To the Editor—Persistent or chronic infection is an essential issue for pathogen-host interaction, disease, and antibiotic-resistant emergence. Persistent carriers are of special public health concern as they represent the reservoirs for spreading the “evolved” pathogen of drug-resistant. In current issue, Marzel et al presents an interesting study to explore the causes and consequences of patients with Salmonella persistent infection [1], albeit defects in study designs and results interpretation.
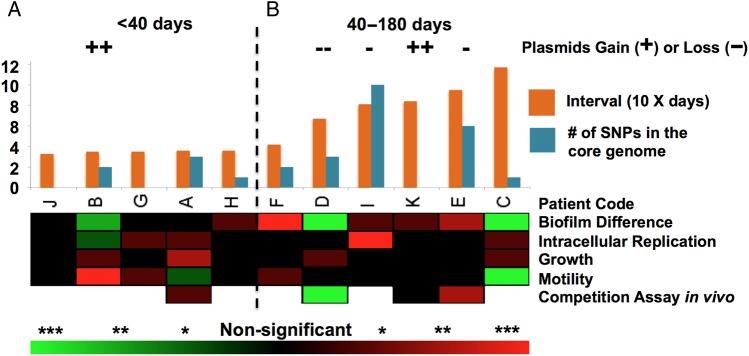
The authors showed limited genetic mutations or variations in Salmonella for persistent infection, and parallel mutations were also not identified between eleven patients. These “negative” results are due to 2 major factors: the short-read method for genomic sequencing and short interval sampling between the persistent strains. The short-read technique proposed by the authors has the power in investigating single nucleotide polymorphisms or indel, although it has limitation to study large structure variations and plasmids. Here, plasmids play an important role for phenotypic changes since multiple gain-or-loss events are observed. However, the plasmids information is underappreciated by using short-read method. The short interval for bacterial sampling, spanning 1–6 months, is another limiting factor. Established genetic mutations in persistent bacteria were usually traced for years, even decades, to be detected [2–5]. Accordingly, an accumulation of genetic variations were identified mostly for persistent strains with interval over 40 days (Figure 1). Moreover, the phenotypic changes between the pair strains for each patient showed no convergent fitness, including biofilm formation, motility, replication abilities within macrophage, and in vivo competition (Figure 1) [1]. These confounding results pose a sharp contrast with strains that had parallel phenotypic and genetic fitness during year interval studies [2–4]. The bacteria within-host substitution rate is around 10−7 per year per site [3], and phenotypic fitness is usually required years; thus it is not surprising that neither fitness mutation nor convergent phenotype were observed in the current investigation.
Figure 1.
Salmonella enterica serovar Typhimurium genetic and phenotypic dynamics during persistent infection. A, Accumulation of genetic mutation, including number of single-nucleotide polymorphisms (SNPs) in the core genome, and variations, such as large genomic elements gain or loss, are correlated with interval time of the isolates. In general, patient samplings interval within 40 days have less genetic changes than samples of interval days over 40. B, The summary results for the phenotypic assays between early and later persistent isolates for each patient. The heatmap shows phenotypic changes between the early and later isolates, red indicates later isolate show an increasing phenotypic fitness when comparing with early isolate, whereas green suggests a decreasing phenotypic fitness when comparing with early isolate. The color density represents if the change between early and later persistent strain is statistically different. *P < .05; **P < .01; ***P < .001.
Additional concerns are regarding bacterial competition assay in a murine infection model. The proposed C3H/HeNHsd mice of 7–8 weeks (adult-age) have the robust immune system and are resistant to infection, even by virulent Salmonella strains. This model cannot resemble current study focusing suckling-age patients (9 out of 11 less than 1 year-old). Furthermore, the pretreated streptomycin model can deplete the ability for Salmonella outgrowth the normal gut microbiota [6], which reduces the sensitivity in current model. Indeed, 2-week-old mice, without antibiotic pretreatment, is the matched infection model for the current study, although younger mice infected by Salmonella usually leads to systemic or lethal infection due to immature microbiota [7, 8]. This information raises the expectation for the role of microbiota in establishing Salmonella persistent infection.
I believe the rational study design is the key to investigate heterogeneous patient of persistent infection, and new evidence, including antibiotic treated intestine showing distinct disease syndromes for Salmonella infections, exposes the knowledge gap on how microbiota regulate different disease pathway including persistent infection [8–10]. Thus, an improved understanding of 3-way interaction in pathogen-host-microbiota axis is critical to develop therapeutic approaches to eliminate pathogen reservoir in patients with persistent infection.
Notes
Financial support. (Grants: NIH: AI098041; USDA: 2013-67015-21285), the University of Pennsylvania Veterinary Center for Infectious Disease, and the Center for Host-Microbial Interactions.
Potential conflict of interest. Author certifies no potential conflicts of interest. The author has submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Marzel A, Desai TP, Goren A et al. Persistent infections by nontyphoidal Salmonella in humans: epidemiology and genetics. Clin Infect Dis 2016; 62:879–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klemm EJ, Gkrania-Klotsas E, Hadfield J et al. Emergence of host-adapted Salmonella enteritidis through rapid evolution in an immunocompromised host. Nat Microbiol 2016; doi:10.1038/nmicrobiol.2015.23. [DOI] [PubMed] [Google Scholar]
- 3.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genetics 2015; 47:57–64. [DOI] [PubMed] [Google Scholar]
- 4.Price EP, Sarovich DS, Mayo M et al. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. mBio 2013; 4:e00388–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golubchik T, Batty EM, Miller RR et al. Within-host evolution of Staphylococcus aureus during asymptomatic carriage. PLoS ONE 2013; 8:e61319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winter SE, Thiennimitr P, Winter MG et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010; 467:426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J Exp Med 2004; 199:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stecher B, Paesold G, Barthel M et al. Chronic Salmonella enterica serovar Typhimurium-induced colitis and cholangitis in streptomycin-pretreated Nramp1+/+ mice. Infect Immun 2006; 74:5047–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiennimitr P, Winter SE, Bäumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012; 15:108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam LH, Monack DM. Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PLoS Pathog 2014; 10:e1004527. [DOI] [PMC free article] [PubMed] [Google Scholar]