Abstract
Background Living organisms are continuously confronted with perturbations, such as environmental changes that include fluctuations in temperature and nutrient availability, or genetic changes such as mutations. While some developmental systems are affected by such challenges and display variation in phenotypic traits, others continue consistently to produce invariable phenotypes despite perturbation. This ability of a living system to maintain an invariable phenotype in the face of perturbations is termed developmental robustness. Biological robustness is a phenomenon observed across phyla, and studying its mechanisms is central to deciphering the genotype–phenotype relationship. Recent work in yeast, animals and plants has shown that robustness is genetically controlled and has started to reveal the underlying mechinisms behind it.
Scope and Conclusions Studying biological robustness involves focusing on an important property of developmental traits, which is the phenotypic distribution within a population. This is often neglected because the vast majority of developmental biology studies instead focus on population aggregates, such as trait averages. By drawing on findings in animals and yeast, this Viewpoint considers how studies on plant developmental robustness may benefit from strict definitions of what is the developmental system of choice and what is the relevant perturbation, and also from clear distinctions between gene effects on the trait mean and the trait variance. Recent advances in quantitative developmental biology and high-throughput phenotyping now allow the design of targeted genetic screens to identify genes that amplify or restrict developmental trait variance and to study how variation propagates across different phenotypic levels in biological systems. The molecular characterization of more quantitative trait loci affecting trait variance will provide further insights into the evolution of genes modulating developmental robustness. The study of robustness mechanisms in closely related species will address whether mechanisms of robustness are evolutionarily conserved.
Keywords: Developmental robustness, species diversity, canalization, insensitivity, plasticity, sensitivity, variance, genotype × phenotype, perturbation, cryptic genetic variation, buffering.
INTRODUCTION
Perturbations represent a constant challenge to living organisms, so it is remarkable how biological systems often continue to produce stable phenotypes in spite of perturbations. Such a property of a biological system to cope with perturbations and ensure an invariant output in the presence of considerable noise is called robustness or insensitivity (Kitano, 2004; Wagner, 2007; Masel and Siegal, 2009). Robustness reflects the degree of variation in biological systems, so it is important for system evolution. As such, it can be very relevant to plant breeding, which aims at uniformity of phenotypic traits. It is also important in medicine since many diseases can be studied in the context of loss of robustness of normal physiological states (Kitano, 2007a). Therefore, understanding the mechanisms and consequences of robustness represents a fundamental problem in biology in general.
Developmental robustness is defined as the state of reduced phenotypic variation under a given perturbation. Low phenotypic variation can be found, for example, among individuals in a population or among different cells and tissues within the same individual. Developmental robustness therefore refers to an observable property that can be quantified. Related terms, such as canalization or developmental buffering, mostly referring to the processes via which robustness is achieved, are often used interchangeably in the literature (Debat and David, 2001). Although the study of robustness has lately attracted a lot of attention (Kitano, 2007b; Masel and Siegal, 2009), the realization of this phenomenon is not new in biology and stems from extending the notion of physiological homeostasis to developmental processes (Debat and David, 2001). C. H. Waddington, who coined the term canalization, first observed ‘that the wild-type of an organism, that is to say, the form which occurs in nature under the influence of natural selection, is much less variable in appearance than the majority of mutant races’ (Waddington, 1942). Waddington famously illustrated the concept of robustness by showing development depicted as a ball rolling down a slope in well-defined grooves, from which it is difficult to displace the developmental process (Waddington, 1957). However, the molecular mechanisms ensuring the consistency of developmental events are only little understood. Recent advances in developmental biology for precise perturbations and phenotyping, including techniques for single-cell manipulations and monitoring, offer unprecedented possibilities for experimental, mechanistic studies of robustness (Raj and van Oudenaarden, 2009; Schmidt et al., 2011)
Robustness and sensitivity represent the extremes of a continuum of possible responses to perturbations. Levels of developmental robustness may differ between animals and plants, with plants being perhaps more sensitive, for example, to environmental signals due to their extended period of post-embryonic morphogenesis and sessile nature. However, the same principles apply when it comes to defining developmental robustness and studying the contribution of genes to stabilization of developmental outcomes. We discuss here some robustness lessons from various other systems, focusing on their potential implications to understanding phenotypic buffering in plants.
CHOOSING THE SYSTEM AND PERTURBATION: ROBUSTNESS ‘OF WHAT’… ‘TO WHAT’
To study biological robustness experimentally, one needs first to define precisely two parameters: what is the system of choice and what is the perturbation the system is facing (Felix and Wagner, 2008). For the case of developmental robustness, the answer to the question ‘what is robust’ lies in specifying what is the developmental trait of interest. For example, a relevant trait could be any quantifiable phenotype related to the development of an organism and at any possible level including gross morphology, cellular characteristics of a given system (i.e. cell fate patterns or number of cells), molecular measurements (i.e protein or gene expression levels), physiological features or behaviour. The trait of interest may be the final phenotype of a system (also known as ‘output’ phenotype) or any intermediate ‘endophenotype’ that can be quantified during the developmental process. It is therefore possible for a given output to be robust despite considerable variation in intermediate processes.
To answer the question ‘to what the system is robust’ one needs to define the exact nature of the perturbation. Perturbations can be internal for an organism or external (Masel and Siegal, 2009). Internal perturbations include uncontrollable stochastic or microenvironmental variation, for instance in the concentration of proteins or variation in gene expression levels among individuals or cells, and genetic variation upon heritable mutation accumulation and recombination. External perturbations are commonly environmental changes such as temperature, nutrient concentration, humidity and photoperiod that are likely to affect growth and development. These different sources of perturbations ultimately lead us to distinguish the types of developmental robustness as microenvironmental, environmental and genetic robustness.
So what makes a good model system to study developmental robustness? A leading paradigm from animal studies over the last few years is the simple cell fate pattern of the nematode vulva, which is an egg-laying and copulatory organ (Fig. 1) (Felix and Barkoulas, 2012). There are many reasons why nematodes, and the vulva in particular, represent good experimental models of developmental robustness. First, robustness studies rely on differences in trait variance and therefore require large sample sizes in order to compare distributions, and Caenorhabditis elegans nematodes are amenable to large sample phenotyping. Secondly, C. elegans strains are nearly isogenic due to their hermaphroditic reproductive mode, which eliminates the confounding effect of mixing together different perturbations, for example background genetic variation and environmental variation when studying robustness. This is in stark contrast to non-isogenic model systems, where each individual has a unique genotype and therefore separating the effects of different types of perturbations on developmental robustness can be a daunting task. Lastly, the nematode C. elegans is the only animal for which the entire cell lineage is known (Sulston and Horvitz, 1977) and the development is highly stereotypical, allowing precise characterization of developmental defects with high accuracy and single-cell resolution. Furthermore, vulval cell fate development in particular has been studied for 30 years now and it is a textbook example of animal tissue patterning. Pursuing questions about the mechanisms of robustness can be greatly facilitated by prior knowledge about the molecular underpinnings of a developmental system and its gene network topology. Although we will not go into details of the actual molecular players of the vulval network, we need to introduce, for the purposes of this review, that the master inducer of vulval patterning is an epidermal growth factor (EGF)-like molecule that is secreted by the dorsally located anchor cell of the somatic gonad (Fig. 1A; Felix and Barkoulas, 2012).
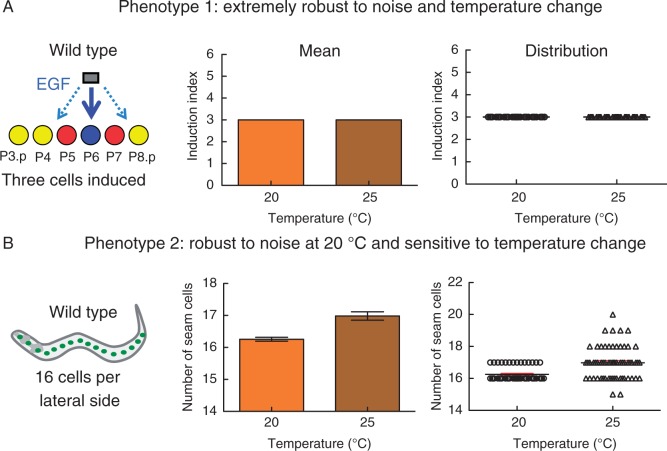
Fig. 1.
Comparison of developmental robustness between two Caenorhabditis elegans nematode tissues. (A) Vulval development in C. elegans involves induction of only three cells (P5.p–P7.p) out of a group of six competent cells, by adopting one of the two following cell fates; the primary cell fate, which is acquired by P6.p (depicted in blue), and the secondary fate, which is adopted by P5.p and P7.p (depicted in red). The remaining cells P3.p, P4.p and P8.p (in yellow) act as a back-up system and can only be induced if P5.p–P7.p fail to do so. An EGF-like molecule is the main inducer of the vulva. It is secreted from the anchor cell of the somatic gonad, which is located dorsally to the Pn.p cells. The average number of induced cells in the population (three in the wild type) can be used as a way to quantify the phenotype (phenotype 1). The induction index is robust to stochastic noise both at 20 °C (standard growth temperature) and at 25 °C. (B) Seam cells are epithelial stem cell-like cells in C. elegans. The number of cells (16 per lateral side) can be used as a quantitative phenotype, and this number is robust at 20 °C but not at 25 °C (phenotype 2). However, seam cell number at 20 °C is less robust compared with the vulval induction index (note the increase in standard error in seam cell number). Note the difference in information that can be inferred from analysing the mean (panels in the centre) and phenotypic distribution (right-hand panels). For example, phenotype 2 distribution is wider at 25 °C, with a bias towards an increase in seam cell number. Data in both (A) and (B) come from our lab after phenotyping 50 animals of the lab reference strain N2.
Developmental robustness is a quantifiable trait, and studies in the vulva have shown that robustness can be high or low. However, robust systems are not infallible, so robustness has its limits. For example, the vulval cell fate pattern is highly robust to stochastic and environmental variation (Fig, 1A; Braendle and Felix, 2008) as well as to standing genetic variation, as nematodes which are genetically distant from, but related to, C. elegans all share the same vulval cell fate pattern (Felix, 2007). However, extensive cell fate phenotyping in the vulva showed that cell fate patterning errors do happen at very low frequency (usually <5 %). Furthermore, studies on the vulva showed that the degree of robustness depends on the exact nature of the perturbation, since the same C. elegans isolate (or ‘accession’ for plant biologists) can be more sensitive to one perturbation than to some others – for example the lab reference strain of C. elegans is more sensitive to food starvation than changes in growth temperature (Braendle and Felix, 2008). The type of developmental errors also depends on the genetic background, as different C. elegans isolates respond differently to distinct perturbations (Braendle and Felix, 2008). Different tissues or cells within an organism may show different degrees of robustness. For example, despite the stereotypical nematode cell patterning, vulval cell induction is more robust to stochastic noise than patterning of some other epithelial cells called seam cells (Fig. 1). This difference in robustness becomes even more pronounced when animals are grown at higher (25 °C) than standard (20 °C) temperature (Fig. 1B). It is of note that the more insensitive a system is, it does not necessarily mean that it is a better model for robustness and, depending on the biological question and approach, a marginal degree of sensitivity may also be beneficial.
Robustness can also be studied theoretically, using computational models that mimic a developmental process. Again, the phenotype of interest could be any developmental phenotype such as segmentation in flies, bacterial chemotaxis or vulval cell fate patterning (Barkai and Leibler, 1997; von Dassow et al., 2000; Ma et al., 2006; Hoyos et al., 2011). Computational models reinforce experimental results and make novel predictions about experimental outcomes. For example, leaf margin patterning in Arabidopsis thaliana relies on PINFORMED1 (PIN1)-mediated auxin maxima that appear sequentially along the margin of the growing leaf. A computational model of leaf margin development predicted that the CUP-SHAPED COTYLEDON2 (CUC2) transcription factor stabilizes these maxima, allowing the consistent generation of leaf margin serrations (Bilsborough et al., 2011). Mathematical models can have different degrees of complexity and abstraction. They often include parameters, such as half-lives of mRNAs or binding rates, whose values are randomly set as they are experimentally hard to quantify. Robustness in this case refers to model output performance upon parameter change over a certain range. For example, a 40-parameter model reconstituting the known topology of the vulval gene network and explaining some key experimental results was challenged to a ten-fold variation in model parameters. It was shown that the model is robust to changes in many but not all parameters, with one point of sensitivity being variation in EGF synthesis (Hoyos et al., 2011). Such theoretical approaches can generate novel experimental predictions about the underlying basis of developmental robustness.
In plants, there is no single system on which scientists have focused efforts in order to quantify systematically robustness to various perturbations. Similar to the nematode vulva, quantifications can be performed for any developmental phenotype of interest such as organ number (i.e. rosette leaf, cauline leaf, flower and branches), organ or tissue size and architecture (i.e. plant height, hypocotyl length and rosette diameter) or developmental timing traits (i.e. flowering time) (Pouteau et al., 2004; Fu et al., 2009). Due to the plastic nature of plants, most of these phenotypes are likely to be more responsive to perturbations and thus more variable than nematode cell patterning. For example, flowering time is sensitive to fluctuating environmental cues, such as the seasonality of flowering times, which is affected by the photoperiod, light intensity and temperature changes acting through the circadian clock (Samach and Coupland, 2000). However, some phenotypes in plants are thought to be quite invariable. One example is the number of cotyledons, which in angiosperms is either one or two, with little variance, and pleiocotyly is a developmental deviant pattern that is rare to find (Conner and Agrawal, 2005). Another example is petal or sepal number in arabidopsis that also shows very little variance (Sieber et al., 2007). We anticipate that advances in high-throughput and automated phenotyping in plants will increase systematic and comparative quantification of developmental phenotypes in different genetic backgrounds and under various perturbations (Tisne et al., 2013; Yang et al., 2013).
MEAN VS. VARIANCE OF DEVELOPMENTAL TRAITS AND TWO-SIDED PHENOTYPIC ERRORS
Most of modern developmental biology is dominated by mean-centric approaches, where phenotypic averages are compared but details about phenotypic distributions in populations are usually ignored (Geiler-Samerotte et al., 2013). However, phenotypic distributions hide valuable information about the individuals of the population. Phenotypic heterogeneity is in fact abundant even within genetically identical individuals (Eldar et al., 2009; Burga et al., 2011), and in many cases this heterogeneity may be critical for cell differentiation, patterning and species evolution (Eldar and Elowitz, 2010; Johnston and Desplan, 2010; Balazsi et al., 2011). For example, bacterial heterogeneity in growth allows some cells to survive antibiotic treatment (Bishop et al., 2007), and growth heterogeneity also contributes to chemoresistance in tumours (Roesch et al., 2010; Sharma et al., 2010). In plants, considerable variability in growth has been found in the leaf epidermis and the meristem (Elsner et al., 2012; Kierzkowski et al., 2012; Uyttewaal et al., 2012), and cell heterogeneity and anisotropic growth were shown in another study to correlate with sepal growth (Schiessl et al., 2012). Cell to cell heterogeneities often arise from stochasticity in gene expression, one example being photoreceptor choice of individual cone cells in mammals (Jacobs, 2009) or the monogenic expression of a single odorant receptor in olfactory sensory neurons (Magklara and Lomvardas, 2013). In arabidopsis, cell to cell variation in FLOWERING LOCUS C (FLC) expression due to silencing may act as a way to register epigenetic memory of cold exposure (Angel et al., 2011).
Analysing phenotypic distributions is very central to studying the genetics of robustness. We argue here that distinguishing between gene effects on trait mean and variance is essential for studying robustness (Fig. 2A). In developmental genetic terms, understanding the mechanisms of robustness entails identifying genes influencing phenotypic variance. So let us consider as a phenotypic example the length of the primary root. In contrast to the classical approach of isolating mutants showing a significant change in the average root length in the population, a targeted robustness screen would rather focus on identifying factors specifically affecting root length variance without affecting the trait mean (Fig. 2B). The main reason for ideally selecting against changes in mean is that mutants tend to be generally more variable than the wild type, as first postulated by (Waddington, 1942), so deviations from the wild-type mean are commonly accompanied by changes in variance. Genuine changes in variance are still possible when the trait mean is different. However, one would have to support that there is unexpectedly high variance for that given phenotypic mean. This involves studying in detail the relationship between mean and variance, which is phenotype dependent (Levy and Siegal, 2008).
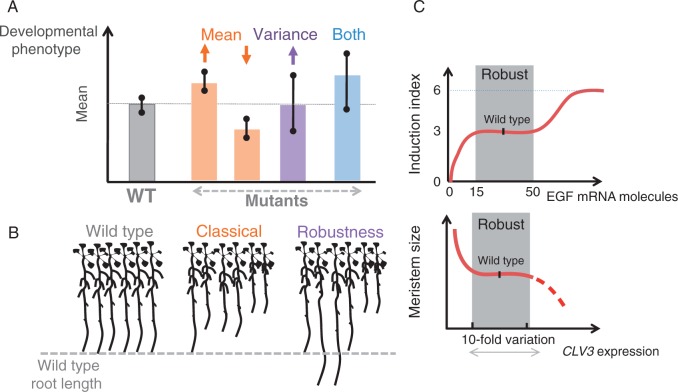
Fig. 2.
Defining robustness genes and robustness to gene expression change. (A) For any quantifiable phenotype, classical developmental mutants are defined herein as those displacing the mean, leading to either an increased or decreased mean, whereas robustness mutants as those increasing the phenotypic variance without much effect on the mean. In practice, the most common case is mutants that do both at the same time sensu Waddington. (B) An example showing a strict robustness defect using root length as the phenotype of interest. (C) Examples illustrating the effect of changing gene expression levels on two different developmental phenotypes: the nematodes’ vulval cell fate induction (upper panel) in response to EGF expression (presented by the number of mRNA molecules quantified in situ) showing that the induction index tolerates a change in expression ranging from 15 to 50 mRNA molecules (Barkoulas et al., 2013). Exposure to <15 mRNA molecules causes hypoinduction, whereas expression of >50 mRNA molecules causes hyperinduction. The lower panel illustrates the example of plant meristem size in response to change in expression level of CLAVATA3 (CLV3) relative to the wild type (100 %). Meristem size is shown to tolerate a ten-fold variation in CLV3 expression (from 33 % to 320 % that of the wild type) (Muller et al., 2006).
An increase in trait variance coupled with changes in the mean can simply arise due to partial penetrance or variable expressivity of mutations, and describing such defects in the light of robustness failure is a common problem in the recent literature. For example, in some robust systems, with one example being the nematode vulva, all mutations affecting developmental patterning show either partial penetrance or variable expressivity, so any change in the mean is intrinsically linked with a change in phenotypic variance (Barkoulas et al., 2013). Moreover, condition-dependent effects on trait mean such as environmental sensitivity of mutations are indicative of genotype × environment interactions, but not good evidence for loss of developmental robustness. Genotype × environment interactions are indeed a common finding, as demonstrated using the yeast knockout library in different culture environments (Hillenmeyer et al., 2008).
It is conceivable that a certain perturbation may lead to a wider phenotypic distribution but would maintain the phenotypic mean if it results in two-sided phenotypic errors within the population. Going back to the root example discussed above, a two-sided phenotypic error would mean that some individuals respond to this perturbation by showing an increase and some others a decrease in root length within the same genotype. Such two-sided errors can be used as a proxy for developmental robustness defects in genetic screens, and their developmental basis is interesting to understand in the context of loss of buffering. However, in some cases, developmental constraints may only allow one-sided phenotypic distributions.
PROBING THE GENETICS OF TRAIT VARIANCE
If developmental robustness evolves under natural selection, there should be loci in the genome that act as suppressors of phenotypic variation. As a consequence, inducing mutations in these particular loci should increase phenotypic variance to a given perturbation. Such trait variance controllers are often described in the literature as ‘phenotypic capacitors’ (Levy and Siegal, 2008) or ‘master regulators’ of robustness when several traits are affected (Lempe et al., 2013). From a developmental genetics point of view, questions arising from the notion of having robustness genes include (a) whether these genes can be found frequently experimentally or are rare; (b) what type of gene products they encode; (c) whether they are specific to the trait and source of variation; and (d) whether they harbour natural genetic variation that could explain within-species differences in phenotypic robustness. We discuss below how recent studies, through a combination of classical genetic screens and quantitative genetics in different systems, have just started providing some answers to these questions.
Deciphering the full spectrum and frequency of robustness genes would require unbiased and systematic screens to reveal all genetic factors shaping phenotypic variation. As argued above, these robustness screens should focus on discovering genes affecting specifically trait variance, rather than phenotypic means. Such screens have not yet been performed in plants or any other multicellular eukaryote, but have been carried out in Saccharomyces cerevisiae (Levy and Siegal, 2008; Rinott et al., 2011; Bauer et al., 2015).
There are a number of messages emerging from these studies in yeast. First, there are a large number of single genes that contribute to developmental robustness. This was anticipated by early theoretical work showing extensive loss of robustness in simulations of single mutants of evolved robust networks (Bergman and Siegal, 2003), predicting that single-gene disruption can be sufficient to result in a robustness breakdown. More recently, Levy and Siegal (2008) identified 300 yeast mutants in a yeast knockout library that exhibit reduced robustness of quantitative morphological markers to stochastic variation, and Rinott et al. (2011) characterized multiple genes buffering cell–cell stochastic variability using reporter gene expression as the phenotypic read-out. These conclusions are supported from studies in flies, where Takahashi used genomic deficiency lines to identify multiple genomic regions harbouring loci that are necessary to buffer wing shape to genetic variation and sensory bristles to environmental variation (Takahashi et al., 2012; Takahashi, 2013).
A second message emerging from the yeast studies involves the molecular identity of phenotypic capacitors, which share the characteristic of being part of highly connected nodes in cellular networks such as chromatin maintenance factors, cell cycle proteins, transcriptional regulators, and components of the stress response (Levy and Siegal, 2008; Rinott et al., 2011). Interestingly, some deletions in genes involved in clathrin-dependent vesicle transport or transcription regulation were found to decrease rather than increase the phenotypic variance in growth rate (Levy et al., 2012). This suggests that some loci in the genome may also act as variance amplifiers for certain phenotypes. A more recent study reported a high proportion of phenotypic stabilizers among essential genes (Bauer et al., 2015). Therefore, the molecular identity of these factors suggests that phenotypic capacitors may be broad regulators of cell homeostasis, buffering many developmental phenotypes at once as a result of their high connectivity with several other cellular gene networks. It is still unclear to what extent some tissue-specific components may also affect system robustness. One example comes from studies in flies where mutations in the transcription factor gene TAILESS have been shown to affect embryo to embryo variability in segmentation gene expression patterns (Janssens et al., 2013).
Single genes acting as phenotypic capacitors have also been identified through candidate gene approaches. The classical example is heat shock protein 90 (HSP-90), which is an ATP-dependent chaperone helping the maturation of a wide range of proteins through its association with various co-chaperones and cofactors (Whitesell and Lindquist, 2005). Impairment of HSP-90 function, first in flies and later in other organisms including plants, revealed a wide range of developmental abnormalities (Rutherford and Lindquist, 1998; Queitsch et al., 2002; Samakovli et al., 2007; Sangster et al., 2008). HSP-90 may contribute to system robustness to standing genetic variation or noise either directly through association with mutant interactors such as kinases, ubiquitin ligases and transcription factors, or indirectly by regulating the activity of signal transduction pathways. However, developmental abnormalities upon HSP-90 inhibition have been mostly studied in a qualitative rather than a quantitative way in the literature, so it is unclear in many cases whether HSP-90 impairment affects specifically trait variance (see Rohner et al., 2013 for a recent exception to this). Another single-gene disruption in plants related to robustness involves the circadian clock regulator EARLY FLOWERING 4 (ELF4). In this case, two-sided phenotypic errors in the circadian clock period have been found in elf4 mutants in arabidopsis (Doyle et al., 2002).
A class of genes thought to be key players in developmental robustness is micro-RNAs (miRNAs) (Hornstein and Shomron, 2006). miRNAs are post-transcriptional regulators of gene expression in both animals and plants (Bartel, 2009; Rubio-Somoza and Weigel, 2011). They function by tuning the expression levels of their target genes, setting up sharp developmental boundaries of differential gene expression. They also participate in feedback and feedforward loops within developmental networks buffering the stochastic expression of their target genes (Wu et al., 2009; Siciliano et al., 2011). One example in arabidopsis involves the mir164 family: mir164abc triple mutants show increased variance in stem internode size as a consequence of derepressing CUC1 and CUC2 gene expression (Sieber et al., 2007). However, miRNAs are often considered as robustness factors simply based on the condition-dependent developmental defects of many miRNA mutants, for example showing phenotypes specifically in one environment and not in another. Once again, we argue that distinguishing the effects on mean and variance is very important in order to determine on a case by case basis whether miRNAs are indeed regulators of developmental robustness sensu stricto.
In plants, the distinction between gene effects on trait mean and variance has mostly been discussed so far in the context of natural variation in robustness. Quantitative trait locus (QTL) mapping approaches have proved successful in identifying effects on trait variance, not just the mean (Weller et al., 1988), and QTL studies and genome-wide association studies have since been pursued in plants including maize and arabidopsis to identify loci affecting specifically trait variance upon a given perturbation (Hall et al., 2007; Ordas et al., 2008; Jimenez-Gomez et al., 2011; Shen et al., 2012). Phenotypes of choice include gross plant morphology, metabolite profiling or gene expression, and the most common perturbation is microenvironmental or environmental variation. Similar to lab-induced mutations, a main message emerging from these studies is that multiple independent genomic regions contribute to natural variation in trait variance, with some of these regions affecting at the same time the trait mean and variance, while others act specifically on one or the other (Hall et al., 2007; Ordas et al., 2008; Jimenez-Gomez et al., 2011). Additionally, some QTLs were found to affect sensitivity in many different phenotypes or to act as ‘hotspots’ in the genome, underlying system-wide buffering of many phenotypic traits (Hall et al., 2007; Fu et al., 2009). However, in very few cases have the causative alleles for trait variance been identified down to the nucleotide level.
One example is a QTL identified for variance of rosette leaf number under long-day photoperiods between the two most widely used A. thaliana accessions, Columbia and Landsberg erecta. This QTL maps closely to the ERECTA (ER) gene, a member of the leucine-rich repeat/receptor-like protein kinases, which has pleiotropic functions in plant development (van Zanten et al., 2009). Interestingly, the effect of ER in this case was found to be allele specific, with only one of the available mutations in ER reproducing the variance defect (Hall et al., 2007). Another example is genetic variation in the ELF3 gene, another core component of the circadian clock and protein interactor of ELF4, which was found to affect differences between the Bayreuth and Shahdara A. thaliana accessions for sensitivity to noise in many phenotypes. Interestingly, the Shahdara ELF3 allele affects noise in a context-dependent manner, increasing trait variance for some phenotypes and decreasing variance for others. This suggests that the exact direction of the effects for genes influencing trait variance may be phenotype dependent. Another good example from yeast revealed the molecular identity of loci controlling trait variance in natural populations. Using five isogenic yeast strains, it was shown that loci related to uracil metabolism and sensing the environment buffer cell to cell stochastic variation in GFP (green fluorescent protein) reporter gene expression (Ansel et al., 2008; Fehrmann et al., 2013).
Findings from single-gene perturbations support the idea that phenotypic robustness can be genotype specific, especially when studying related or interconnected traits (Bauer et al., 2015). However, robustness can also be trait specific. Extensive phenotyping of cell morphology and intracellular organization in wild yeast isolates revealed that most strains show trait-specific noise variation, although some strains can be globally variable for many phenotypes. The genetic diversity of the globally variable strains suggested multiple evolutionary transitions to high global variance under different ecological pressures (Yvert et al., 2013)
MECHANISMS AND EVOLUTION
The identification of single genes buffering developmental phenotypes raises the question of how robustness is mechanistically achieved. Developmental robustness is linked to functional redundancy, which ensures trait stability in the face of perturbations by providing back-up opportunities for a given system. Redundancy in biological systems can be found at many different levels, such as in cells, genes and regulatory elements (Wagner, 2007). For example, in the developmental context of the vulva, a common phenotypic error is mis-centring of the anchor cell above the P5.p cell upon environmental variation, whereas normally the anchor cell is located above P6.p. However, three competent cells P(3,4,8).p provide back-up cell redundancy, and such mis-centring is buffered without leading to phenotypic consequences. Gene redundancy can provide mutational robustness when a gene duplicate can substitute for a mutated paralogue or when gene duplicates show different sensitivities to environmental factors such as temperature (Hsiao and Vitkup, 2008; Keane et al., 2014). Further focusing down at the nucleotide level, redundancy of regulatory elements such as transcriptional enhancers ensures insensitivity of gene expression programmes to macroenvironmental perturbations (Frankel et al., 2010; Perry et al., 2010)
The mechanistic basis of robustness lies not only in redundant parts, but also in the distribution and connections of parts within a system. In this case, several components of a system contribute to the flow of information and thus system function. Distributed robustness is very common in metabolic and developmental networks (Felix and Wagner, 2008). Network topology including feedback or feedforward regulatory loops and signalling pathway cross-talk are important for developmental robustness (Posadas and Carthew, 2014). In plants, multiple interconnected feedback loops are important for the stability of the circadian clock (Mas and Yanovsky, 2009). For example, work on the arabidopsis circadian clock showed that the feedback regulatory loop between the LIGHT-REGULATED WD1 (LWD1) and PSEUDO-RESPONSE REGULATOR9 (PRR9) is important for the robustness of the circadian rhythm, which is variable in lwd1;lwd2 double mutant plants under continuous dark conditions (Y. Wang et al., 2011). Feedback regulation has been shown in many different systems to result in threshold-like system behaviour and thus to increase output stability to stochastic, environmental and standing genetic variation (Becskei and Serrano, 2000; Ramsey et al., 2006; Shinar et al., 2007; Denby et al., 2012).
Phenotypic variance may be explained through variation in gene expression across individuals. Variable gene expression can arise upon many different genetic perturbations. For example, overexpression of the chromatin remodelling SWI/SNF2-type ATPase AtCHR23 in arabidopsis leads to increased variation in gene expression between individual plants (Folta et al., 2014). Continuous variation in gene expression may propagate as a bimodal output for another downstream gene, and this was shown to be the underlying basis of partial penetrance for some intestinal mutations in C. elegans (Raj et al., 2010). However, biological systems can be robust to a range of changes in gene dosage (Acar et al., 2010; Barkoulas et al., 2013). For example, vulva cell fate patterning is sensitive to changes in the level of EGF-like signalling and exhibits two distinct thresholds: one below which the vulva is underinduced and another above which the vulva is overinduced (Barkoulas et al., 2013). These boundaries of the robustness of cell induction to EGF expression variation were determined at single-molecule resolution by quantitative in situ hybridization in C. elegans (Fig. 2C; Barkoulas et al., 2013). In plants, Müller and colleagues addressed what is the range of variation in CLAVATA3 (CLV3) expression that the meristem can buffer without changing its size (Muller et al., 2006). The authors used CLV3 promoter deletion derivatives to modulate the expression levels of CLV3 and showed that shoot and flower meristem size is robust to a ten-fold change (Muller et al., 2006) (Fig. 2C).
Are changes in phenotypic variance adaptive? Developmental robustness is just an observable property, so a lack or low levels of phenotypic variation does not necessarily imply that this is the product of selection. It may arise neutrally because of non-linearity between parameters and phenotypic effects in biological systems resulting in robustness plateaux (Lynch, 2007) (Fig. 2C). It may also arise pleiotropically due to selection for another phenotype or due to selection for robustness to another perturbation. The latter is because it has been shown that, at least in some cases, there is similarity between the responses to two different types of variation. For example, alleles selected for environmental canalization may also be responsible for genetic canalization (Meiklejohn and Hartl, 2002). To address experimentally whether a certain phenotype is maintained under stabilizing selection in the lab, mutation accumulation lines are very useful, which are constructed in self-fertile or hermaphrodite species by continuing with a random single individual for many generations, thus minimizing the effect of selection. Such lines were used in C. elegans to show that the high degree of robustness of the vulval cell fate pattern is likely to be maintained under selection, as it rapidly breaks down upon random mutation accumulation (Braendle et al., 2010).
The genetic basis of trait variance suggests that natural selection may act to optimize phenotypic variation within a population. Is it better for a system to be robust or sensitive to perturbations? A high degree of developmental robustness and so low phenotypic variation in the population may in some cases be beneficial for it to withstand various perturbations. However, phenotypic plasticity and high phenotypic variation can also be key in order to cope with environmental challenges or spark evolutionary innovation. Therefore, depending on the phenotype of interest and the ecological circumstances, natural selection may act either to stabilize or to destabilize phenotypic traits. A recent example concerning gene expression compared the effects of natural polymorphisms in the promoter of the glucose metabolism gene TDH3 within 85 S. cerevisiae strains with those of random point mutations in this promoter (Metzger et al., 2015). This study suggested that selection on gene expression noise has had a greater impact on sequence variation than selection on mean expression levels, highlighting that purifying selection constrains variation in TDH3 expression among isogenic individuals (Metzger et al., 2015). It is important in the future to better link phenotypic variation with fitness. Phenotypic capacitors identified in genetic screens in yeast represent highly connected nodes in cellular networks and network hubs that are probably enriched for pleiotropic effects (Costanzo et al., 2010). This suggests that increased phenotypic variation in such mutant backgrounds may only come as a side effect due to a broader reduction in fitness (G. Z. Wang et al., 2011). This is not, however, a general conclusion since increased morphological variation in yeast was not found to correlate with a decrease in fitness (Bauer et al., 2015).
Robust systems are still adaptable and they do evolve by accumulating cryptic genetic variation (Paaby and Rockman, 2014). This is abundant genetic variation that is normally buffered, so it is silent at the phenotypic level, but can be revealed upon system perturbation such as experimental introgression of mutations or cell ablations (Milloz et al., 2008). For example, in the case of HSP-90-mediated buffering, functional impairment of this chaperone pharamcologically, or perhaps by temperature in the wild, leads to background-dependent pleiotropic defects in develepment (Rutherford and Lindquist, 1998). The release of cyptic genetic variation in the form of phenotypic variation can be enriched by selection, allowing adaptation to new environments (Rohner et al., 2013).
CONCLUSIONS
By drawing on findings in animals and yeast, we discuss here how studies on plant developmental robustness may benefit from strict definitions of what is the developmental system of choice and what is the relevant perturbation. They will also benefit from a clear distinction between gene effects on trait mean and trait variance. Such a distinction has been discussed in the context of plant quantitative genetics but very little in the plant development field. Recent advances in quantitative developmental biology and high-throughput phenotyping now allow the design of targeted genetic screens to identify genes amplifying or restricting developmental trait variance and study how variation propagates across different phenotypic levels in biological systems. The molecular characterization of more QTLs affecting trait variance will provide further insights into the evolution of genes modulating developmental robustness. The study of robustness mechanisms in closely related species will address whether mechanisms of robustness are evolutionarily conserved.
ACKNOWLEDGEMENTS
We thank Miltos Tsiantis and Jie Song for comment, and Marie-Anne Félix for valuable discussions. Research on developmental robustness in the Barkoulas lab is supported by a Biotechnology and Biological Sciences Research Council award (BB/L021455/1).
LITERATURE CITED
- Acar M, Pando BF, Arnold FH, Elowitz MB, van Oudenaarden A. 2010. A general mechanism for network–dosage compensation in gene circuits. Science 329: 1656–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M. 2011. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476: 105–108. [DOI] [PubMed] [Google Scholar]
- Ansel J, Bottin H, Rodriguez-Beltran C, et al. 2008. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genetics 4: e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, Collins JJ. 2011. Cellular decision making and biological noise: from microbes to mammals. Cell 144: 910–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkai N, Leibler S. 1997. Robustness in simple biochemical networks. Nature 387: 913–917. [DOI] [PubMed] [Google Scholar]
- Barkoulas M, van Zon JS, Milloz J, van Oudenaarden A, Felix MA. 2013. Robustness and epistasis in the C. elegans vulval signaling network revealed by pathway dosage modulation. Developmental Cell 24: 64–75. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CR, Li S, Siegal ML. 2015. Essential gene disruptions reveal complex relationships between phenotypic robustness, pleiotropy, and fitness. Molecular Systems Biology 11: 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Serrano L. 2000. Engineering stability in gene networks by autoregulation. Nature 405: 590–593. [DOI] [PubMed] [Google Scholar]
- Bergman A, Siegal ML. 2003. Evolutionary capacitance as a general feature of complex gene networks. Nature 424: 549–552. [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, et al. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proceedings of the National Academy of Sciences, USA 108: 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AL, Rab FA, Sumner ER, Avery SV. 2007. Phenotypic heterogeneity can enhance rare-cell survival in ‘stress-sensitive’ yeast populations. Molecular Microbiology 63: 507–520. [DOI] [PubMed] [Google Scholar]
- Braendle C, Felix MA. 2008. Plasticity and errors of a robust developmental system in different environments. Developmental Cell 15: 714–724. [DOI] [PubMed] [Google Scholar]
- Braendle C, Baer CF, Felix MA. 2010. Bias and evolution of the mutationally accessible phenotypic space in a developmental system. PLoS Genetics 6: e1000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burga A, Casanueva MO, Lehner B. 2011. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature 480: 250–253. [DOI] [PubMed] [Google Scholar]
- Conner JK, Agrawal AA. 2005. Mechanisms of constraints: the contributions of selection and genetic variance to the maintenance of cotyledon number in wild radish. Journal of Evolutionary Biology 18: 238–242. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, et al. 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. 2000. The segment polarity network is a robust developmental module. Nature 406: 188–192. [DOI] [PubMed] [Google Scholar]
- Debat V, David P. 2001. Mapping phenotypes: canalization, plasticity and developmental stability. Trends in Ecology and Evolution 16: 555–561. [Google Scholar]
- Denby CM, Im JH, Yu RC, Pesce CG, Brem RB. 2012. Negative feedback confers mutational robustness in yeast transcription factor regulation. Proceedings of the National Academy of Sciences, USA 109: 3874–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, et al. 2002. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77. [DOI] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. 2010. Functional roles for noise in genetic circuits. Nature 467: 167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Chary VK, Xenopoulos P, et al. 2009. Partial penetrance facilitates developmental evolution in bacteria. Nature 460: 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner J, Michalski M, Kwiatkowska D. 2012. Spatiotemporal variation of leaf epidermal cell growth: a quantitative analysis of Arabidopsis thaliana wild-type and triple cyclinD3 mutant plants. Annals of Botany 109: 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann S, Bottin-Duplus H, Leonidou A, et al. 2013. Natural sequence variants of yeast environmental sensors confer cell-to-cell expression variability. Molecular Systems Biology 9: 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA. 2007. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Current Biology 17: 103–114. [DOI] [PubMed] [Google Scholar]
- Felix MA, Barkoulas M. 2012. Robustness and flexibility in nematode vulva development. Trends in Genetics 28: 185–95. [DOI] [PubMed] [Google Scholar]
- Felix MA, Wagner A. 2008. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity (Edinburgh) 100: 132–140. [DOI] [PubMed] [Google Scholar]
- Folta A, Severing EI, Krauskopf J, et al. 2014. Over-expression of Arabidopsis AtCHR23 chromatin remodeling ATPase results in increased variability of growth and gene expression. BMC Plant Biology 14: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJ, Bouwmeester H, et al. 2009. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nature Genetics 41: 166–167. [DOI] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Bauer CR, Li S, Ziv N, Gresham D, Siegal ML. 2013. The details in the distributions: why and how to study phenotypic variability. Current Opinion in Biotechnology 24: 752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Dworkin I, Ungerer MC, Purugganan M. 2007. Genetics of microenvironmental canalization in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104: 13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. 2006. Canalization of development by microRNAs. Nature Genetics 38: S20–S24. [DOI] [PubMed] [Google Scholar]
- Hoyos E, Kim K, Milloz J, et al. 2011. Quantitative variation in autocrine signaling and pathway crosstalk in the Caenorhabditis vulval network. Current Biology 21: 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao TL, Vitkup D. 2008. Role of duplicate genes in robustness against deleterious human mutations. PLoS Genetics 4: e1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH. 2009. Evolution of colour vision in mammals. Philosophical Transactions of the Royal Society B: Biological Sciences 364: 2957–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens H, Crombach A, Wotton KR, et al. 2013. Lack of tailless leads to an increase in expression variability in Drosophila embryos. Developmental Biology 377: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Gomez JM, Corwin JA, Joseph B, Maloof JN, Kliebenstein DJ. 2011. Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genetics 7: e1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Desplan C. 2010. Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annual Review of Cell and Developmental Biology 26: 689–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane OM, Toft C, Carretero-Paulet L, Jones GW, Fares MA. 2014. Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae. Genome Research 24: 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzkowski D, Nakayama N, Routier-Kierzkowska AL, et al. 2012. Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335: 1096–1099. [DOI] [PubMed] [Google Scholar]
- Kitano H. 2004. Biological robustness. Nature Reviews Genetics 5: 826–837. [DOI] [PubMed] [Google Scholar]
- Kitano H. 2007a. A robustness-based approach to systems-oriented drug design. Nature Reviews Drug Discovery 6: 202–210. [DOI] [PubMed] [Google Scholar]
- Kitano H. 2007b. Towards a theory of biological robustness. Molecular Systems Biology 3: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J, Lachowiec J, Sullivan AM, Queitsch C. 2013. Molecular mechanisms of robustness in plants. Current Opinion in Plant Biology 16: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. 2008. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biology 6: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. 2012. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biology 10: e1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The evolution of genetic networks by non-adaptive processes. Nature Reviews Genetics 8: 803–813. [DOI] [PubMed] [Google Scholar]
- Ma WZ, Lai LH, Qi OY, Tang C. 2006. Robustness and modular design of the Drosophila segment polarity network. Molecular Systems Biology 2: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Lomvardas S. 2013. Stochastic gene expression in mammals: lessons from olfaction. Trends in Cell Biology 23: 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Yanovsky MJ. 2009. Time for circadian rhythms: plants get synchronized. Current Opinion in Plant Biology 12: 574–579. [DOI] [PubMed] [Google Scholar]
- Masel J, Siegal ML. 2009. Robustness: mechanisms and consequences. Trends in Genetics 25: 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL. 2002. A single mode of canalization. Trends in Ecology and Evolution 17: 468–473. [Google Scholar]
- Metzger BP, Yuan DC, Gruber JD, Duveau F, Wittkopp PJ. 2015. Selection on noise constrains variation in a eukaryotic promoter. Nature 521: 344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milloz J, Duveau F, Nuez I, Felix MA. 2008. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes and Development 22: 3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. 2006. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. The Plant Cell 18: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordas B, Malvar RA, Hill WG. 2008. Genetic variation and quantitative trait loci associated with developmental stability and the environmental correlation between traits in maize. Genetics Research 90: 385–395. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Rockman MV. 2014. Cryptic genetic variation: evolution’s hidden substrate. Nature Reviews Genetics 15: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Current Biology 20: 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posadas DM, Carthew RW. 2014. MicroRNAs and their roles in developmental canalization. Current Opinion in Genetics and Development 27: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau S, Ferret V, Gaudin V, et al. 2004. Extensive phenotypic variation in early flowering mutants of Arabidopsis. Plant Physiology 135: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. 2009. Single-molecule approaches to stochastic gene expression. Annual Review of Biophysics 38: 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. 2010. Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey SA, Smith JJ, Orrell D, et al. 2006. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nature Genetics 38: 1082–1087. [DOI] [PubMed] [Google Scholar]
- Rinott R, Jaimovich A, Friedman N. 2011. Exploring transcription regulation through cell-to-cell variability. Proceedings of the National Academy of Sciences, USA 108: 6329–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, et al. 2010. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, et al. 2013. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 342: 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D. 2011. MicroRNA networks and developmental plasticity in plants. Trends in Plant Science 16: 258–264. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342. [DOI] [PubMed] [Google Scholar]
- Samach A, Coupland G. 2000. Time measurement and the control of flowering in plants. Bioessays 22: 38–47. [DOI] [PubMed] [Google Scholar]
- Samakovli D, Thanou A, Valmas C, Hatzopoulos P. 2007. Hsp90 canalizes developmental perturbation. Journal of Experimental Botany 58: 3513–3524. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Lee HN, et al. 2008. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 105: 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K, Kausika S, Southam P, Bush M, Sablowski R. 2012. JAGGED controls growth anisotropy and coordination between cell size and cell cycle during plant organogenesis. Current Biology 22: 1739–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U. 2011. Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biology 9: e1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Pettersson M, Ronnegard L, Carlborg O. 2012. Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genetics 8: e1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinar G, Milo R, Martinez MR, Alon U. 2007. Input output robustness in simple bacterial signaling systems. Proceedings of the National Academy of Sciences, USA 104: 19931–19935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D. 2011. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nature Communications 4: 2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. 2007. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental Biology 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Takahashi KH. 2013. Multiple capacitors for natural genetic variation in Drosophila melanogaster. Molecular Ecology 22: 1356–1365. [DOI] [PubMed] [Google Scholar]
- Takahashi KH, Okada Y, Teramura K. 2012. Deficiency screening for genomic regions with effects on environmental sensitivity of the sensory bristles of Drosophila melanogaster. Evolution 66: 2878–2890. [DOI] [PubMed] [Google Scholar]
- Tisne S, Serrand Y, Bach L, et al. 2013. Phenoscope: an automated large-scale phenotyping platform offering high spatial homogeneity. The Plant Journal 74: 534–544. [DOI] [PubMed] [Google Scholar]
- Uyttewaal M, Burian A, Alim K, et al. 2012. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1957. The strategy of the genes. George Allen & Unwin. [Google Scholar]
- Wagner A. 2007. Robustness and evolvability in living systems. Princeton, NJ: Princeton University Press. [Google Scholar]
- Wang GZ, Liu JA, Wang W, Zhang HY, Lercher MJ. 2011. A gene’s ability to buffer variation is predicted by its fitness contribution and genetic interactions. PLoS One 6: e17650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, Wu SH. 2011. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. The Plant Cell 23: 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JI, Soller M, Brody T. 1988. Linkage analysis of quantitative traits in an interspecific cross of tomato (Lycopersicon esculentum×Lycopersicon pimpinellifolium) by means of genetic-markers. Genetics 118: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. 2005. HSP90 and the chaperoning of cancer. Nature Reviews Cancer 5: 761–772. [DOI] [PubMed] [Google Scholar]
- Wu CI, Shen Y, Tang T. 2009. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Research 19: 734–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Duan L, Chen G, Xiong L, Liu Q. 2013. Plant phenomics and high-throughput phenotyping: accelerating rice functional genomics using multidisciplinary technologies. Current Opinion in Plant Biology 16: 180–187. [DOI] [PubMed] [Google Scholar]
- Yvert G, Ohnuki S, Nogami S, et al. 2013. Single-cell phenomics reveals intra-species variation of phenotypic noise in yeast. BMC Systems Biology 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Snoek LB, Proveniers MC, Peeters AJ. 2009. The many functions of ERECTA. Trends in Plant Science 14: 214–218. [DOI] [PubMed] [Google Scholar]