Abstract
Background and Aims Brassicaceae is one of the most diversified families in the angiosperms. However, most species from this family exhibit a very similar floral bauplan. In this study, we explore the Brassicaceae floral morphospace, examining how corolla shape variation (an estimation of developmental robustness), integration and disparity vary among phylogenetically related species. Our aim is to check whether these floral attributes have evolved in this family despite its apparent morphological conservation, and to test the role of pollinators in driving this evolution.
Methods Using geometric morphometric tools, we calculated the phenotypic variation, disparity and integration of the corolla shape of 111 Brassicaceae taxa. We subsequently inferred the phylogenetic relationships of these taxa and explored the evolutionary lability of corolla shape. Finally, we sampled the pollinator assemblages of every taxon included in this study, and determined their pollination niches using a modularity algorithm. We explore the relationship between pollination niche and the attributes of corolla shape.
Key Results Phylogenetic signal was weak for all corolla shape attributes. All taxa had generalized pollination systems. Nevertheless, they belong to different pollination niches. There were significant differences in corolla shape among pollination niches even after controlling for the phylogenetic relationship of the plant taxa. Corolla shape variation and disparity was significantly higher in those taxa visited mostly by nocturnal moths, indicating that this pollination niche is associated with a lack of developmental robustness. Corolla integration was higher in those taxa visited mostly by hovering long-tongued flies and long-tongued large bees.
Conclusions Corolla variation, integration and disparity were evolutionarily labile and evolved very recently in the evolutionary history of the Brassicaceae. These floral attributes were strongly related to the pollination niche. Even in a plant clade having a very generalized pollination system and exhibiting a conserved floral bauplan, pollinators can drive the evolution of important developmental attributes of corolla shape.
Keywords: corolla shape, phenotypic integration, canalization, robustness, plant–pollinator interactions, Brassicaceae, floral morphospace, geometric morphometrics, phenotypic disparity
INTRODUCTION
Phenotypic integration is the coordinated variation of morphological traits within functional modules (Olson and Miller, 1958; Pigliucci and Preston, 2004). Trait covariation and phenotypic integration may be the consequences of natural selection acting to improve the functioning of those modules, a phenomenon called functional integration (Pigliucci, 2003; Armbruster et al., 2014; Klingenberg, 2014). Phenotypic integration may also be the mere consequence of architectural and developmental processes prompting covariation among traits (Herrera et al., 2002; Armbruster et al., 2014; Klingenberg, 2014).
Angiosperm flowers are complex structures that perform an essential function in plants, namely reproduction (Thomson, 1983; Harder and Barrett, 2006; Willmer, 2011). In animal-pollinated plants, flowers are functional modules composed of integrated units that work together to attract pollinators and boost their pollination effectiveness. Their efficacy in enhancing plant reproduction depends on the coordinated functioning of their elements (Córdoba and Cocucci, 2011). Due to its direct association with plant fitness, it is widely assumed that floral integration has been optimized by selection (Berg, 1960; Armbruster et al., 2004; Ordano et al., 2008). Berg (1960), in her seminal study, showed that plant species with specialized pollination systems exhibit ‘correlation pleiades’ or modules of integrated traits. Following these ideas, subsequent studies have reported that self-compatible plants display weaker floral integration than self-incompatible species (Anderson and Busch, 2006), that species with specialized pollination exhibit greater floral integration than those with generalized pollination (Pérez et al., 2007; Rosas-Guerrero et al., 2011; Ellis et al., 2014; Gómez et al., 2014), and that the type of pollinators may determine the magnitude of phenotypic integration of the flowers (Pérez-Barrales et al., 2007, 2014; González et al., 2015). This plethora of studies indicates that pollinators may select not only for floral traits but also for floral integration (Nattero et al., 2011).
The functioning of integrated complex traits may be hindered by the occurrence of phenotypic variation and trait imprecision (Hansen et al., 2006; Young, 2006; Pélabon et al., 2012). Natural selection, being an optimizing mechanism, tends to increase the accuracy of complex traits by, among other sources, decreasing their variation and increasing their precision (Bell, 1997; Hansen et al., 2006). Although empirical evidence remains scarce, it seems that the magnitude of phenotypic variation in floral traits, a manifestation of the lack of developmental robustness, is in several plant species a consequence of their interaction with pollinators (Williams & Conner, 2001; Armbruster et al., 2009a, b; Rosas-Guerrero et al., 2011; Pélabon et al., 2012). A relaxation of the selection imposed by pollinators can cause an increase in the magnitude of floral variation (Galen, 1996; Williams and Conner, 2001).
An essential but still unsolved key question is how phenotypic variation and integration is expressed at a macroevolutionary scale (Geber, 2013; Goswami et al., 2014). In particular, it is still unknown whether these attributes affect the morphological disparity of the members of a given clade (Geber, 2013; Goswami et al., 2014). Morphological disparity, estimated as the phenotypic distinctness of a form in a given morphospace (Eble, 2004), is an estimate of the magnitude of the phenotypic divergences of taxa (Erwin, 2007). In fact, morphological disparity has been recently used to estimate floral morphological divergence (Chartier et al., 2014). Unfortunately, empirical information on the magnitude of variation of morphological disparity across plant taxa and how this variation is related to pollination remains scarce, although it would help to reveal how pollinators mediate the divergence in floral shapes.
The main goal of this study is to investigate the role of pollinators in the evolution of phenotypic integration, disparity and variation of the Brassicaceae corolla shape. Several features make Brassicaceae an especially useful plant family to investigate this question. Brassicaceae is one of the most widespread families worldwide. It comprises more than 3700 species, about 340 genera and 25 tribes, including economically important crops (e.g. Brassica, Raphanus, Eruca), weeds (e.g. Capsella, Lepidium, Sisymbrium, Thlaspi), ornamentals (e.g. Hesperis, Erysimum, Lobularia, Matthiola) and one of the most celebrated model species, Arabidopsis thaliana (L.) Heynh. (Al-Shehbaz et al., 2006; Warwick et al., 2006; Couvreur et al., 2010; Al-Shehbaz, 2012). Brassicaceae is considered a ‘model family’ for evolutionary developmental studies (Beilstein et al., 2008). Despite being one of the most diversified families in the angiosperms, most species exhibit a very conserved and distinctive flower displaying a cruciform corolla (i.e. four petals arranged in the form of a cross) (Heywood et al., 2007; Franzke et al., 2011). Nevertheless, it is possible to find departures in shape from this typical crucifer floral bauplan in some genera (Endress, 1992). For example in a few genera such as Teesdalia and Iberis the outer petals are radiate and larger than the inner petals, while the petals are absent in Pringlea and a few species from Lepidium and Coronopus (Heywood et al., 2007). Furthermore, not only have floral traits but also floral integration has been found to vary between Brassicaceae species in several genera, such Streptanthus, Brassica, Raphanus and Erysimum (Murren et al., 2002; Anderson and Busch, 2006; Penrod, 2010; Gómez et al., 2014). Another distinctive feature of Brassicaceae is its generalized pollination system. Most crucifers are pollinated by a high diversity of pollinators belonging to many disparate functional groups. Despite this generalized pollination system, both diversity and type of pollinators varies among species even within the same genus (Gómez et al., 2015). The specific goals of the current study are: (1) to quantify the morphological disparity, variation and integration of the corolla shape in Brassicaceae; (2) to examine how corolla shape variation, disparity and integration has evolved along the phylogeny of this family; and (3) to determine the role of pollinators in the evolution of these corolla shape attributes.
MATERIAL AND METHODS
Study species
We have studied 111 Brassicaceae species and subspecies belonging to 30 genera and 16 tribes (Supplementary Data, Table S1). We have included species belonging to all main phylogenetic lineages identified by Couvreur et al. (2010), in order to cover as much as possible the Brassicaceae evolutionary diversity. Their flowers were polysymmetric, disymmetric or monosymetric, as well as tetradynamous, with four inner and long stamens and two outer and short stamens located between the adaxial and abaxial petals. From each species, we have tried to study at least two populations located in different areas of their distribution range to avoid any local effect on our results, although this was not possible in all cases (Table S1).
Phylogenetic relationships among studied Brassicaceae
We inferred a phylogenetic hypothesis for the species included in our data set using a supermatrix approach (Bailey et al., 2006; Couvreur et al., 2010). This supermatrix was assembled by concatenating available GenBank sequences for nine commonly used markers in Brassicaceae phylogenetics (Table S2). First, we surveyed in GenBank for the most frequent markers for our species and two outgroup species from two families considered the closest relative to Brassicaceae: Cleome spinosa (Cleomaceae) and Capparis hastata (Capparaceae). We downloaded the sequences available for nine markers of these species: ITS and phyA of the nuclear genome, ndhF, matK, rbcL, intergenetic spacer trnL-trnF, intergenetic spacer trnT-trnL and intergenetic spacer rpl32-trnL of the plastid genome, and nad4 intron 1 of the mitochondrial genome. To minimize missing data, we concatenated sequences from different species of the same genus if evidence for monophyly of the genus existed in the literature (Table S2). This procedure has been previously followed in supermatrix approaches (Springer et al., 2004) as well as in phylogenetic inference of Brassicaceae evolutionary relationships (Couvreur et al., 2010).
All sequences were aligned using MAFFT (Katoh and Standley, 2013). Alignments were improved by removing poorly aligned or ambiguous positions. We trimmed all the alignments using ‘automated 1’ settings in trimAl software (Capella-Gutiérrez et al., 2009). We used the trimAl 1.3 version available at the Phylemon 2.0 server (http://phylemon.bioinfo.cipf.es, Sánchez et al., 2011). All trimmed alignments were concatenated resulting in a matrix of 10 534 sites and 87 taxa. This matrix was analysed using a maximum-likelihood (ML) approach as is implemented in RAxML 8 (Stamatakis, 2014). RAxML implements a very fast and efficient heuristic search and rapid bootstrap heuristic search. We used the RAxML web-server program available at the CIPRES portal (Miller et al., 2010) in which 1000 bootstrap replicates were performed. We kept the best ML phylogenetic hypothesis found by RAxML, collapsing those nodes with a bootstrap support lower than 75 %.
Some taxa in our data set did not have any sequence available at GenBank. We grafted these taxa to their genus when monophyly was previously confirmed. In those cases where monophyly was not confirmed, we looked for those species that the literature suggests as being the closest relative. These species were added to their genus using functions in R package ‘phytools’ (Revell, 2012).
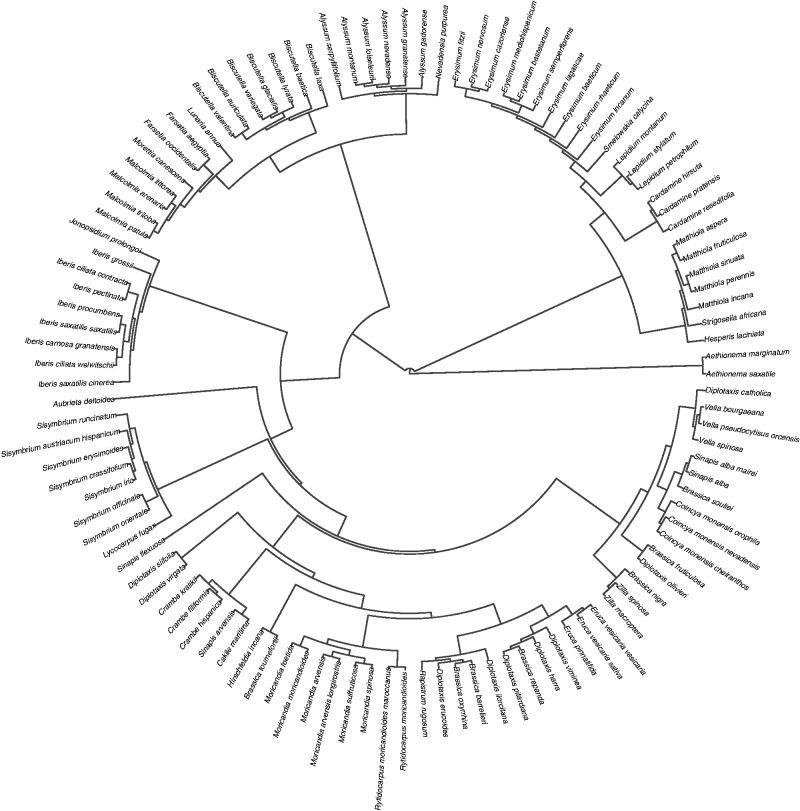
Our final tree presented some polytomies as a result of low bootstrap support for some clades, and also due to the procedure we followed to incorporate not previously sequenced species grafting them into their genus. We created a set of 100 trees in which polytomies were randomly resolved. All the analyses were performed under this set of trees, thus incorporating the phylogenetic uncertainty. In addition, we ultrametrized the consensus tree using the function compute.brlen from R package ‘ape’ (Paradis et al., 2004). Figure 1 shows the phylogenetic relationships between the taxa used in this study.
Fig. 1.
Phylogenetic relationships of the Brassicaceae taxa included in this study.
Corolla-shape variation, disparity and integration
Shape variation in the corolla of the Brassicaceae was studied by means of geometric morphometric tools using a landmark-based methodology (Zelditch et al., 2012). For this, we selected flowers at anthesis of each of 111 Brassicaceae species included in this study (totalling 7336 flowers; see Table S1 for information on sample size per species) and took a digital photograph of the front view and planar position. Intra-individual variation in corolla shape was reduced by taking the photo always at the same floral phenophase. We considered only flowers having unbent petals. However, some species recurrently display odd shapes with bent petals. We included these specimens because we were interested in the attractive function of corolla shape. In addition, we have previously shown in Erysimum mediohispanicum that intraspecific corolla shape variation is much lower than inter-individual and inter-population variation (Gómez and Perfectti, 2010), a pattern that it is probably shared by most Brassicaceae species.
We defined 32 co-planar landmarks covering the corolla shape and using midrib, primary and secondary veins and petal extremes and connections (Gómez and Perfectti, 2010). In Brassicaceae, adaxial and abaxial petals are the inner and outer petals, respectively (Busch and Zachgo, 2007). We identified the adaxial and abaxial petals of our study species by determining the relative position of the petals with respect to the flowering stalks and the location of the short stamens (Gómez and Perfectti, 2010). From the two-dimensional coordinates of landmarks, we extracted shape information and computed the generalized orthogonal least-squares Procrustes averages using the generalized Procrustes analysis (GPA) superposition method. We performed a principal component analysis (PCA) to explore variation in corolla shape across Brassicaceae species, obtaining the shape of the corollas at the ends of the range of variability along the first three principal components.
Corolla shape variation was estimated as the Procrustes variance of observations in each taxon (Young, 2006). Procrustes variance quantifies the average dispersion of data points around the mean shape. This metric measures the variety of forms of a taxon (Zelditch et al., 2012). Corolla shape variance is obtained as the sum of the variances across all coordinates in shape space (the trace of the covariance matrix) or, equivalently, the sum of all eigenvalues in the PCA. We used the intra-populational magnitude of shape variance, to avoid any effect due to intraspecific geographical variation in corolla shape. So, for each taxon with specimens from more than one population, we calculated the shape variance of each population and get the average value as the shape variation of that taxon.
Corolla shape disparity was estimated by partial static disparity, a metric indicating the contribution that a particular taxon makes to the overall disparity of the morphospace generated by the pool of studied species (Zelditch et al., 2012). We first calculated the Procrustes distance between the mean shape of each taxon and the grand mean of the whole pool of taxa. We then calculated the partial static disparity as the squared Procrustes distances of each specimen to the mean shape of the respective group or, equivalently, the sum of the sample variances of all Procrustes coordinates, divided by the number of taxa studied minus one (Zelditch et al., 2012). Because partial disparities are additive, we expressed the contribution of each taxon to total disparity as a percentage (Zelditch et al., 2012).
Corolla shape integration was computed as the relative variance of eigenvalues of the covariance matrix of Procrustes coordinates per plant species, using the original units of squared Procrustes distance (Young, 2006). One aspect of integration is that variation is concentrated in one or a few of the available dimensions (Klingenberg, 2013). As a consequence, there will be one or a few large and many small eigenvalues for the covariance matrix of integrated data, whereas eigenvalues of the covariance matrix will be more homogeneous for data lacking integration. To control for among-species differences in sampling size, we re-scaled the relative variance of eigenvalues by the total shape variance and the number of dimensions (Young, 2006). By doing this, corolla shape integration ranges between 0 and 1, and can be interpreted as the percentage of integration regarding the maximum possible integration. This index is thus directly comparable to other integration indices found using different approaches.
All these analyses were performed in MorphoJ (Klingenberg, 2011) and in the R package ‘geomorph’ (Adams and Otarola-Castillo, 2013).
Evolution of corolla shape variation, disparity and integration
We tested the evolutionary lability of corolla shape by calculating its phylogenetic signal and examining how floral traits changed along the phylogeny. The phylogenetic signal was tested using Pagel’s λ (Pagel, 1999; Freckleton et al., 2002). To test phylogenetic signal, we compared a model generating an ML estimate of Pagel’s λ for floral traits with a model constraining λ to 0. A significant departure from the model with λ = 0 would indicate phylogenetic correlation (Freckleton et al., 2002). We illustrated the phylogenetic signal of the corolla-shape attributes by means of traitgrams (Ackerly, 2009). Traitgrams arrange species along a continuous trait axis (the x-axis) and connect them with their underlying phylogenetic tree (time on the y-axis) (Münkemüller et al., 2012). Internal node positions correspond to ancestral states obtained by ML. Node depths reflect phylogenetic branch lengths (Ackerly, 2009). The phylogenetic signals and traitgrams were obtained using the R package ‘phytools’ (Revell, 2012).
Ancestral state reconstruction of the corolla shape attributes was done using the ‘ace’ function implemented in the R package ‘ape’ (Paradis et al., 2004). This function estimates ancestral character states, and the associated uncertainty. We included phylogenetic uncertainty by doing this analysis with the set of 100 trees.
Evolutionary allometry of corolla shape variation, integration and disparity
We estimated the allometry of corolla shape attributes by determining their correlation with corolla size and shape. For this, in each of 20 individuals per taxon, in addition to quantifying corolla shape, we also quantified corolla size, as the distance in millimetres between the apical edges of two opposite petals. This is an estimate of the overall size of the Brassicaceae corolla (Gómez et al., 2006). Furthermore, we also measured the corolla tube length, as the distance in millimetres between the corolla tube aperture and the base of the sepals.
We explored the correlated evolution between corolla shape attributes and corolla size using phylogenetic generalized least square (PGLS) models (Grafen, 1989; Martins and Hansen, 1997). This analysis optimizes the phylogenetic signal while performing the analysis (Revell, 2010). PGLSs were performed using the R package ‘caper’ (Orme, 2013).
Pollinator diversity and pollination niches
We conducted flower visitor counts in 1–10 populations per plant species (Table S1). We visited the populations during the peak of the bloom, always at the same phenological stage and between 1100 and 1700 h. Insects were identified in the field, and some specimens were captured for further identification in the laboratory. We recorded only those insects contacting anthers or stigma and making legitimate visits at least during part of their foraging at flowers. We did not record those insects only eating petals or thieving nectar without making a legitimate visit. We grouped the insects visiting the flowers of the studied species into functional groups. Here, ‘functional group’ is defined as those insects that interact with the flowers in a similar manner (Fenster et al., 2004). We used criteria of similarity in body length, proboscis length, morphological match with the flower, foraging behaviour and feeding habits (Gómez et al., 2015). Table S3 describes the 35 functional groups used in this study and Table S4 shows the distribution of these functional groups among the studied Brassicaceae taxa.
We described the diversity of the flower visitor fauna of the studied plants at functional group levels, using two complementary indices: (1) pollinator richness (Sobs), calculated as the overall number of functional groups recorded in the flowers of each plant taxon – to control for sampling effort, we divided the observed Sobs by the number of flower visitors recorded; (2) pollinator diversity, calculated as Hurlbert’s PIE, the probability that two randomly sampled insects from the community pertain to two different functional groups. This is an evenness index that incorporates the frequency of visitation of pollinators and combines the dominance and abundance of the species. These indices were generated using the ‘addpart’ function in R package ‘stratigraph’ (Green, 2012).
We determined the occurrence of different pollination niches in our studied populations using bipartite modularity, a complex-network metric. We constructed a weighted bipartite network including only the Brassicaceae taxa with pollinator data. In this network, we pooled the data from the different conspecific populations. We subsequently determined the modularity level of this network by using the QuanBiMo algorithm (Dormann and Strauss, 2014). This method uses a simulated annealing Monte Carlo approach to find the best division of taxa into modules. A maximum of 1010 Markov chain Monte Carlo steps with a tolerance level = 10–10 were used in 100 iterations, retaining the iterations with the highest likelihood value as the optimal modular configuration. We tested whether our network was significantly more modular than random networks by running the same algorithm in 100 random networks, with the same linkage density as the empirical one (Guimerà and Amaral, 2005). Modularity significance was tested for each individual iteration by comparing the empirical versus the random modularity indices by means of a z-score test (Dormann and Strauss, 2014). After testing the modularity of our network, we determined the number of modules using the approach proposed by Newman (2004). We subsequently identified the pollinator functional groups defining each module and the plant species that were ascribed to each module. Modularity analysis was performed using R package ‘bipartite’ (Dormann et al., 2008).
We explored the correlated evolution between corolla shape attributes and pollinators using PGLS models as explained above. We performed a separate model for each attribute of corolla shape disparity and integration.
RESULTS
Variation in corolla shape variation, disparity and integration
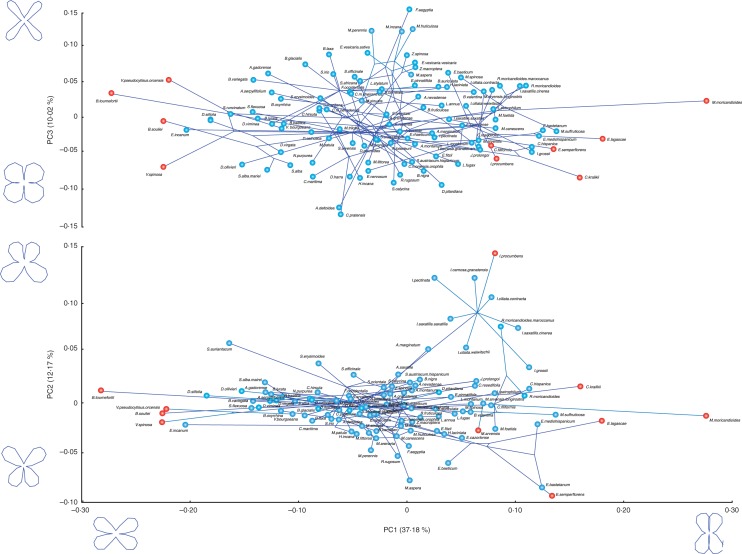
We found ample between-species variation in corolla shape. The first PC explained 37 % of the variation in shape and was related to changes in the parallelism of the petals. Species receiving positive scores of this component displayed corollas with the two adaxial petals parallel and the two abaxial petals also in the same direction, while species having negative scores displayed corollas with the abaxial and adaxial petals divergent (Fig. 2). The second PC explained 12 % of the variation in shape and was related to changes in corolla monosymmetry. Species with positive scores of this PC displayed corollas with an overdevelopment of lower petals, whereas those species with negative scores had corollas with overdeveloped upper petals (Fig. 2). The third component explained 10 % of the variation in shape, and was related to changes in petal width. Species with positive scores had narrow petals, whereas species with negative scores had wide petals (Fig. 2). The remaining PCs explained very low variation in corolla shape (Table S5).
Fig. 2.
Phylomorphospace of the first three PC axes showing the relationship between the phylogeny and corolla shapes for the Brassicaceae taxa included in this study. Indicated in red is the position in the shape space of the ten species receiving the highest values of corolla shape partial static disparity.
The average shape variation of the studied taxa was 0·0025 ± 0·001 (mean ± 1 s.e.), ranging between 0·01 in some Biscutella species and 0·067 in Farsetia aegyptia (Fig. 3, Table S6). Average partial static disparity was 0·009 ± 0·001, and ranged between 0·0007 in Sisymbrium orientale and 0·042 in Moricandia moricandioides (Fig. 3, Table S6). This low magnitude indicates that an average taxon contributes less than 1 % to the overall disparity of the entire group of taxa. Moreover, the taxon contributing most contributed only 4·2 % to the overall disparity. The ten taxa with highest disparity values were M. moricandioides, Iberis procumbens, Vella pseudocytisus orcensis, V. spinosa, Brassica souliei, B. tournefortii, Crambe kralikii, Erysimum lagascae, M. arvensis and E. semperflorens (Fig. 2).
Fig. 3.
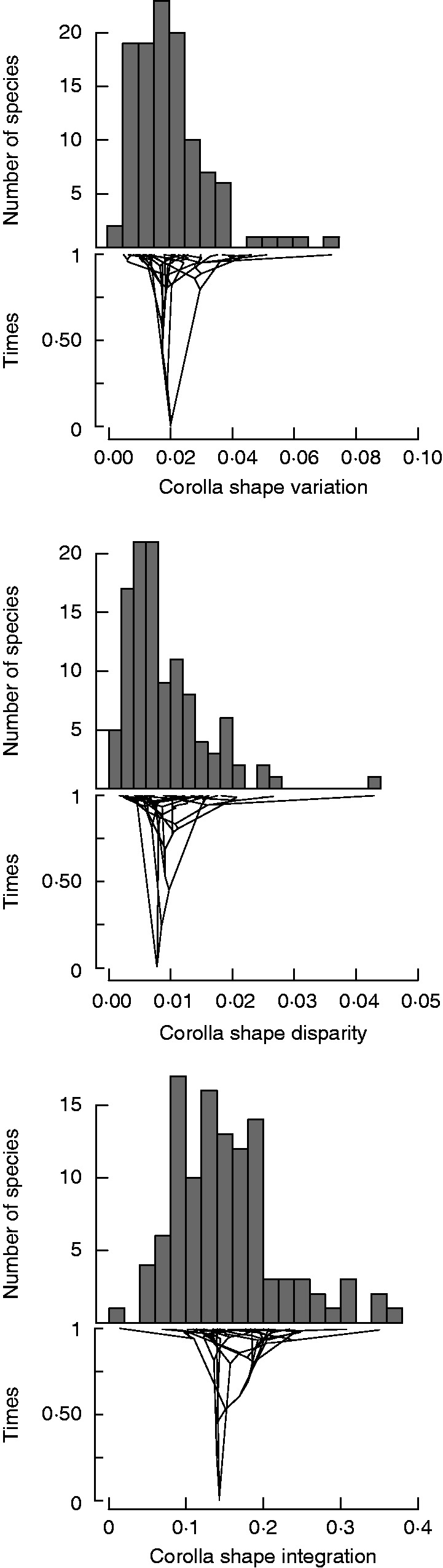
Frequency distribution of the four estimates of corolla shape variation, disparity and integration. Below the histograms are traitgrams depicting the association between the phylogenetic relationships and the values of the corolla shape variation, disparity and integration.
The across-taxa average corolla shape integration was low (0·182 ± 0·006), representing only 15 % of the maximum possible integration. Corolla shape integration ranged between 0·081 and 0·363 in Sisymbrium orientale and Brassica souliei, respectively (Fig. 3, Table S6). Corolla shape integration was marginally and positively related to corolla shape variation (1·133 ± 0·578, F = 3·84, P = 0·054, λ = 0·083; PGLS models), whereas it was not related to partial static disparity (0·767 ± 1·014, t = 0·76, P = 0·451, λ = 0; PGLS models).
Evolutionary lability of corolla shape variation, disparity and integration
We found significant phylogenetic signal for corolla shape variation (Pagel’s λ = 0·644, P = 0·0001). In contrast, the phylogenetic signal was not significant for corolla shape disparity (Pagel’s λ = 0·001, P = 0·900) or integration (Pagel’s λ = 0·279, P = 0·623). This outcome was strongly consistent across the set of 100 phylogenetic trees (Table S7). Accordingly, the traitgrams showed many crossings in the values of these three corolla shape attributes (Fig. 3).
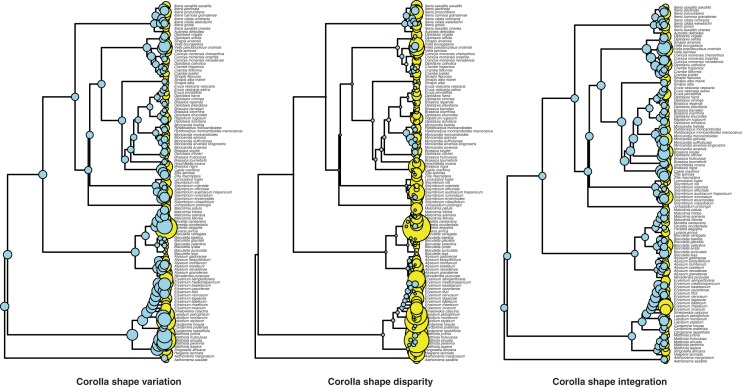
The reconstruction analysis suggests that the ancestral corollas of the studied Brassicaceae taxa had intermediate values of variation (0·026 ± 0·0001), disparity (0·009 ± 0·0003) and integration (0·174 ± 0·002; see Table S8 for the values of the most recent common ancestor obtained in the 100 phylogenetic trees, and Supplementary Data 1 for the values obtained in the whole set of internal nodes). Most changes in these three traits seem to have occurred very recently (Fig. 4).
Fig. 4.
Reconstruction of the corolla shape variation, disparity and integration using the ML method. The diameter of the spheres is proportional to the value of each variable. Internal nodes are shown blue, and extant Brassicaceae taxa in yellow.
Evolutionary allometry of corolla shape variation, disparity and integration
There was no phylogenetic correlation between corolla size and the variation, disparity or integration of corolla shape (Table 1). In contrast, there was a significant correlation between corolla shape integration and corolla tube length, plant species with longer corolla tubes having corollas with more integrated shapes. There was also a significant positive relationship between corolla tube length and corolla shape variation, species with deeper corollas having more variable shapes (Table 1).
Table 1.
Outcome of the PGLS models testing the relationship between the variation, disparity and integration of corolla size and corolla tube length; significant values appear in bold type
| Corolla size |
Corolla tube length |
|||||
|---|---|---|---|---|---|---|
| Estimate ± 1 s.e. | F1,109 | P | Estimate ± 1 s.e. | F1,109 | P | |
| Corolla shape variation | –0·006 ± 0·003 | 1·67 | 0·097 | 0·001 ± 0·000 | 2·36 | 0·020 |
| Corolla shape disparity | –0·001 ± 0·003 | 0·31 | 0·758 | 0·002 ± 0·002 | 1·02 | 0·307 |
| Corolla shape integration | –0·002 ± 0·002 | 0·49 | 0·623 | 0·006 ± 0·003 | 2·09 | 0·039 |
Pollinator diversity and pollination niches
We recorded 39 088 floral visits from 33 pollinator functional groups (Tables S1 and S3; we did not record visits by earwigs or lacewings). In general, the plant taxa included in this study had a diverse pollinator assemblage. The mean number ( ± 1 s.e.) of functional groups visiting each plant taxon was 11·6 ± 0·6, whereas the across-taxa mean Hurlbert’s PIE index was 0·61 ± 0·03. There was a large difference among plants in the diversity of their pollinator assemblages. The richness of functional groups ranged between one in several Matthiola species and Hesperis laciniata, which were visited exclusively by nocturnal moths, and more than 20 in several taxa from the genera Sinapis, Brassica, Erysimum and Cakile. Similarly, PIE values ranged between 0 for Matthiola and more than 0·90 for Cakile maritima, Erysimum baeticum, Iberis saxatilis and Erysimum mediohispanicum.
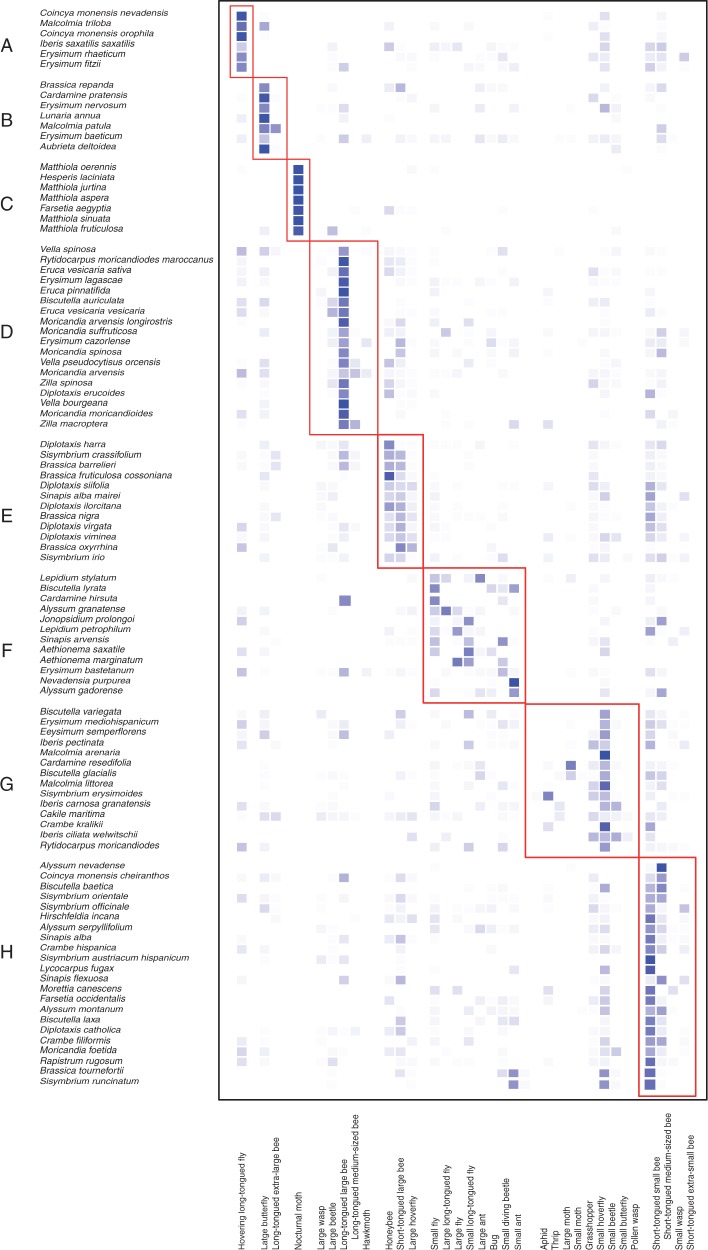
The network between the Brassicaceae species and the pollinator functional groups was significantly modular (Modularity = 0·477 ± 0·001, all P-values < 0·0001, n = 100 iterations; Table S9). Modularity analysis detected eight pollination niches (niches A–H in Fig. 5). Plants from niche A were visited mostly by hovering long-tongued flies (i.e. Bombylius spp.), plants from niche B were visited mostly by butterflies and extra-large long-tongued bees (mostly Bombus spp.), plants from niche C were visited by nocturnal moths, plants from niche D were visited mostly by long-tongued large bees (mostly Anthophoridae), plants from niche E were mainly visited by short-tongued large bees and honeybees, plants from niche F were visited mostly by flies and ants, plants from niche G were visited mostly by small beetles and plants from niche H were visited mostly by small bees (Fig. 5).
Fig. 5.
Plot showing the classification of the studied species into different pollination niches, according to the analysis of bipartite modularity QuanBiMo. The intensity of the colours indicates the relative abundance of each flower visitor’s functional group per plant taxon.
There was between-niche difference in the generalization degree (Sobs: F = 3·15, d.f. = 7,89, P < 0·005; Hurlbert’s PIE: F = 9·91, d.f. = 7,89, P < 0·0001; PGLS models). Plants belonging to niche C were visited by pollinator assemblages with very low diversity (Fig. S1), composed mostly of nocturnal moths (Fig. 5). In the other extreme, plants from niche E had highly diverse pollinator assemblages (Fig. S1). Plants from the remaining niches had pollinator assemblages with intermediate diversities (Fig. S1).
Relationship between corolla shape and pollinators
We did not find any significant relationship between pollinator diversity and corolla shape variation, disparity or integration (Table 2). In contrast, we found significant between-niches differences in all these three attributes for corolla shape (Table 2). Plants from niche C had significantly higher corolla shape variation than plants from any other niche (Fig. 6). Corolla shape disparity was significantly higher in plants from niches C, D and G (Fig. 6). Corolla shape integration was significantly higher in plants from niches A, D and E and significantly smaller in plants from niche C (Fig. 6).
Table 2.
Outcome of the PGLS models testing the relationship between pollinators and corolla shape variation, disparity and integration; significant values are in bold type
|
Sobs |
Hurlbert’s PIE |
Pollination niche |
||||||
|---|---|---|---|---|---|---|---|---|
| Estimate ± 1 s.e. | F1,95 | P | Estimate ± 1 s.e. | F1,95 | P | F7,89 | P | |
| Corolla shape variation | 0·000 ± 0·000 | 0·60 | 0·456 | –0·005 ± 0·004 | 1·59 | 0·210 | 4·61 | 0.0002 |
| Corolla shape disparity | –0·001 ± 0·003 | 0·04 | 0·950 | –0·001 ± 0·002 | 0·09 | 0·760 | 2·41 | 0.024 |
| Corolla shape integration | –0·000 ± 0·000 | 0·41 | 0·523 | 0·001 ± 0·024 | 0·03 | 0·857 | 3·07 | 0.006 |
Fig. 6.
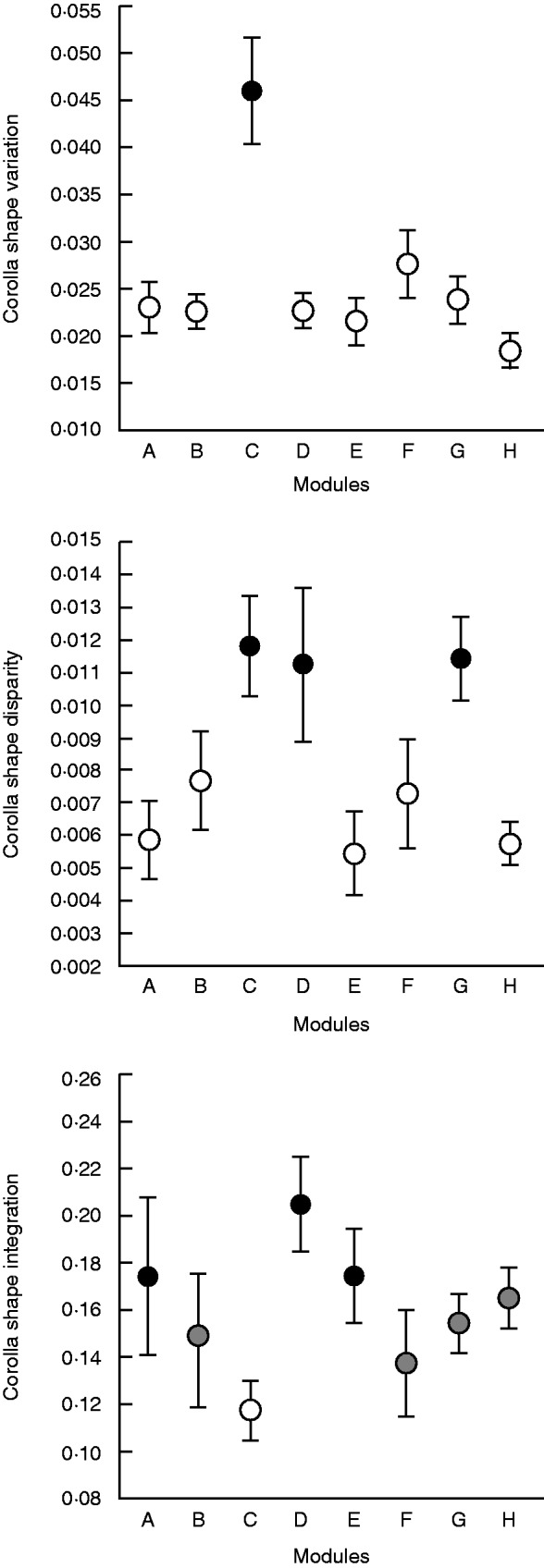
Between-pollination niche differences in corolla shape variation, disparity and integration. Pollination niches indicated by full circles are significantly different at α= 0·05 according to phylogenetic post-hoc tests from pollination niches indicated by open circles. Niches indicated by grey circles are not significantly different from any other pollination niche. Error bars are ± 1 s.e.
DISCUSSION
The low magnitude of corolla shape disparity and integration in Brassicaceae
The magnitude of integration found in this study for the shape of the corollas was lower than those reported in other plants. The average across-taxa corolla shape integration was 15 % of the maximum possible integration, whereas the taxa having maximum integration had only 36 % of the maximum possible integration. These values are much lower than those found in plant species belonging to other families (Pérez et al., 2007; Ordano et al., 2008; Rosas-Guerrero et al., 2011; Ishii and Harder, 2012; González et al., 2015; but see Pérez-Barrales et al., 2014). Several concurrent reasons may cause the low integration values observed in Brassicaceae corollas.
Brassicaceae have heterochlamydeous flowers with separate and unfused sepals and petals (Taiyan et al., 2001; Singh, 2010). This architectural pattern favours the independent movements and arrangement of the different organs of the perianth, both during ontogeny of the flower and once the flower is fully developed as a consequence of external environmental factors. This is no doubt a main architectural and non-functional factor determining the low integration observed in Brassicaceae corollas.
Low corolla shape integration may also be a consequence of the pollination systems of the species included in this study. Most Brassicaceae species were extremely generalist in their interactions with pollinators. The number and diversity of insects visiting their flowers was very high even within plant populations. Due to their differences in morphology, foraging behaviour and preference pattern, this diversity of pollinators surely prompts the occurrence of conflicting selection on corolla shape (Galen and Cuba, 2001; Medel et al., 2003; Gómez et al., 2008). Under these circumstances, the local coexistence of several pollinator functional groups will negate the selection on any single type of corolla, a scenario preventing the evolution of highly integrated corollas (Gómez et al., 2014).
The magnitude of corolla shape integration found in our study was even lower than those reported for other Brassicaceae species (Murren et al., 2002; Anderson and Busch, 2006; Penrod, 2010; Edwards and Weinig, 2011). For example, Penrod (2010) found that Brassica nigra floral integration ranged between 0·40 and 0·56. In contrast, the magnitude of B. nigra corolla shape integration found in our study was 0·32. We believe that these discrepancies may be partially related to the analytical procedure used to obtain the integration indices. An important difference between our study and most other studies on floral integration is that we have studied a trait related to organ shape rather than to organ size and we have thereby used tools from geometric morphometrics to explore the integration of corolla shape, whereas most other studies have calculated floral integration from standard linear measurements. Because variation in each linear measurement contains a considerable degree of floral size variation, linear measurements tend to be integrated by this shared variation of size. This component of variation can capture a large part of the variance because it is generally associated with between-individual variation in resource acquisition (Torices and Méndez, 2014), and it is thereby the one generating a very strong integration in most standard studies of morphological integration. By concentrating exclusively on shape variation and removing size variation as part of the Procrustes superimposition, we removed this component of phenotypic variation, a procedure probably contributing to the observed low corolla shape integration for shape. Our estimates of phenotypic integration, although low, can be considered robust estimations after excluding biases due to between-individual differences in resource availability. This reasoning suggests that phenotypic integration will presumably be lower in analyses of shape than in analyses that characterize morphology by the sizes of floral parts. In this respect, the magnitude of floral integration found in this study is similar to that found in other geometric morphometric studies, studying both Brassicaceae species (Gómez et al., 2014) and other disparate organisms (Young, 2006; Gómez-Robles and Polly, 2012).
Corolla shape disparity was also very low for the studied Brassicaceae taxa. In fact, average corolla shape disparity was less than 1 %, and the plant having the most distinctive corolla, Moricandia moricandioides, contributed only 4·2 % of the overall disparity of corolla shapes. Because static disparity measures within-species variation as a fraction of the total disparity, these observed low values indicate that most variation in corolla shape occurs among taxa rather than within taxa. This suggests that taxa are relatively constant, but have markedly diverged from each other at similar rates. Low corolla shape disparity may occur because the corolla shapes of different taxa have diverged along contrasting axes of the morphospace. Some species, such as M. moricandioides and Erysimum lagascae, have corollas with parallel petals and high positive values in PC1. Other taxa, such as Vella species, have corollas with very open petals and high negative values of PC1. Some other taxa, such as Farsetia aegyptia and species of Matthiola and Eruca, have corollas with extremely narrow petals and high positive values of PC3. Finally, taxa belonging to the genus Iberis have zygomorphic corollas with high positive values of PC2. Under this scenario, there is not a large group of taxa with very similar corollas coexisting with a small subset of species having distinctive corollas and thereby exhibiting high values of corolla shape disparity. In contrast, the study system is composed of a large number of taxa with distinctive corollas but exploring the floral morphospace along different directions. There is large among-taxon variation in corolla shape even in this plant family characterized by a conserved floral bauplan (Franzke et al., 2011).
Evolutionary lability of the variation, disparity and integration of Brassicaceae corolla shape
The phylogenetic signal was not significant for the integration and disparity of Brassicaceae corolla shape. Low phylogenetic signal may be caused by measurement errors related to the uncertainty of the phylogenetic relationships of the taxa included in the analyses (Blomberg et al., 2003; Ives et al., 2007). However, because we built the phylogeny using several nuclear, mitochondrial and plastidial genes and performed the analysis using a set of 100 trees, we think that the observed low phylogenetic signal is actually evidence of evolutionary lability in Brassicaceae floral integration and disparity (Losos, 2008). It seems that the floral disparity and integration of a given Brassicaceae taxon is not associated with the floral integration and disparity displayed by its relatives. The evolution of floral disparity and integration seems not to be strongly constrained by phylogenetic inertia in Brassicaceae. This pattern is clearly illustrated in the traitgrams, which show many lines crossing. Evolutionary lability is frequent in floral traits (Beardsley et al., 2003; Smith et al., 2008; Roncal et al., 2012; Alcantara and Lohmann 2010; McEwen and Vamosi, 2010; Muchhala et al., 2014), and it has been recently found for the phenotypic integration of Erysimum corolla shape (Gómez et al., 2014). As pointed out by Franzke et al. (2011), most traits in Brassicaceae exhibit substantial homoplasy.
We have found, however, significant phylogenetic signal for corolla shape variation. Intraspecific variation in corolla shape could be a consequence, at least partially, of the loss of canalization and the presence of developmental noise. Under this perspective, intraspecific variation can be a manifestation of the lack of developmental robustness. Although flowers appear to show more stable development than other plant organs (Sherry and Lord, 1996; Evans and Marshall, 1996), they also undergo some degree of developmental noise. We presume that taxa showing high values of corolla shape variation will be those being more susceptible to developmental noise, probably due to relaxation of the stabilizing selection for canalization and developmental robustness in those lineages. In contrast, taxa with low corolla shape variation will be those showing stronger developmental robustness. The ability to buffer developmental noise has a genetic basis in many organisms (Gavrilets and Hasting, 1994). Canalization thereby could result from evolution, whether adaptive or non-adaptive (Flatt, 2005). Our result suggests that the evolution of developmental robustness in Brassicaceae is influenced by phylogenetic relatedness, with plants belonging to the same lineages having a similar ability to cope with developmental noise.
The role of pollinators in the evolution of corolla shape variation, disparity and integration in Brassicaceae
Our study has found ample across-taxa variability in corolla shape variation, disparity and integration. Indeed, corolla shape integration varied among the crucifer taxa included in this study by more than two orders of magnitude, between 0·002 and 0·363, whereas corolla shape disparity varied over one order of magnitude, from 0·07 to 4·2 %. Variation in corolla shape has been reported for some Brassicaceae, such as Lepidium (Bowman et al., 1999), Erysimum (Gómez et al., 2014) and several genera from the tribe Brassiceae (Takahata, 2009). Similarly, variation in floral integration has been also reported among Brassicaceae species belonging to the genera Brassica (Murren et al., 2002), Leavenworthia (Anderson and Busch, 2006) and Erysimum (Gómez et al., 2014). Knowing which factors are associated with this pervasive variation will surely help to discover the causes driving the evolution of floral integration and disparity in Brassicaceae. Several non-exclusive reasons suggest that pollinators may play a role in the evolution of corolla shape variation, disparity and integration at higher taxonomic level.
First, the observed evolutionary lability in corolla integration and disparity may be a consequence of the selection exerted by pollinators. Although several genetic and ecological factors cause evolutionary lability (Losos, 2008; Bell, 2010; Cooper et al., 2010; Crisp and Cook, 2012), low phylogenetic signal can be produced by punctuated divergent selection, a process occurring when daughter lineages face different selective scenarios and evolve to two different optima after every bifurcation (Revell et al., 2008; Ackerly, 2009). Under this scenario, low phylogenetic signal is expected when there is recurrent short-term variation in pollinator-mediated selection (Beardsley et al., 2003; Ornelas et al., 2007; Wilson et al., 2007; Harder and Johnson, 2009). Gómez et al. (2014) suggested that low phylogenetic signal in Erysimum corolla shape integration is associated with recurrent shifts in pollination niches.
Second, our reconstruction analysis suggests that the changes in Brassicaceae corolla shape occurred very recently. So, the reconstructed corollas of the deep internal nodes were all very similar, whereas most changes in integration and disparity appeared in shallow internal nodes. This evolutionary pattern suggests that forces acting at the ecological time scale, rather than forces associated with deep phylogenetic relationships, may have driven the observed evolution in corolla shape integration and disparity.
Third, the absence of any correlation between floral integration or disparity and corolla size indicates that no allometric effect influences the evolution of these characters in our studied taxa. Allometry is a factor that may greatly affect phenotypic integration (Klingenberg and Marugán-Lobón, 2013). In fact, allometry has been proposed as the factor fuelling interspecific changes in character shape in the absence of any adaptive mechanism (Herrera, 1992). Under these circumstances, we presume that the evolution of corolla shape integration and disparity, not being associated with allometric effects, is prompted by the effect of some selective forces.
Finally, and most importantly, we found a significant phylogenetic association between floral integration and disparity and pollinators. Contrary to previous studies (Pérez-Barrales et al., 2007; Pérez et al., 2007; Gómez et al., 2014), we have not found any association between corolla integration and pollinator diversity. This result is probably a consequence of having included in our study very disparate plant taxa belonging to different lineages. The effect of pollinator diversity on corolla shape integration may be only evident within plant lineages. However, we did find a significant association between pollination niche and corolla shape variation, disparity and integration of each plant taxon. This outcome suggests that the type of pollinator visiting the flowers, rather than the diversity, has a major effect on the evolution of floral integration and disparity (Pérez-Barrales et al., 2014; González et al., 2015). This is a remarkable finding, as the effect of pollinators on phenotypic integration is expected to be more intense for those traits related to the efficiency of pollen transfer (anthers, pistils, stigmas) than for traits related to the attraction of pollinators (Ordano et al., 2008).
Corolla shape variation was significantly higher in those taxa belonging to a pollination niche where nocturnal moths are the main floral visitors (niche C in our study). Furthermore, we found that plants from niche C had a significantly higher corolla shape disparity, suggesting that plant taxa pollinated mostly by moths not only have highly variable but also very distinctive corollas. This pollination niche comprises species from the genus Matthiola, as well as Hesperis laciniata and Farsetia aegyptia. These species are characterized as having flowers where petals are arranged in a variety of positions, sometimes forming very odd shapes (Fig. 7), and this variance occurs not only between individuals but also within individuals (authors’ pers. obs.). We presume that because the main pollinators of these taxa are nocturnal insects, selection on corolla shape developmental robustness and canalization is weak, resulting in high shape variation. Great variation in corolla shape also occurs in other Brassicaceae species presumably pollinated by moths, such as Hesperis tristis (Lovell, 1902; Faegri and van der Pijl, 1979) or those from the tribe Schizopetalae (Toro-Núñez et al., 2015). Note that all these species do not close their corollas during the day, in contrast to other moth-pollinated species; thus, the shapes studied here are the actual shapes of the corollas of these species.
Fig. 7.
Intrapopulation variation in the corolla shape of Farsetia aegyptia. Photographs were taken during January 2015 at Petra (Jordan).
Corolla integration was higher in those plants belonging to pollination niches composed of hovering long-tongued flies mostly belonging to the genus Bombylius (niche A in this study) and of long-tongued large bees mostly belonging to the genus Anthophora (niche D). These two types of insects have been shown to be very effective pollinators of several species of Brassicaceae (Motten, 1986; Kuchmeister et al., 1995; Kastinger and Weber, 2001; Ollerton et al., 2007; Gómez et al., 2010; Lay et al., 2011; Fernández et al., 2012). In addition, their behaviour at flowers differs from that displayed by most other Brassicaceae pollinators. They use hovering to approach the flowers, frequently touching the petals only slightly and using the entire corolla as landing platforms. Hovering is energetically costly (Heinrich, 1993), and these insects probably try to decrease this cost by changing their behaviour at flowers. For example, beeflies, which collect nectar while hovering but stand on their second and third pairs of legs to decrease the cost of hovering, prefer to visit rounded Erysimum mediohispanicum corollas because they can use this type of corolla as an efficient landing platform (Gómez et al., 2008). By choosing very specific corolla shapes, we presume that beeflies have been selecting for more integrated corollas. According to the current results, the anthophorid bees have presumably prompted the evolution of more integrated corollas in Brassicaceae as well. It is remarkable that these two types of pollinators are long-tongued insects, visiting the Brassicaceae flowers mostly to consume nectar. This can explain the significant and consistent relationship found across Brassicaceae taxa between corolla integration and the depth of their corolla tube. Pollinators are not only driving the evolution of corolla integration but also its correlational evolution with corolla tube length.
CONCLUSION
This study shows that corolla adaptive evolution is even possible in a plant family having a very conserved floral bauplan. In fact, it seems that the shape of the Brassicaceae corolla has, at least partially, evolved as a consequence of the selective pressures exerted by some pollinators. This adaptive process has resulted in multiple convergences and divergences along the phylogeny of Brassicaceae, causing corolla shape to be a trait more evolutionarily labile than traditionally thought. Extending this study to species from other lineages and inhabiting distant geographical areas would provide precious information on how pollinators propel the successful colonization of the floral morphospace in Brassicaceae.
Supplementary Data
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: List of the Brassicaceae species and subspecies included in this study. Table S2: GenBank numbers for taxa used in the super matrix analysis. Table S3: Description of the functional groups of the insects visiting the flowers of the studied species. Table S4: Relative abundance of each floral visitor functional group in each Brassicaceae species included in this study. Table S5: Outcome of the PCA on the superimposed specimens. Table S6: Corolla shape variation, disparity and integration, corolla size and pollinator diversity of the Brassicaceae species and subspecies included in this study. Table S7: Outcome of the phylogenetic signal analysis. Table S8: Corolla shape reconstruction of the most recent common ancestor of the phylogeny used in this study. Table S9: Outcome of the modularity. Figure S1: Relationship between pollinator diversity and pollination niches. Supplementary Data 1: Values obtained in the whole set of internal nodes.
ACKNOWLEDGEMENTS
We are deeply grateful to Jordi Bosch (UAB-CREAF, Barcelona, Spain), Elena Amat and Pablo Vargas (RJB-CSIC, Madrid, Spain), Alfredo Valido and Ignasi Bartomeus (EBD-CSIC, Seville, Spain), Rubén Alarcon (The University of Arizona, Tucson, USA), Milhail Garbuzov (University of Sussex, Sussex, UK), David Cuerda (Biologist, Natural Park of Cazorla, Segura y las Villas, Consejería de Medio Ambiente, Junta de Andalucía, Jaén, Spain), Sandra García de Lucas (Jardín Botánico Torre del Vinagre, Consejería de Medio Ambiente, Junta de Andalucía), Luis Navarro (University of Vigo, Vigo, Spain), Silvia Santamaría, Adrián Escudero, José María Iriondo, Carlos Lara, Javier Morente, Marcos Méndez, Raúl Garcia-Camacho, Luis Giménez-Benavides and Rubén Milla (URJC, Madrid , Spain) (URJC, Madrid, Spain), Sharon Strauss (University of California at Davis, Davis, USA), Jeffrey Conner (Michigan State University, Hickory Corners, USA), Dani Lucas-Barbosa (Wageningen University, Wageningen, The Netherlands), Rocio Castro-Urgal and Anna Traveset (IMEDEA-CSIC, Esporles, Spain), M. Ángeles Marcos García (Research Institute CIBIO, Universidad de Alicante, Spain), Theodora Petanidou (University of the Aegean, Mytilene, Greece), Olivia Norfolk (University of Nottingham, Nottingham, UK), Natacha Chacoff (CONICET, Mendoza, Argentina), Carolina L. Morales (CONICET, Bariloche, Argentina), Yedra García (CIDE-CSIC, Valencia, Spain), Lindsay Zink, Mark Wonneck and Ralph Cartar (University of Calgary, Calgary, Canada) and Jochen Fründ (Georg-August-Universität, Göttingen, Germany) for sharing information on Brassicaceae flower visitors. We also want to thank several taxonomists who kindly identified insect specimens: L. A. Aguado Martín, M. A. Alonso Zarazaga, M. Baena, J. Bosch, M. Carles-Tolrà, R. Constantin, S. Fernández Gayubo, M. Goula, F. Gusenleitner, J. Háva, P. Leblanc, M. A. Marcos, J. C. Otero, A. Sánchez Ruiz, A. Sánchez Terrón, M. Schwarz, A. Tinaut, F. Vallhonrat and D. Ventura. The Ministerio de Medio Ambiente and Consejerías de Medio Ambiente of Andalucía, Castilla y León, Cataluña, and Aragón, as well as the Sierra Nevada National Park Headquarter, granted permission to work in several protected areas of Spain. This study was partially supported by grants from the Spanish MCyT (CGL2013-47558-P and CGL2012-34736, including FEDER funds), and Junta de Andalucía (P07-RNM-02869 and P11-RNM-7676).
LITERATURE CITED
- Ackerly D. 2009. Conservatism and diversification of plant functional traits: evolutionary rates versus phylogenetic signal. Proceedings of the National Academy of Sciences USA 106: 19699–19706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DC, Otarola-Castillo E. 2013. geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods in Ecology and Evolution 4: 393–399. [Google Scholar]
- Al-Shehbaz I. 2012. A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954. [Google Scholar]
- Al-Shehbaz IA, Beilstein MA, Kellogg EA. 2006. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Systematics and Evolution 259: 89–120. [Google Scholar]
- Alcantara S, Lohmann LG. 2010. Evolution of floral morphology and pollination system in Bignonieae (Bignoniaceae). American Journal of Botany 97: 782–796. [DOI] [PubMed] [Google Scholar]
- Anderson IA, Busch JW. 2006. Relaxed pollinator-mediated selection weakens floral integration in self-compatible taxa of Leavenworthia (Brassicaceae). American Journal of Botany 93: 860–867. [DOI] [PubMed] [Google Scholar]
- Armbruster WC, Pélabon C, Hansen TF, Mulder CPH. 2004. Floral integration, modularity, and accuracy: Distinguishing complex adaptations from genetic constraints. In: Pigliucci M, Preston K, eds. Phenotypic integration: studying the ecology and evolution of complex phenotypes. New York: Oxford University Press, 23–49. [Google Scholar]
- Armbruster WC, Hansen TF, Pélabon C, Pérez-Barrales R, Maad J. 2009a. The adaptive accuracy of flowers: measurement and microevolutionary patterns. Annals of Botany 103: 1529–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster WC, Pélabon C, Hansen TF, Bolstad GH. 2009b. Macroevolutionary patterns of pollination accuracy: a comparison of three genera. New Phytologist 183: 600–617. [DOI] [PubMed] [Google Scholar]
- Armbruster WC, Pélabon C, Bolstad GH, Hansen TF. 2014. Integrated phenotypes: understanding trait covariation in plants and animals. Philosophical Transactions of the Royal Society of London B 369: 20130245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CD, Koch MA, Mayer M, et al. 2006. Toward a global phylogeny of the Brassicaceae. Molecular Biology and Evolution 23: 2142–2160. [DOI] [PubMed] [Google Scholar]
- Beardsley PM, Yen A, Olmstead RG. 2003. AFLP phylogeny of Mimulus section Erythranthe and the evolution of hummingbird pollination. Evolution 57: 1397–1410. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA. 2008. Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. American Journal of Botany 95: 1307–1327. [DOI] [PubMed] [Google Scholar]
- Bell G. 1997. The basics of selection. New York: Chapman and Hall. [Google Scholar]
- Bell G. 2010. Fluctuating selection: the perpetual renewal of adaptation in variable environment. Philosophical Transactions of the Royal Society of London B 365: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RL. 1960. The ecological significance of correlation pleiades. Evolution 14: 171–180. [Google Scholar]
- Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57: 717–745. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Brüggemann H, Lee JY, Mummenhoff K. 1999. Evolutionary changes in floral structure within Lepidium L. (Brassicaceae). Internation Journal of Plant Science 160: 917–929. [DOI] [PubMed] [Google Scholar]
- Busch A, Zachgo S. 2007. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proceedings of the National Academy of Sciences USA 104: 16714–16719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier M, Jabbour F, Gerber S, et al. 2014. The floral morphospace – a modern comparative approach to study angiosperm evolution. New Phytologist 204: 841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N, Jetz W, Freckleton RP. 2010. Phylogenetic comparative approaches for studying niche conservatism. Journal of Evolutionary Biology 23: 2529–2539. [DOI] [PubMed] [Google Scholar]
- Córdoba S, Cocucci A. 2011. Flower power: its association with bee power and floral functional morphology in papilionate legumes. Annals of Botany 108: 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TLP, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. 2010. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Molecular Biology and Evolution 27: 55–71. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Cook LG. 2012. Phylogenetic niche conservatism: what are the underlying evolutionary and ecological causes? New Phytologist 196: 681–694. [DOI] [PubMed] [Google Scholar]
- Dormann CF, Strauss R. 2014. A method for detecting modules in quantitative bipartite networks. Methods in Ecology and Evolution 5: 90–98. [Google Scholar]
- Dormann CF, Gruber B, Fruend J. 2008. Introducing the bipartite Package: Analysing Ecological Networks. R news 8: 8–11. [Google Scholar]
- Eble GJ. 2004. The macroevolution of phenotypic integration. In: Pigliucci M, Preston K, eds. Phenotypic integration: studying the ecology and evolution of complex phenotypes. New York: Oxford University Press, 253–273. [Google Scholar]
- Edwards CE, Weinig C. 2011. The quantitative-genetic and QTL architecture of trait integration and modularity in Brassica rapa across simulated seasonal settings. Heredity 106: 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis AG, Brockington SF, de Jager ML, Mellers G, Walker RH, Glover BJ. 2014. Floral trait variation and integration as a function of sexual deception in Gorteria diffusa. Philosophical Transactions of the Royal Society of London B 369: 20130563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 1992. Evolution and floral diversity: the phylogenetic surroundings of Arabidopsis and Antirrhinum. International Journal of Plant Sciences 153: S106–S122. [Google Scholar]
- Erwin D. 2007. Disparity: morphological pattern and developmental context. Paleontology 50: 57–73. [Google Scholar]
- Evans AS, Marshall M. 1996. Developmental instability in Brassica campestris (Cruciferae): fluctuating asymmetry of foliar and floral traits. Journal of Evolutionary Biology 9: 717–736 [Google Scholar]
- Faegri K, van der Pijl L. 1979. The principles of pollination ecology. Oxford: Pergamon Press. [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Annual Review of Ecology and Systematics 35: 375–403 [Google Scholar]
- Fernández JD, Bosch J, Nieto-Ariza B, Gómez JM. 2012. Pollen limitation in a narrow endemic plant: geographical variation and driving factors. Oecologia 170: 421–431. [DOI] [PubMed] [Google Scholar]
- Flatt T. 2005. The evolutionary genetics of canalization. The Quartely Review of Biology 80: 287–316. [DOI] [PubMed] [Google Scholar]
- Franzke A, Lysak M, Al-Shehbaz I, Koch M, Mummenhoff K. 2011. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Science 16: 108–116. [DOI] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. American Naturalist 160: 712–726. [DOI] [PubMed] [Google Scholar]
- Galen C. 1996. Rates of floral evolution: adaptation to bumblebee pollination in an alpine wildflower, Polemonium viscosum. Evolution 50: 120–125. [DOI] [PubMed] [Google Scholar]
- Galen C, Cuba J. 2001. Down the tube: pollinators, predators, and the evolution of flower shape in the alpine skypilot, Polemonium viscosum. Evolution 55: 1963–1971. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Hasting A. 1994. A quantitative-genetic model for selection on developmental noise. Evolution 48: 1478–1486. [DOI] [PubMed] [Google Scholar]
- Geber S. 2013. On the relationship between the macroevolutionary trajectories of morphological integration and morphological disparity. PLoS One 8: e63913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JM, Perfectti F. 2010. Evolution of complex traits: the case of Erysimum corolla shape. International Journal of Plant Sciences 171: 987–998. [Google Scholar]
- Gómez JM, Perfectti F, Camacho JPM. 2006. Natural selection on Erysimum mediohispanicum flower shape: insights into the evolution of zygomorphy. The American Naturalist 168: 531–545. [DOI] [PubMed] [Google Scholar]
- Gómez JM, Bosch J, Perfectti F, Fernández JD, Abdelaziz M, Camacho JPM. 2008. Spatial variation in selection on corolla shape in a generalist plant is promoted by the preference patterns of its local pollinators. Proceedings of the Royal Society B 275: 2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JM, Abdelaziz M, Lorite J, Muñoz-Pajares AJ, Perfectti F. 2010. Changes in pollinator fauna cause spatial variation in pollen limitation. Journal of Ecology 98: 1243–1252. [Google Scholar]
- Gómez JM, Perfectti F, Klingenberg CP. 2014. The role of pollinator diversity in the evolution of corolla-shape integration in a pollination-generalist plant clade. Philosophical Transactions of the Royal Society of London B 369: 20130557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JM, Perfectti F, Abdelaziz M, Lorite J, Muñoz-Pajares AJ, Valverde J. 2015. Evolution of pollination niches in a generalist plant clade. New Phytologist 205: 440–453. [DOI] [PubMed] [Google Scholar]
- Gómez-Robles A, Polly PD. 2012. Morphological integration in the hominid dentition: evolutionary, developmental and functional factors. Evolution 66: 1024–1043. [DOI] [PubMed] [Google Scholar]
- González A, Murúa M, Pérez F. 2015. Floral integration and pollinator diversity in the generalized plant-pollinator system of Alstroemeria ligtu (Alstroemeriaceae). Evolutionary Ecology 29: 63–75. [Google Scholar]
- Goswami A, Smaers JB, Soligo C, Polly PD. 2014. The macroevolutionary consequences of phenotypic integration: from development to deep time. Philosophical Transactions of the Royal Society of London B 369: 20130254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafen A. 1989. The phylogenetic regression. Philosophical Transactions of the Royal Society of London B 326:119–157. [DOI] [PubMed] [Google Scholar]
- Green WA. 2012. Stratigraph: Toolkit for the plotting and analysis of stratigraphic and palaeontological data. R package version 0.64. https://cran.r-project.org/web/packages/stratigraph/index.html [Google Scholar]
- Guimerà R, Amaral LAN. 2005. Functional cartography of complex metabolic networks. Nature 433: 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TF, Carter AJR, Pélabon C. 2006. On adaptive accuracy and precision in natural populations. American Naturalist 168: 168–181. [DOI] [PubMed] [Google Scholar]
- Harder LD, Barrett SC. 2006. Ecology and evolution of flowers. Oxford: Oxford University Press. [Google Scholar]
- Harder LD, Johnson SD. 2009. Darwin’s beautiful contrivances: evolutionary and functional evidence for floral adaptation. New Phytologist 183: 530–545. [DOI] [PubMed] [Google Scholar]
- Heinrich B. 1993. The hot-blooded insects. New York: Harvard University Press. [Google Scholar]
- Herrera CM. 1992. Interspecific variation in fruit shape: allometry, phylogeny, and adaptation to dispersal agents. Ecology 73: 1832–1841. [Google Scholar]
- Herrera CM, Cerdá X, García MB, et al. 2002. Floral integration, phenotypic covariance structure and pollinator variation in bumblebee-pollinated Helleborus foetidus. Journal of Evolutionary Biology 15: 108–121. [Google Scholar]
- Heywood VH, Brummitt RK, Culham A, Seberg O. 2007. Flowering plant families of the world. Richmond: Royal Botanical Gardens Kew. [Google Scholar]
- Ishii HS, Harder LD. 2012 Phenological associations of within- and among-plant variation in gender with floral morphology and integration in protandrous Delphinium glaucum. Journal of Ecology 100: 1029–1038. [Google Scholar]
- Ives AR, Midford PE, Garland T. 2007. Within-species variation and measurement error in phylogenetic comparative methods. Systematic Biology 56: 252–270. [DOI] [PubMed] [Google Scholar]
- Kastinger C, Weber A. 2001. Bee-flies (Bombylius spp., Bombyliidae, Diptera) and the pollination of flowers. Flora 196: 3–25. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. 2011. MorphoJ: an integrated software package for geometric morphometrics. Molecular Ecology Resources 11: 353–357. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. 2013. Cranial integration and modularity: insights into evolution and development from morphometric data. Hystrix 24: 43–58. [Google Scholar]
- Klingenberg CP. 2014. Studying morphological integration and modularity at multiple levels: concepts and analysis. Philosophical Transactions of the Royal Society of London B 369: 20130249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP, Marugán-Lobón J. 2013. Evolutionary covariation in geometric morphometric data: analyzing integration, modularity, and allometry in a phylogenetic context. Systematic Biology 62: 591–610. [DOI] [PubMed] [Google Scholar]
- Kuchmeister H, Schmida A, Gottsberger G. 1995. Phenology and pollination ecology of the desert plant Moricandia nitens (Brassicaceae) in the Negev, Israel. Advances in GeoEcology 28: 157–171. [Google Scholar]
- Lay CR, Linhart YB, Diggle PK. 2011. The good, the bad and the flexible: plant interactions with pollinators and herbivores over space and time are moderated by plant compensatory responses. Annals of Botany 108: 749–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JH. 1902. The colors of northern polypetalous flowers. American Naturalist 36: 203–242. [Google Scholar]
- Losos JB. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecology Letters 11: 995–1007. [DOI] [PubMed] [Google Scholar]
- Martins EP, Hansen TF. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. American Naturalist 149: 646–667. [Google Scholar]
- McEwen JR, Vamosi JC. 2010. Floral colour versus phylogeny in structuring subalpine flowering communities. Proceedings of the Royal Society of London Series B 277: 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medel R, Botto-Mahan C, Kalin-Arroyo M. 2003. Pollinator-mediated selection on the nectar guide phenotype in the Andean monkey flower, Mimulus luteus. Ecology 84: 1721–1732. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop, GCE 2010. [Google Scholar]
- Motten AF. 1986. Pollination ecology of the spring wildflower community of a temperate deciduous forest. Ecological Monographs 56: 21–42. [Google Scholar]
- Muchhala N, Johnsen S, Smith SD. 2014. Competition for hummingbird pollination shapes flower color variation in Andean Solanaceae. Evolution 68: 2275–2286. [DOI] [PubMed] [Google Scholar]
- Münkemüller T, Lavergne S, Bzeznik B, Dray S, Jombart T, Schiffers K, Thuiller W. 2012. How to measure and test phylogenetic signal. Methods in Ecology and Evolution 3: 743–756. [Google Scholar]
- Murren CJ, Pendleton N, Pigliucci M. 2002. Evolution of phenotypic integration in Brassica (Brassicaceae). American Journal of Botany 89: 655–663. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. 2004. Analysis of weighted networks. Physical Review E70: 056131. [DOI] [PubMed] [Google Scholar]
- Nattero J, Malerba R, Medel R, Cocucci A. 2011. Factors affecting pollinator movement and plant fitness in a specialized pollination system. Plant Systematics and Evolution 296: 77–85. [Google Scholar]
- Ollerton J, Grace J, Smith K. 2007. Pollinator behaviour and adaptive floral colour change in Anthophora alluadii (Hymenoptera- Apidae) and Erysimum scoparium (Brassicaceae) on Tenerife. Entomologia Generalis 29: 253–268. [Google Scholar]
- Olson EC, Miller RL. 1958. Morphological integration. Chicago: University of Chicago Press. [Google Scholar]
- Ordano M, Fornoni J, Boege K, Domínguez CA. 2008. The adaptive value of phenotypic floral integration. New Phytologist 179: 1183–1192. [DOI] [PubMed] [Google Scholar]
- Orme D. 2013. Caper: comparative analysis of phylogenetics and evolution in R. R package version 0.5. http://CRAN.R-project.org/package=caper. [Google Scholar]
- Ornelas JF, Ordano M, De-Nova AJ, Quintero ME, Garland T., Jr 2007. Phylogenetic analysis of interspecific variation in nectar of hummingbird-visited plants. Journal of Evolutionary Biology 20: 1904–1917. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401: 877–884. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pélabon C, Armbruster WC, Hansen TF, Bolstad GH, Pérez-Barrales R. 2012. Adaptive accuracy and adaptive landscape. In: Svensson EI, Calsbeek R, eds. The adaptive landscape in evolutionary biology. Oxford: Oxford University Press, 150–168. [Google Scholar]
- Penrod M. 2010. Floral phenotypic integration of eight Brassicaceae species from native and non-native enviroments. MS thesis, Charleston College. [Google Scholar]
- Pérez F, Arroyo MTK, Medel R. 2007. Phylogenetic analysis of floral integration in Schizanthus (Solanaceae): does pollination truly integrate corolla traits? Journal of Evolutionary Biology 20: 1730–1738. [DOI] [PubMed] [Google Scholar]
- Pérez-Barrales R, Arroyo J, Armbruster WS. 2007. Differences in pollinator faunas may generate geographic differences in floral morphology and integration in Narcissus papyraceus (Amaryllidaceae). Oikos 116: 1904–1918. [Google Scholar]
- Pérez-Barrales R, Simón-Porcar VI, Santos-Gally R, Arroyo J. 2014. Phenotypic integration in style dimorphic daffodils (Narcissus, Amaryllidaceae) with different pollinators. Philosophical Transactions of the Royal Society of London B 369: 21030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. 2003. Phenotypic integration: studying the ecology and evolution of complex phenotypes. Ecology Letters 6: 265–272. [Google Scholar]
- Pigliucci M, Preston K. 2004. Phenotypic integration: studying the ecology and evolution of complex phenotypes. New York: Oxford University Press. [Google Scholar]
- Revell LJ. 2010. Phylogenetic signal and linear regression on species data. Methods in Ecology and Evolution 1: 319–329. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Revell LJ, Harmon LJ, Collar DC. 2008. Phylogenetic signal, evolutionary process, and rate. Systematic Biology 57: 591–601. [DOI] [PubMed] [Google Scholar]
- Roncal J, Henderson A, Borchsenius F, Cardoso SRS, Balslev H. 2012. Can phylogenetic signal, character displacement, or random phenotypic drift explain the morphological variation in the genus Geonoma (Arecaceae)? Biological Journal of the Linnean Society 106: 528–539. [Google Scholar]
- Rosas-Guerrero V, Quesada M, Armbruster WS, Pérez-Barrales R, Smith SD. 2011. Influence of pollination specialization and breeding system on floral integration and phenotypic variation in Ipomoea. Evolution 65: 350–364. [DOI] [PubMed] [Google Scholar]
- Sánchez R, Serra F, Tárraga J, et al. 2011. Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Research 39: 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry RA, Lord EM. 1996. Developmental stability in flowers of Clarkia tembloriensis (Onagraceae). Journal of Evolutionary Biology 9: 911–930 [DOI] [PubMed] [Google Scholar]
- Singh G. 2010. Plant systematics, an integrated approach, 3rd edn Boca Raton: CRC Press. [Google Scholar]
- Smith SD, Ane C, Baum DA. 2008. The role of pollinator shifts in the floral diversification of Iochroma (Solanaceae). Evolution 62: 793–806. [DOI] [PubMed] [Google Scholar]
- Springer MS, Scally M, Madsen O, De Jong WW, Douady CJ, Stanhope MJ. 2004. The use of composite taxa in supermatrices. Molecular Phylogenetics and Evolution 30: 883–884. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiyan Z, Lianli L, Guang Y, Al Shehbaz IA. 2001. Brassicaceae (Cruciferae). In Flora of China, Vol. 8 Beijing: Science Press and Missouri Botanical Garden, 1–193. [Google Scholar]
- Takahata Y. 2009. Floral variation in the subtribe Brassicinae with special reference to pollination strategies and pollen-ovule ratios. In: Gupta SK, ed. Biology and breeding of Crucifers. Boca Raton, FL: CRC Press, 69–78. [Google Scholar]
- Thomson J. 1983. Component analysis of community-level interactions in pollination systems. In: Jones CE, Little RJ, eds. Handbook of experimental pollination biology. New York: Van Nostrand Reinhold, 451–460. [Google Scholar]
- Torices R, Méndez M. 2014. Resource allocation to inflorescence components is highly integrated despite differences between allocation currencies and sites. International Journal of Plant Sciences 175: 713–723. [Google Scholar]
- Toro-Nuñez O, Al-Shehbaz IA, Mort ME. 2015. Phylogenetic study with nuclear and chloroplast data and ecological niche reveals Atacama (Brassicaceae), a new monotypic genus endemic from the Andes of the Atacama Desert, Chile. Plant Systematics and Evolution 301: 1377–1396. [Google Scholar]
- Warwick SI, Francis A, Al-Shehbaz I. 2006. Brassicaceae: check-list and database on CD-ROM. Plant Systematics and Evolution 259: 249–258. [Google Scholar]
- Williams J, Conner JK. 2001. Sources of variation in floral traits in wild radish, Raphanus raphanistrum (Brassicaceae). American Journal of Botany 88: 1577–1581. [PubMed] [Google Scholar]
- Willmer P. 2011. Pollination and floral ecology. Princeton University Press, Princeton, USA. [Google Scholar]
- Wilson P, Wolfe AD, Armbruster WS, Thomson JD. 2007. Constrained lability in floral evolution: counting convergent origins of hummingbird pollination in Penstemon and Keckiella. New Phytologist 176: 883–890. [DOI] [PubMed] [Google Scholar]
- Young NM. 2006. Function, ontogeny and canalization of shape variance in the primate scapula. Journal of Anatomy 209: 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch ML, Swiderski DL, Sheets HD. 2012. Geometric morphometrics for biologists, a primer, 2nd edn. London: Academic Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.