osterhaus SR, Karp PH, Welsh MJ, McCray PB., Jr A genomic signature approach to rescue ΔF508-cystic fibrosis transmembrane conductance regulator biosynthesis and function. American journal of respiratory cell and molecular biology. 2014;51:354–362. doi: 10.1165/rcmb.2014-0007OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hu G, Agarwal P. Human disease-drug network based on genomic expression profiles. PLoS One. 2009;4:e6536. doi: 10.1371/journal.pone.0006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Iorio F, Rittman T, Ge H, Menden M, Saez-Rodriguez J. Transcriptional data: a new gateway to drug repositioning? Drug Discov Today. 2013;18:350–357. doi: 10.1016/j.drudis.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Science translational medicine. 2011;3:96ra76–96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen B, Greenside P, Paik H, Sirota M, Hadley D, Butte A. Relating Chemical Structure to Cellular Response: An Integrative Analysis of Gene Expression, Bioactivity, and Structural Data Across 11,000 Compounds. CPT: pharmacometrics & systems pharmacology. 2015;4:576–584. doi: 10.1002/psp4.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Iskar M, Zeller G, Blattmann P, Campillos M, Kuhn M, Kaminska KH, Runz H, Gavin AC, Pepperkok R, van Noort V. Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Molecular systems biology. 2013;9:662. doi: 10.1038/msb.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Parkkinen JA, Kaski S. Probabilistic drug connectivity mapping. BMC bioinformatics. 2014;15:113. doi: 10.1186/1471-2105-15-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yao L, Zhang Y, Li Y, Sanseau P, Agarwal P. Electronic health records: Implications for drug discovery. Drug Discovery Today. 2011:10. doi: 10.1016/j.drudis.2011.05.009. doi. [DOI] [PubMed] [Google Scholar]
- 139.Xu H, Aldrich MC, Chen Q, Liu H, Peterson NB, Dai Q, Levy M, Shah A, Han X, Ruan X. Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. Journal of the American Medical Informatics Association. 2014:amiajnl–2014-002649. doi: 10.1136/amiajnl-2014-002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maggiora GM. On outliers and activity cliffs why QSAR often disappoints. Journal of chemical information and modeling. 2006;46:1535–1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 141.Huang Y-F, Yeh H-Y, Soo V-W. Inferring drug-disease associations from integration of chemical, genomic and phenotype data using network propagation. BMC medical genomics. 2013;6:S4. doi: 10.1186/1755-8794-6-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chindelevitch L, Ziemek D, Enayetallah A, Randhawa R, Sidders B, Brockel C, Huang ES. Causal reasoning on biological networks: interpreting transcriptional changes. Bioinformatics. 2012;28:1114–1121. doi: 10.1093/bioinformatics/bts090. [DOI] [PubMed] [Google Scholar]
- 143.Schmidt BJ, Papin JA, Musante CJ. Mechanistic systems modeling to guide drug discovery and development. Drug Discov Today. 2013;18:116–127. doi: 10.1016/j.drudis.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 146.Argmann C, Dobrin R, Heikkinen S, Auburtin A, Pouilly L, Cock TA, Koutnikova H, Zhu J, Schadt EE, Auwerx J. Ppargamma2 is a key driver of longevity in the mouse. PLoS Genet. 2009;5:e1000752. doi: 10.1371/journal.pgen.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 148.Lum PY, Derry JM, Schadt EE. Integrative genomics and drug development. Pharmacogenomics. 2009;10:203–212. doi: 10.2217/14622416.10.2.203. [DOI] [PubMed] [Google Scholar]
- 149.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- 150.Pinto JP, Machado RS, Xavier JM, Futschik ME. Targeting molecular networks for drug research. Front Genet. 2014;5:160. doi: 10.3389/fgene.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chang RL, Xie L, Xie L, Bourne PE, Palsson BO. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput Biol. 2010;6:e1000938. doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fan S, Geng Q, Pan Z, Li X, Tie L, Pan Y, Li X. Clarifying off-target effects for torcetrapib using network pharmacology and reverse docking approach. BMC Syst Biol. 2012;6:152. doi: 10.1186/1752-0509-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li Z, Wang RS, Zhang XS. Two-stage flux balance analysis of metabolic networks for drug target identification. BMC Syst Biol. 2011;5(Suppl 1):S11. doi: 10.1186/1752-0509-5-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang B, Horvath S. A General Framework for Weighted Gene Co-Expression Network Analysis. Statistical Applications in Genetics and Molecular Biology. 2005;4 doi: 10.2202/1544-6115.1128. Available at: // http://www.degruyter.com/view/j/sagmb.2005.4.issue-1/sagmb.2005.4.1.1128/sagmb.2005.4.1.1128.xml. [DOI] [PubMed] [Google Scholar]
- 156.Barabási A-L. Scale-free networks: a decade and beyond. science. 2009;325:412. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 157.Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Daly R, Shen Q, Aitken S. Learning Bayesian networks: approaches and issues. The knowledge engineering review. 2011;26:99–157. [Google Scholar]
- 160.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, GuhaThakurta D, Sieberts SK, Monks S, Reitman M, Zhang C. An integrative genomics approach to infer causal associations between gene expression and disease. Nature genetics. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sima C, Hua J, Jung S. Inference of Gene Regulatory Networks Using Time-Series Data: A Survey. Current Genomics. 2009;10:416–429. doi: 10.2174/138920209789177610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dai W, Liu X, Gao Y, Chen L, Song J, Chen D, Gao K, Jiang Y, Yang Y, Chen J. Matrix Factorization-Based Prediction of Novel Drug Indications by Integrating Genomic Space. Computational and Mathematical Methods in Medicine. 2015:2015. doi: 10.1155/2015/275045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cobanoglu MC, Liu C, Hu F, Oltvai ZN, Bahar I. Predicting Drug–Target Interactions Using Probabilistic Matrix Factorization. Journal of Chemical Information and Modeling. 2013;53:3399–3409. doi: 10.1021/ci400219z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li R, Dong Y, Kuang Q, Wu Y, Li Y, Zhu M, Li M. Inductive matrix completion for predicting adverse drug reactions (ADRs) integrating drug–target interactions. Chemometrics and Intelligent Laboratory Systems. 2015;144:71–79. [Google Scholar]
- 165.Bell RM, Koren Y, Volinsky C. The BellKor solution to the Netflix prize. 2007 [Google Scholar]
- 166.Töscher A, Jahrer M. The bigchaos solution to the netflix prize 2008. Netflix Prize, Report. 2008 [Google Scholar]
- 167.Piotte M, Chabbert M. The pragmatic theory solution to the netflix grand prize. Netflix prize documentation. 2009 [Google Scholar]
- 168.Bennett J, Lanning S. The netflix prize. Proceedings of KDD cup and workshop. 2007 [Google Scholar]
- 169.Chi EC, Zhou H, Chen GK, Del Vecchyo DO, Lange K. Genotype imputation via matrix completion. Genome research. 2013;23:509–518. doi: 10.1101/gr.145821.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Saporito MS, Lipinski CA, Reaume AG. Phenotypic in vivo screening to identify new, Unpredicted indications for existing drugs and drug candidates. In: Barratt MJ, Frail DE, editors. Drug repositioning: Bringing new life to shelved assets and existing drugs. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2012. pp. 253–290. [Google Scholar]
- 171.Ho JC, Ghosh J, Steinhubl SR, Stewart WF, Denny JC, Malin BA, Sun J. Limestone: High-throughput candidate phenotype generation via tensor factorization. Journal of biomedical informatics. 2014;52:199–211. doi: 10.1016/j.jbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li L, Cheng W-Y, Glicksberg BS, Gottesman O, Tamler R, Chen R, Bottinger EP, Dudley JT. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Science translational medicine. 2015;7:311ra174–311ra174. doi: 10.1126/scitranslmed.aaa9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes, Brain and Behavior. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]