Abstract
Background and aims Phenotypic diversification of flowers is frequently attributed to selection by different functional groups of pollinators. During optimization of floral phenotype, developmental robustness to genetic and non-genetic perturbations is expected to limit the phenotypic space available for future evolutionary changes. Although adaptive divergence can occur without altering the basic developmental programme of the flower (ontogenetic scaling hypothesis), the rarity of reversion to ancestral states following adaptive radiations of pollination syndromes suggests that changes in the ancestral developmental programme of the flower are common during such evolutionary transitions. Evidence suggests that flower diversification into different pollination syndromes in the Loasoideae genus Caiophora took place during a recent adaptive radiation in the central Andes. This involved transitions from bee to hummingbird and small rodent pollination. The aim of this work was to examine if the adaptive radiation of pollination syndromes in Caiophora occurred through ontogenetic scaling or involved a departure from the ontogenetic pattern basal to this genus.
Methods We used geometric morphometric variables to describe the shape and size of floral structures taking part in the pollination mechanism of Loasoideae. This approach was used to characterize the developmental trajectories of three species basal to the genus Caiophora through shape–size relationships (ontogenetic allometry). We then tested if the shape–size combinations of these structures in mature flowers of derived Caiophora species fall within the phenotypic space predicted by the development of basal species.
Key Results Variation in the size and shape of Caiophora flowers does not overlap with the pattern of ontogenetic allometry of basal species. Derived bee-, hummingbird- and rodent-pollinated species had divergent ontogenetic patterns of floral development from that observed for basal bee-pollinated species.
Conclusions The adaptive radiation of Caiophora involved significant changes in the developmental pattern of the flowers, rejecting the ontogenetic scaling hypothesis.
Keywords: Loasaceae, subfam. Loasoideae, Caiophora, Loasa, Blumenbachia, adaptive radiation, pollination syndrome, reversion, ontogenetic allometry, developmental robustness, ontogenetic scaling
INTRODUCTION
Flower size expresses a tremendous range of variation (>1000-fold), from flowers of less than 1 mm to flowers of nearly 1 m in diameter (Davis et al., 2008). Substantial variation in flower size has been attributed to different pollination strategies (Davis et al., 2008) and can be observed even within related lineages that experienced adaptive radiations (e.g. Whittall and Hodges, 2007; Givnish et al., 2009). There is consistent evidence of pollinator-mediated selection on flower size (Harder and Johnson, 2009) and shape (Benitez-Vieyra et al., 2006; Gómez et al., 2006, 2014). Nevertheless, we are still far from understanding how the overwhelming diversity of floral designs arose from a conserved Bauplan.
Among plants, advances in evo-devo have revealed the presence of mechanisms promoting stability (robustness) during flower development, for example robustness of gene interactions (Lenser et al., 2009; Alvarez-Buylla et al., 2010) and robustness of the patterning of tissue development (Breuninger and Lenhard, 2010). Robust flower development may be important for two reasons. First, in plant species with pollination systems specialized on a functional group of pollinators (e.g. dependent on hummingbird pollination) (Ollerton et al., 2007), optimization of the mechanical fit with the pollinator’s behaviour and morphology is expected to promote the evolution of developmental stability and high phenotypic integration of the flower (Berg, 1960; Pérez-Barrales et al., 2007; Pélabon et al., 2011). Second, the stability of early flower developmental stages may be crucial for the performance at later stages (Rice, 1990; Arthur, 2011). As floral development can be described as a phenotypic continuum in which many genetic and hormonal determinants are shared (Dornelas et al., 2012) and developing tissues interact influencing each other mechanically (Endress, 2006), cascading effects may be expected. Despite the observed strong developmental stability of the flower due to its functional role in reproduction (Pélabon et al., 2011), floral diversification is a central aspect of angiosperm evolution (van der Niet and Johnson, 2012; Rosas-Guerrero et al., 2014). While regulatory (pleiotropic) genes are known to have significant effects on the stability of flower development (Juenger et al., 2000; Li et al., 2012), mutations affecting this pleiotropic control of development are likely to enable flower diversification (e.g. Hermann and Kuhlemeier, 2011).
There are two ways in which morphological diversification can arise from an ancestral developmental trajectory. On the one hand, provided that the ancestral developmental trajectory is allometric, i.e. that shape changes during development with the increase in size, evolution can take place without a rearrangement of the shape–size relationship that characterizes floral growth in the ancestral species. In this case, evolutionary changes constitute proportional changes in size and shape, as predicted by the ontogenetic scaling hypothesis (Fig. 1, situations A and B) (Gould, 1977; Klingenberg, 1998). Changes in the time or rate of development (heterochrony) along the developmental pathway described through the shape–size relationship of the ancestral ontogeny can explain diversification under the ontogenetic scaling hypothesis (Fig. 1) (Klingenberg, 1998; Gould, 2002, Ch. 10). On the other hand, evolutionary changes can involve departures from the ancestral developmental trajectory by changes in either the intercept and/or the slope of the shape–size relationship (Fig. 1, situations C and D) (Klingenberg, 1998). During adaptive diversification, developmental robustness will be expressed as proportional changes in size and shape. In contrast, diversification of shape independently of size would indicate either low ancestral developmental robustness or a disruption of robustness because of strong directional selection.
Fig. 1.
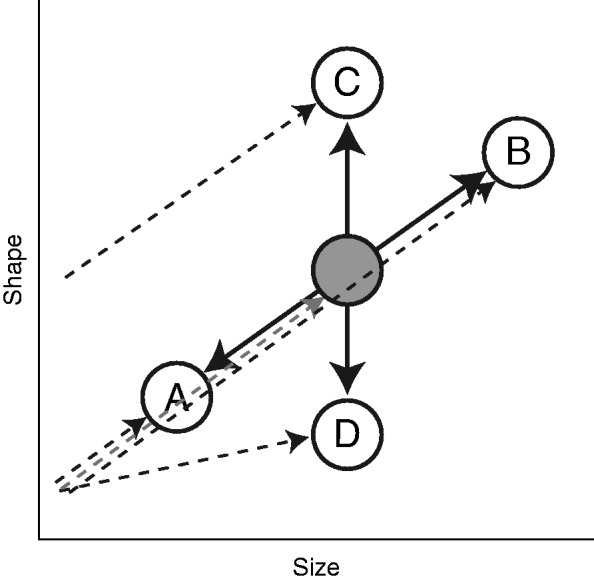
Schematic representation of the evolution of a morphological structure following or not following the ontogenetic scaling expectation. The grey circle represents the hypothetical shape–size configuration in the ancestral lineage at a given developmental stage (e.g. the adult). A, B, C and D are shape–size configurations of the same developmental stage in derived species. Thick arrows connecting the ancestral and derived species indicate the direction of the morphological changes shown by each species. Dotted single headed arrows indicate the developmental trajectory leading to the adult stage in each species: ancestral developmental trajectory (grey), derived developmental trajectories (black). Species A and B evolved through ontogenetic scaling (following the ancestral pattern of ontogenetic allometry); species C and D experienced a divergence from the ancestral developmental trajectory.
Adaptive radiations are exciting natural settings to examine how changes to ancestral developmental trajectories allow rapid selection-driven morphological diversification (Schluter, 2000). Following the ontogenetic scaling hypothesis, flower adaptive divergence can occur without significant modifications to the basic developmental programme of the flower (Fig. 1). This can take place either through the extension or through the earlier interruption of the ancestral developmental programme; changes in the rate at which the ancestral developmental programme unfolds can also allow for this kind of divergence (e.g. Guerrant, 1982; Armbruster, 2012). As the developmental trajectory remains the same, these processes would not imply the loss of the ancestral pattern of developmental robustness. The fact that reversion to the ancestral pollination syndrome or sexual system following an adaptive radiation is rare, however, suggests that this may not generally be the case among flowering plants (Whittall and Hodges, 2007; Barrett, 2013). Developmental changes to the flower, involving the loss of the ancestral shape–size ontogenetic relationship, may explain why reversals to the ancestral state are limited. Although some evidence suggests ontogenetic scaling to be involved in the diversification of animal and plant architecture (e.g. Maie et al., 2007; Miller et al., 2008; Olson et al., 2009), this hypothesis has not yet been tested for flower diversification.
The genus Caiophora C.Presl. (Loasaceae, subfam. Loasoideae) recently diversified in the central Andes in the context of an adaptive radiation from a presumably bee-pollinated ancestor (Ackermann, 2012). Caiophora includes not only bee-pollinated species, but also several hummingbird-pollinated species (Ackermann & Weigend, 2006) and even a species pollinated by small rodents (Cocucci and Sérsic, 1998). The aim of this study was to determine whether the diversification of Caiophora supports the ontogenetic scaling hypothesis of proportional changes in flower size and shape (Fig. 1, situations A and B) or if it involved changes in shape–size relationships through deviation from the direction of the ancestral ontogenetic trajectory (Fig. 1, situations C and D). Using geometric morphometrics we first described the morphology of mature flowers in all analysed species, and then described the ontogenetic trajectory of floral development in basal species of the subfamily (which are expected to represent the ancestral condition) and finally tested if the morphology of the mature flowers in different derived Caiophora species overlapped with the direction of the developmental trajectory predicted by basal species. If the morphology of mature flowers in Caiophora overlaps with the direction of the developmental trajectory in basal species (Fig. 1, situations A and B), flower diversification in Caiophora probably took place by ontogenetic scaling, conserving the ancestral developmental programme. In contrast, if the morphology of the mature flowers does not overlap with the direction of the developmental trajectory of the basal species (Fig. 1, situations C and D), evolution of flower morphology probably involved a rearrangement of the ancestral developmental programme.
METHODS
Study system
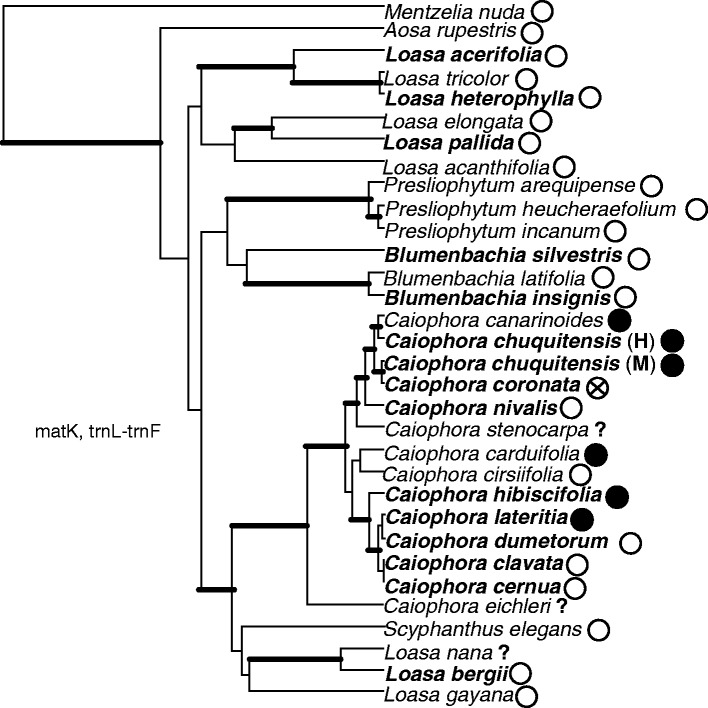
Loasaceae subfam. Loasoideae is a monophyletic and mostly Neotropical angiosperm subfamily. Species in Loasa Adans., Blumenbachia Schrad. and Scyphanthus Sweet. (basal genera to Caiophora) are only pollinated by bees, while the genera Nasa Weigend. and Caiophora present bee and hummingbird pollination (Ackermann, 2012). A single species of this last genus, Caiophora coronata (Gillies ex Arnott) Hook. & Arn., is pollinated by small rodents (Cocucci and Sérsic, 1998). The origin of Caiophora is dated to the late Oligocene (Schenk and Hufford, 2010) and coincides with the arrival of hummingbirds in South America (McGuire et al., 2014). Additionally, species can hybridize and give rise to viable descendants in only a few generations, which suggest recent divergence between Caiophora species (Ackermann et al., 2008). As adaptive radiations are characterized by rapid diversification linked to novel ecological opportunities, for example the presence of a new pollinator in a given environment (Simpson, 1953), these pieces of evidence suggest that Caiophora diversified in the context of a recent adaptive radiation, potentially triggered or facilitated by the arrival of hummingbirds in South America. No reversion seems to have taken place during the evolution of pollination strategies in Caiophora (Fig. 2)
Fig. 2.
Phylogenetic relationships among Loasoideae (Loasaceae) species. The topology of the phylogeny is the maximum-clade-credibility tree of a Bayesian analysis performed with Hufford (2005) and our own matK and trnL–trnF plastid sequences. Branch lengths are proportional to evolutionary time. The branches with posterior probabilities above 0·95 and the species selected for this study are in bold type. Pollination syndromes are indicated: white = bee pollination, black = hummingbird pollination, strikethrough circle = small rodent pollination. The species for which the pollination strategy is unknown are indicated with ‘?’.
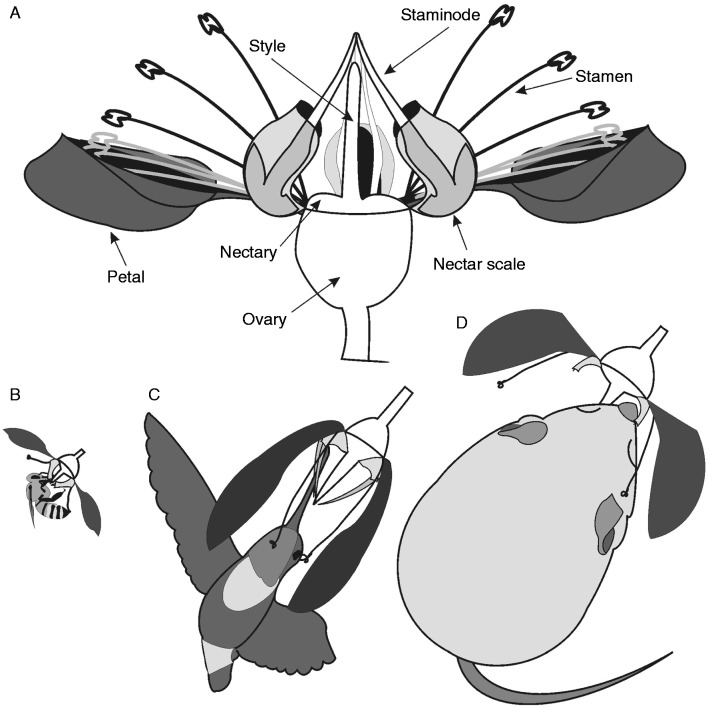
Loasoideae flowers are actinomorphic, with free hood-shaped petals enclosing the stamens (Fig. 3A). Flowers present an additional androecium-derived flower whorl, which is not present in other angiosperm lineages (Brown and Kaul, 1981; Hufford, 2003). This whorl is composed of five nectar scales, each of which is the result of three merged stamens. Each nectar scale encloses two staminodes, derived from the modification of two stamens, protruding from above the nectar scale (Fig. 3A). Variation in the size and shape of the corolla and the staminodial complex, i.e. the unit composed of the nectar scale and the two protruding staminodes, has been attributed to selection mediated by different pollinators (Ackermann and Weigend, 2006; Weigend, et al., 2010; Ackermann, 2012; M. Ackermann and M. Weigend, University of Bonn, pers. comm.). Flowers are reported to be small in bee-pollinated Loasoideae species, and increasingly larger in hummingbird- and rodent-pollinated species (Ackermann and Weigend, 2006). Corolla size is positively correlated with the amount of sugar in the nectar of Caiophora (Ackermann, 2012). Bee-pollinated species have small pendulous flowers, which require the pollinator to land and hold onto the flower by grappling the nectar scales. Corollas are remarkably open in these flowers, making the nectar scales visible and easy to grasp (Fig. 3B) (Weigend, 2004). Hummingbird-pollinated species have narrow and large corollas (Fig. 3C). Staminodes are remarkably long in hummingbird-pollinated species; by guiding the pollinator’s beak towards the nectar container, these staminodes ensure the contact of the hummingbird head with fertile flower structures (Fig. 3C) (M. Weigend, pers. comm.). In the bee-pollinated flowers, pollinator manipulation of the staminodial complex activates a stamen release mechanism. This mechanism is activated when the bee inserts its proboscis between the scale and the two protruding staminodes (which block the nectar scale) and moves the scale outwards to access the nectar (Weigend et al., 2010). No function has been reported for the staminodial complex in the rodent-pollinated species C. coronata, besides the function of the nectar scale as a nectar container. This leaves no functional role for the staminodes, and may explain why these structures are remarkably reduced in this species (Fig. 3D; M. Weigend, pers. comm.).
Fig. 3.
Loasoideae flowers and pollination syndromes. (A) Schematic representation of a typical Loasoideae flower, indicating the name of each floral structure. (B) Bee-pollinated flower; (C) hummingbird-pollinated flower; (D) flower of Caiophora coronata, a rodent-pollinated species (note the reduced staminodes).
Sampling
The species included in this study belong to the genera Loasa, Blumenbachia and Caiophora. The sampling includes between six and 25 mature flowers in anthesis of 14 Loasoideae species (eight belonging to Caiophora and the remaining six to Loasa and Blumenbachia; Supplementary Data Table S1). A single population was sampled per species. We sampled individuals depending on availability of plant material in the field, and retained a single mature flower per individual [except for Caiophora cernua (Griseb.) Urb & Gilg ex Kurtz. for which we sampled ten mature flowers from the single available individual]. The sampling covers all pollination strategies reported for Caiophora. For Loasa acerifolia Dombey ex Juss., Blumenbachia silvestris Poepp. and Blumenbachia insignis Schrad., we collected four flower buds in addition to the mature flower, covering for each individual a range of flower bud diameter from approx. 3 to 10 mm. The sample sizes for mature flowers and flower buds in these three species were as follows: 24 mature flowers and 96 flower buds (Loasa acerifolia); 14 mature flowers and 56 flower buds (Blumenbachia silvestris); 22 mature flowers and 88 flower buds (Blumenbachia insignis). Samples were kept in 70 % EtOH and later dissected, with its structures being photographed using a Leica M420 stereomicroscope (Wetzlar, Germany). The petal and the staminodial complex were photographed in lateral view.
Data collection
The shape and size of the corolla and the staminodial complex were characterized using geometric morphometric tools as both structures contribute to the pollination mechanism of Loasoideae. We used petal shape as an estimator of corolla shape because it is easier to follow developmental changes in the shape of the petal than in the corolla. Petals with a more expanded base correspond to more open corollas, whereas the opposite can be said for narrow corollas (Fig. 3B, C).
We used the program tpsDig (Rohlf, 2009) to plot landmarks and semi-landmarks on the petal (Supplementary Information Fig. S1A) and on the staminodial complex (Fig. S1B) of mature flowers and developing buds of Loasa acerifolia, Blumenbachia silvestris and Blumenbachia insignis.
Landmark coordinates were also obtained for mature flowers in the remaining Loasa species (Loasa heterophylla Hook & Arn., Loasa pallida Gillies ex Arn. and Loasa bergii Hieron.) and for the mature flowers of all Caiophora species selected for this study.
Morphometric analysis
To extract shape information, we applied a Procrustes fit using the program MorphoJ (Klingenberg, 2011). We also computed the centroid size (in centimetres) of each analysed structure as a measure of size. Two data sets were created. The first contained developmental data of basal species including the Procrustes coordinates and the centroid size of Loasa acerifolia, Blumenbachia insignis and Blumenbachia silvestris; the second contained the Procrustes coordinates and the centroid size of mature flowers in all sampled species of Loasa, Blumenbachia and Caiophora.
A first set of morphometric analyses examined whether there is an association between the shape and the size of mature flowers in different Loasoideae species and whether variation in mature flower size and shape is associated with different pollination strategies. To characterize allometry in Loasoideae, we used a multivariate regression model (Monteiro, 1999), in which the variables describing the shape of each flower structure (Procrustes coordinates) were regressed against the log-transformed centroid size. We used the log-transformed centroid size instead of the raw centroid size of each structure because this yields a more linear relationship between size and shape for evolutionary or ontogenetic allometry (Klingenberg et al., 2012). The resulting regression scores summarize the variation in Procrustes coordinates that is the most closely associated with size in each structure (Drake and Klingenberg, 2008). This method is widely used in geometric morphometric studies to characterize the allometry of plant parts and usually achieves a fairly good fit of the data to a straight-line relation (Klingenberg et al., 2012; Silva et al., 2012; Viscosi et al., 2012).
To visualize the evolutionary history of changes in flower size and shape, we projected a phylogeny of Loasoideae (Fig. 2; Supplementary Information Text file S1) into the scatter plots of the log-transformed centroid size and the regression scores (a type of graph sometimes called phylomorphospace; Klingenberg and Ekau, 1996; Sidlauskas, 2008). The positions of internal nodes were determined according to the ancestral reconstruction of those variables using the phytools package in R (Revell, 2012).
Differences in mature flower morphology when comparing species with different pollination strategies were also examined with phylomorphospaces. We separated phylomorphospaces for the petal and for the staminodial complex, using the two first principal components of the Procustes coordinates of each structure. We used a phylogenetic MANOVA to test for differences in the size and in the shape of flower structures between bee- and hummingbird-pollinated species (the single rodent-pollinated species was excluded from the analysis). The log-transformed centroid size of both structures and the principal components used in the phylomorphopaces were included in this analysis. Phylogenetic MANOVA was done with the aov.phylo function (Garland et al., 1993) of the geiger package in R (Harmon et al., 2008), for each flower structure separately. The significance level was assessed using 10 000 iterations.
After describing the size and the shape of mature flowers in different Loasoideae species, we tested if ontogenetic scaling was involved in flower diversification in Caiophora. In a first step, we tested if the size and the shape of flower structures were significantly associated along the ontogenetic trajectory of species basal to Caiophora, i.e. we tested for significant ontogenetic allometry in basal species (Klingenberg, 1998). We assumed basal species to represent the ancestral condition in Loasoideae. The developmental trajectories of the petal and the staminodial complex of Loasa acerifolia, Blumenbachia insignis and Blumenbachia silvestris were estimated using a multivariate regression of Procrustes coordinates against the log-transformed centroid size (Monteiro, 1999). To obtain a combined estimate of ontogenetic allometry from all three species, we used a pooled within-species regression (Klingenberg et al., 2012). The developmental trajectories of the three basal Loasoideae species were reconstructed using the morphometric data of different developmental flower stages. To assess the assumption of linear allometric trajectories, we inspected plots of the regression scores (Drake and Klingenberg, 2008) against log-transformed centroid size. As a more formal test, we also performed a multivariate quadratic regression of shape on size (pooled within species) for both the petals and the staminodial complex. We assessed statistical significance by a Goodall’s (1991) F-test of the predicted sums of squares from the quadratic versus linear regressions.
To test whether derived species of Caiophora evolved by ontogenetic scaling (A or B in Fig. 1) or in other ways such as lateral transposition of growth trajectories (C in Fig. 1) or changes of ontogenetic allometry (D in Fig. 1), the ontogenetic regression model of basal species was used to perform an allometric size correction for mature flowers from all the taxa. For this purpose, regression residuals were computed by using the vector of regression coefficients for the common estimate of ontogenetic allometry in the three basal Loasoideae species. If differences among taxa evolved by ontogenetic scaling, by extending or truncating the ancestral ontogenetic trajectory, such differences among taxa should vanish in the residuals (in Fig. 1, taxa A and B would have identical residual shapes after such a correction, even though they have very different sizes). By contrast, differences among taxa that evolved by lateral shifts of ontogenetic trajectories or by changes in ontogenetic allometry would persist even after such an allometric correction (in Fig. 1, taxa C and D have residuals that differ from each other and from those of taxa A and B).
Residuals corrected for ontogenetic allometry were obtained by applying the regression vectors from the ontogenetic regression analysis to shape data of mature flowers, which removes the effect of any ontogenetic scaling from the differences among species. These residuals were then used in a canonical analysis (CVA), to display the shape differences among taxa optimally and in reduced dimensionality. Using the residual CVA plot we explored the overlap of the 90 % confidence ellipses of the mature flowers of different species and used the multi-response permutation procedure (MRPP) of within- versus among-group dissimilarities implemented in the vegan package of R (Oksanen et al., 2015) to test for the overlap of the shape residuals in Caiophora with the shape residuals in the Loasa–Blumenbachia group. The MRPP of within- versus among-group dissimilarities is a permutation test that allows determination of whether distances among specimens in two different groups (distances in the CVA space in this study) are significantly larger than distances among specimens within those groups. The groups considered for the hypothesis test were: (a) basal species vs. Caiophora; (b1) basal species vs. Caiophora bee-pollinated species; (b2) basal species vs. Caiophora hummingbird-pollinated species; (b3) basal species vs. the Caiophora rodent-pollinated species. Ten thousand permutations were made for each comparison.
RESULTS
Inter-specific shape–size associations in Loasoideae (evolutionary allometry)
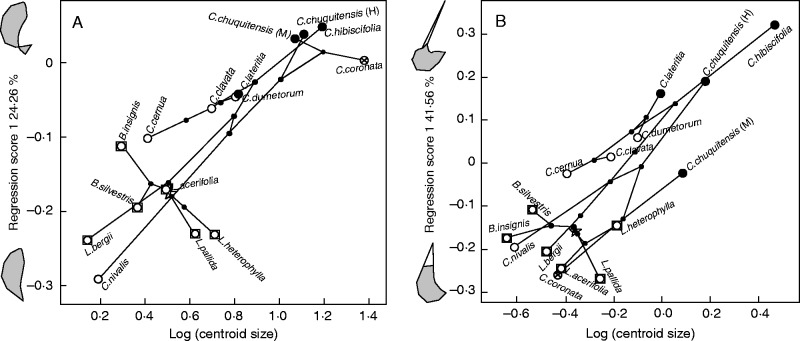
Evolutionary allometry of the petal and the staminodial complex is significant in Loasoideae (P < 0·01 and P < 0·001, respectively). In total, 24·26 % of interspecific petal shape variation is associated with variation in petal size. Small petals with a more expanded base are found in all species basal to Caiophora (Fig. 4A). With the exception of Caiophora nivalis Lillo and C. cernua, which evolved towards smaller petal sizes, Caiophora species have larger petals with a more retracted base. This tendency is particularly accentuated in Caiophora species pollinated by hummingbirds and small rodents (Fig. 4A).
Fig. 4.
Phylomorphospaces describing evolutionary allometry in (A) the petal and (B) the staminodial complex. The percentage of shape variation explained by size is shown along with the shape variants associated with different sizes. The root of the phylogenetic tree is indicated with a star. Basal species are indicated with a square. Pollination strategies are indicated: white = bee pollination, black = hummingbird pollination, strikethrough circle = small rodent pollination.
For the staminodial complex, 41·56 % of interspecific shape variation is size-dependent. Small staminodial complexes, with a staminode slightly protruding from above the nectar scale, are found in species basal to Caiophora (Fig. 4B). The staminodial complex increased its size and acquired more protruding staminodes in most of the Caiophora species, which is particularly striking in hummingbird-pollinated species. This trend was reversed during the evolution of the rodent-pollinated species, C. coronata (Fig. 4B).
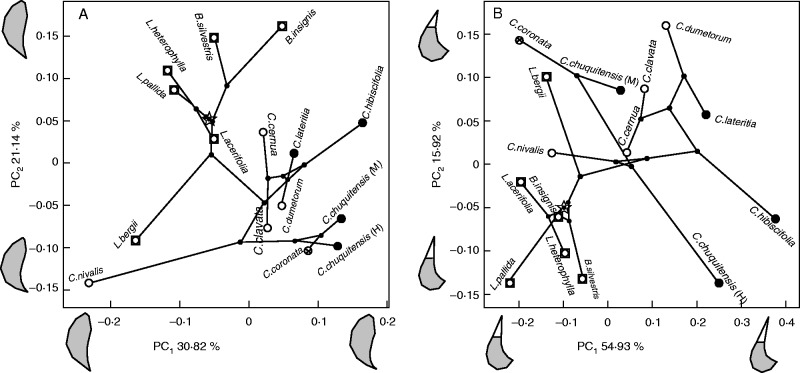
Pollinator-dependent variation in flower shape and size
The first two principal components of petal Procustes coordinates explain 51·96 % of inter-specific variation in petal shape (Fig. 5A); the first two principal components of Procustes coordinates of the staminodial complex explain 70·85 % of inter-specific shape variation (Fig. 5B). With the exception of L. bergii, all species basal to Caiophora are clustered together and occupy a narrow region of the flower morphospace. The evolution of Caiophora and its more closely related basal species, L. bergii, involved a departure from this cluster and the colonization of a wider region of flower morphospace (Fig. 5). Variation in shape, in general, corresponds to the variation in flower morphology expected under different pollination strategies. The petal base is more retracted in the hummingbird-pollinated species (which corresponds to a narrow corolla) (Fig. 5A) and the staminodial complex tends to have a more protruding staminode (Fig. 5B). The petal tends to have an elongated base in the bee-pollinated species (corresponding to an open corolla) (Fig. 5A) and the staminode protrudes slightly from above the nectar scale (Fig. 5B). Petals in the rodent-pollinated species have a retracted base (Fig. 5A) and the staminode almost does not protrude from above the nectar scale (Fig. 5B).
Fig. 5.
Phylomorphospaces describing the shape of (A) the petal and (B) the staminodial complex in mature flowers of different Loasoideae species. The percentage of variation explained by each principal component is shown, along with the shape transformation of the structure along the axis. The root of the phylogenetic tree is indicated with a star. Pollination strategies are indicated: white = bee pollination, black = hummingbird pollination, strikethrough circle = small rodent pollination.
The shape and the size of the two analysed flower structures differ significantly between bee- and hummingbird-pollinated species in Loasoideae (petal: Wilk’s λ = 0·31456, approx. F1,12 = 7·2635, P < 0·01, phylogenetic P < 0·05; staminodial complex: Wilk’s λ = 0·30262; approx. F1,12 = 7·6815, P < 0·01, phylogenetic P < 0·05). Flower structures are significantly larger in hummingbird- than in bee-pollinated species (Fig. 4), the petal base is significantly more retracted and staminodes protrude significantly more from above the nectar scale (Fig. 5).
Ontogenetic shape–size associations in species basal to Caiophora (ontogenetic allometry)
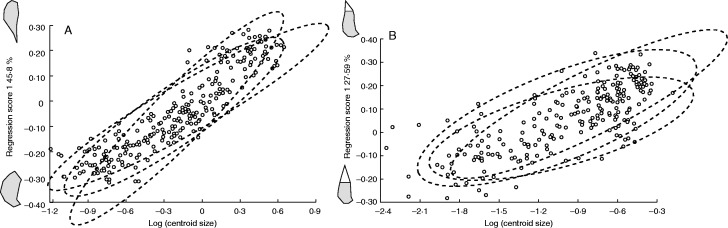
Ontogenetic allometry was statistically significant (P < 0·0001) in both the petal and the staminodial complex, and size accounted for 45·8 and 27·59 % of the total variation in shape, respectively. Plots of regression scores appear linear on a log scale (Fig. 6) and the predicted sums of squares from quadratic regressions exceeded those from linear regressions only marginally and this difference was not statistically significant either for the petals (Goodall’s F48,24 = 1·09, P = 0·41) or for the staminodial complex (Goodall’s F40,20 = 1·13, P = 0·39). Linear multivariate regression therefore provides a satisfactory approximation of the growth trajectories. Petal growth in the basal species can be characterized by an expansion of the petal base (Fig. 6A), in correspondence with a progressive flower opening; while the growth of the staminodial complex can be characterized by the nectar scale overgrowing the staminode (Fig. 6B) ending in the typical melithophilous configuration.
Fig. 6.
Ontogenetic allometry of (A) the petal and (B) the staminodial complex. Dotted ellipses represent 90 % confidence ellipses for shape–size configurations along the ontogeny of Loasa acerifolia, Blumenbachia silvestris and Blumenbachia insignis. Ontogenetic changes in the shape of these structures are schematically represented.
Ontogenetic scaling hypothesis test
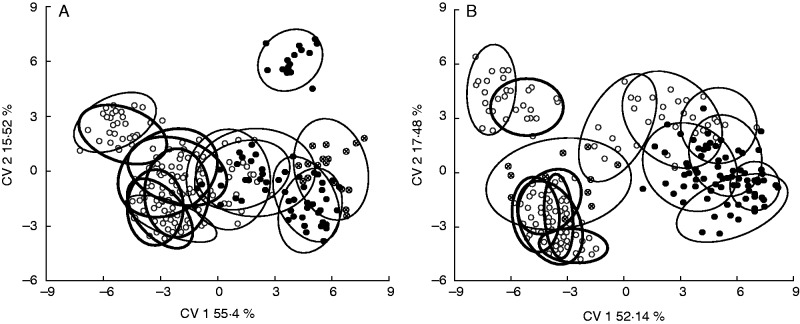
The 90 % confidence ellipses of residuals corresponding to basal bee-pollinated species constitute a cluster in the space defined by the first two CV axes (Fig. 7). For a single basal species, L. bergii, residuals of the staminodial complex depart from this cluster along CV2 (Fig. 7B). The residuals of the two floral structures in derived hummingbird-pollinated species do not overlap with residuals of basal bee-pollinated species (except for the petal in a single derived species, Caiophora lateritia (Hook.) Benth.) (Fig. 7). Residuals corresponding to the two structures differ between basal and derived bee-pollinated species (Fig. 7). The petal residuals of the rodent-pollinated species do not overlap with the residual configuration of the basal bee-pollinated species (Fig. 7A), but this is not the case with the staminodial complex (Fig. 7B). All the MRPP tests were statistically significant at the conventional 5 % level (P < 0·0001 in comparisons a, b1, b2 and b3).
Fig. 7.
Graphical output of the CVAs performed for (A) the petal and (B) the staminodial complex. The 90 % confidence ellipses for the residual configuration of each species were added. Bold ellipses correspond to the species basal to Caiophora. Each small circle represents an individual. Pollination strategies are indicated: white = bee pollination, black = hummingbird pollination, strikethrough circle = small rodent pollination.
DISCUSSION
Results indicate that during the radiation of Caiophora, floral development in bee-, hummingbird- and rodent-pollinated species probably followed ontogenetic trajectories (shape–size relationship during development) that differ from that of basal bee-pollinated species. Furthermore, residuals of derived species presenting new pollination strategies tend to cluster together and occur well beyond the range of residuals of basal bee-pollinated species. This last pattern adds to the expectation that evolutionary transitions in pollination strategies exert, at least in part, an effect on floral development. The pattern uncovered in the present study rejects the ontogenetic scaling hypothesis of proportional changes in the developmental shape–size relationship during the diversification of Caiophora and suggests that robustness of floral development did not condition the evolution of flower morphology during the radiation of this genus. In species basal to Caiophora, between 27 and 46 % of the variation in flower shape during flower ontogeny was significantly related to flower size. If the ancestor of Caiophora had similar values, the remaining size-independent shape variation may have been a source of variability on which selection could act, thus allowing the diversification of flower morphology beyond ancestral ontogenetic allometry. Notice that changes in trait proportions during the diversification of Caiophora are found not only in the developmental shape–size relationship of individual flower structures, but also in the relationship between the size of the petal and the size of the staminodial complex: although flowers in most Caiophora species appear to be scale-upped versions of flowers in basal species (the petals and the staminodial complexes are larger in this genus), a remarkable decoupling of the relationship between the size of the petal and the size of the staminodial complex seems to have taken place in C. coronata, the single rodent-pollinated species in Loasoideae. While this species has extremely large petals, the size of the staminodial complex is comparable to the smaller size of this structure in basal bee-pollinated species.
As opposed to the dramatic changes in flower proportions that seem to have accompanied diversification in Caiophora, no significant developmental rearrangements appear to have affected the evolution of flower morphology in bee-pollinated species basal to this genus. Notice that the residuals of these species cluster closely together. The residuals of L. bergii, the studied species most closely related to Caiophora, tend to depart slightly from the main cluster of basal species, suggesting that the departure from the ancestral shape–size relationship may have began shortly before the radiation of Caiophora. The evolutionary pattern of interspecific variation in the shape of mature flowers, according to which flower shape in L. bergii departs from the shape of the remaining basal species, also suggests a developmental alteration shortly before the radiation of Caiophora.
Interestingly, the size and shape configurations suitable for hummingbird pollination (a narrow and large corolla and a large staminodial complex with protruding staminodes) are not available along the direction of ontogenetic allometry in species basal to Caiophora. Here, petals have a retracted base and staminodes protrude remarkably from above the nectar scale only during early developmental stages. A decoupling between the developmental programmes of flower size and shape during the evolution of Caiophora may have facilitated the evolution of ‘overdeveloped’ size and ‘underdeveloped’ shape in hummingbird-pollinated flowers. The recombination of extant developmental programmes in unprecedented ways during adaptive radiations is proposed to be an important mechanism giving rise to novel phenotypic traits (West-Eberhard, 2005).
Although the ancestral pattern of ontogenetic allometry was not conserved during the diversification in Caiophora, evolutionary allometry is significant in Loasoideae and may reflect a functional association between flower size and shape. Evolutionary allometry in Loasoideae reflects a shape–size relationship that is, roughly speaking, opposed to the shape–size relationship along the ontogeny of basal species: species with larger flowers present shapes corresponding to less developed stages along the ancestral developmental trajectory. As mentioned before, this shape–size combination may represent an adaptation to hummingbird pollination, while small flowers with ‘fully’ developed shapes may represent an adaptation to bee pollination. Although some Loasoideae species are pollinated either only by bees or only by hummingbirds, long-tongued bees seem to also contribute to pollination in some hummingbird-pollinated Caiophora species (Ackermann et al., 2008). Thus, the pattern of evolutionary allometry observed in Loasoideae may reflect the evolutionary tuning by pollinators, with species specialized either on bee or on hummingbird pollination represented by extreme values in the direction of evolutionary allometry and species with mixed pollination strategies occupying intermediate positions (Shoval et al., 2012).
Robustness is a property of biological systems that allows the maintenance of performance in spite of internal (genetic) and external (environmental) perturbations (Ciliberti et al., 2007). During development, robustness warrants the stability of the phenotype to attain a particular design promoting fitness (Waddington, 1942; Jablonka & Lamb, 2006). The flower has been historically viewed as a strongly integrated and developmentally stabilized plant organ (Berg, 1960; Pélabon et al., 2011). Despite variation in flower integration having been linked to adaptations, for example the degree of generalization of the pollination system (Rosas-Guerrero et al., 2010; Gómez et al., 2014) and type of breeding (Rosas-Guerrero et al., 2010) and mating system (Fornoni et al., 2016), the role of morphological floral integration and developmental stability during the radiation of pollination syndromes remains unknown. In this study we support the expectation of changes in the developmental trajectory of functional floral organs during the diversification of pollination syndromes. Our field observations that some individuals of two species of Caiophora display aberrant flower morphologies (something we did not observe in populations of Loasa and Blumenbachia species) suggests a relaxation of developmental robustness during the diversification of Caiophora, which may have enabled the diversification of developmental trajectories reported in our study. Moreover, there is preliminary evidence pointing to the remarkable responsiveness of flower morphology in Caiophora species grown under different temperatures in greenhouse conditions (Ackermann, 2012).
Evolutionary innovations related to the appearance of the hummingbird pollination syndrome involved changes in flower colour, nectar composition, and changes in flower shape and design (Stebbins, 1970; Rausher, 2008). A common pattern behind this transition is rarity of reversal from hummingbird to bee pollination, suggesting that some kind of constraint reduces the opportunities for reversible evolution. The absence of genetic variation (Williams, 1966), phyletic heritage, the nature of selection acting on pollination and mating systems, developmental–genetic constraints (Barrett, 2013) and the kind of mutational changes (Rausher, 2008) are among the explanations that have been proposed to account for irreversible evolution among pollination syndromes. Our results suggest that changes in the ancestral ontogenetic trajectory of flower shape and size may also contribute to strong directionality and irreversibility during the evolution of pollination syndromes. It has been proposed that flower traits lost during the evolution of a lineage are unlikely to be regained in the descendants in the same form in which they existed in recent ancestors, the probability of regaining those traits being inversely proportional to the complexity of the trait (Stebbins, 1974; but see Goldberg and Igic, 2008).
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: sampled localities, principal pollinators and sample sizes of the species in this study. Figure S1: photographs of (A) the petal showing the petal flap and (B) the staminodial complex indicating the location of landmarks and semi-landmarks.
ACKNOWLEDGEMENTS
We thank Maximilian Weigend and Markus Ackermann for sharing their knowledge on the pollination mechanism of Loasoideae; the two anonymous reviewers for their comments on previous versions of the manuscript; and Martin Diller, Alicia Sérsic, Salvador Marino, Jorge Strelin, Adrian Strelin and Leonardo Torielli for assistance in the field. We also thank the Botanical Gardens Bonn, Germany and Guillermo Amico for providing plant material. J.F. thanks the Dirección General de Asuntos del Personal Académico (DGAPA) – Universidad Nacional Autónoma de México for financial support during a sabbatical stay at the Universidad Nacional de Córdoba – Instituto Multidisciplinario de Biología Vegetal – Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina). This work was supported by the Fondo para la Investigación Científica y Tecnológica [PICT 1553 (2012)] awarded to A.A.C. S.B.-V. and A.A.C. are researchers from the Consejo Nacional de Investigaciones Científicas y Técnicas. M.S. was supported by a graduate fellowship from the Consejo Nacional de Investigaciones Científicas y Técnicas.
LITERATURE CITED
- Ackermann M. 2012. Studies on systematics, morphology and taxonomy of Caiophora and reproductive biology of Loasaceae and Mimulus (Phrymaceae). PhD Thesis, Free University of Berlin, Germany. [Google Scholar]
- Ackermann M, Weigend M. 2006. Nectar, floral morphology and pollination syndrome in Loasaceae subfam. Loasoideae (Cornales). Annals of Botany 98: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M, Achatz M, Weigend M. 2008. Hybridization and crossability in Caiophora (Loasaceae, subfam. Loasoideae): are interfertile species and inbred populations results of a recent radiation? American Journal of Botany 95: 1109–1121. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Azpeita E, Barrio R, Benítez M, Padilla-Longoria P. 2010. From ABC genes to regulatory networks, epigenetic landscapes and flower morphogenesis: making biological sense of theoretical approaches. Seminars in Cell and Developmental Biology 21: 108–117. [DOI] [PubMed] [Google Scholar]
- Armbruster WS. 2012. Floral paedomorphy leads to secondary specialization in pollination of Madagascar Dalechampia (Euphorbiaceae). Evolution 67: 1196–1203. [DOI] [PubMed] [Google Scholar]
- Arthur W. 2011. Developmental bias and constraint. In: W Arthur, ed. Evolution. A developmental approach. Oxford: Blackwell Publishing, 200–217. [Google Scholar]
- Barrett CHB. 2013. The evolution of plant reproductive systems. How often are transitions irreversible? Proceedings of the Royal Society B 280: 20130913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Vieyra SM, Medina M, Cocucci AA. 2006. Variable selection patterns on the labellum shape of Geoblasta pennicillata, a sexually deceptive orchid. Journal of Evolutionary Biology 22: 2354–2362. [DOI] [PubMed] [Google Scholar]
- Berg RL. 1960. The ecological significance of correlation Pleiades. Evolution 14: 171–180. [Google Scholar]
- Breuninger H, Lenhard M. 2010. Control of tissue and organ growth in plants. Current Topics in Developmental Biology 91: 185–220. [DOI] [PubMed] [Google Scholar]
- Brown DK, Kaul RB. 1981. Flower structure and mechanism in Loasaceae. American Journal of Botany 68: 361–372. [Google Scholar]
- Ciliberti S, Martin OC, Wagner A. 2007. Innovation and robustness in complex regulatory gene networks. Proceedings of the National Academy of Sciences of the United States of America. 34: 13591–13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci AA, Sérsic AN. 1998. Evidence of rodent pollination in Caiophora coronata (Loasaceae). Plant Systematics and Evolution 211: 113–128. [Google Scholar]
- Davis CD, Endress PK, Baum D. 2008. The evolution of flower gigantism. Current Opinion in Plant Biology 11: 49–57. [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Maistro Patreze C, Angenent GC, Immink RGH. 2012. MADS: the missing link between identity and growth?. Trends in Plant Sciences 16: 90–97. [DOI] [PubMed] [Google Scholar]
- Drake AG, Klingenberg CP. 2008. The pace of morphological change: historical transformation of skull shape in St. Bernard dogs. Proceedings of the Royal Society of London, B Biological Sciences 275: 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 2006. Angiosperm floral evolution: morphological developmental framework. Advances in Botanical Research 4: 1–61. [Google Scholar]
- Fornoni J, Ordano M, Pérez-Ishiwara R, Boege K, Domínguez CA. 2016. A comparison of floral integration between selfing and outcrossing species: a meta-analysis. Annals of Botany 117: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland Jr. T, Dickerman AW, Janis CM, Jones JA. 1993. Phylogenetic analysis of covariance by computer simulation. Systematic Biology 42: 265–292. [Google Scholar]
- Givnish TJ, Barfuss MHJ, Van Ee B, Riina R, Schulte K, Horres R. 2009. Lobeliads (Asterales: Campanulaceae) origin, adaptive radiation and diversification of the Hawaiian. Philosophical Transactions of the Royal Society B 276: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EE, Igic B. 2008. On phylogenetic tests of irreversible evolution. Evolution 62: 2727–2741. [DOI] [PubMed] [Google Scholar]
- Gómez JM, Perfectti F, Camacho PM. 2006. Natural selection on Erysimum mediohispanicum flower shape: insights into the evolution of zygomorphy. The American Naturalist 168: 531–545. [DOI] [PubMed] [Google Scholar]
- Gómez JM, Perfectti F, Klingenberg CP. 2014. The role of pollinator diversity in the evolution of corolla-shape integration in a pollination-generalist plant clade. Philosophical Transactions of the Royal Society B 369: 20130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall CR. 1991. Procrustes methods in the statistical analysis of shape. Journal of the Royal Statistical Society B 53: 285–239. [Google Scholar]
- Gould SJ. 1977. Ontogeny and phylogeny. Cambridge, MA: Harvard University Press. [Google Scholar]
- Gould SJ. 2002. The structure of evolutionary theory. Cambridge, MA: The Belknap Press of Harvard University Press. [Google Scholar]
- Guerrant Jr. EO. 1982. Neotenic evolution of Delphinium nudicaule (Ranunculaceae): a hummingbird-pollinated larkspur. Evolution 36: 699–712. [DOI] [PubMed] [Google Scholar]
- Harder LD, Johnson SD. 2009. Darwin’s beautiful contrivances: evolutionary and functional evidence for floral adaptation. New Phytologist 183: 530–545. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24: 129–131. [DOI] [PubMed] [Google Scholar]
- Hermann K, Kuhlemeier C. 2011. The genetic architecture of natural variation in flower morphology. Current Opinion in Plant Biology 14: 60–65. [DOI] [PubMed] [Google Scholar]
- Hufford L. 2003. Homology and developmental transformation: models for the origins of the staminodes of Loasaceae Subfamily Loasoideae . International Journal of Plant Sciences 164: 409–439. [Google Scholar]
- Hufford L. 2005. A phylogenetic analysis of Loasaceae, subfamily Loasoideae based on plastid DNA sequences. International Journal of Plant Sciences 166: 289–300 [Google Scholar]
- Jablonka E, Lamb MJ. 2006. Evolution in four dimensions: genetic, epigenetic, behavioral, and symbolic variation in the history of life. Cambridge, MA: The MIT Press. [Google Scholar]
- Juenger T, Purugganan M, Mackay TFC. 2000. Quantitative trait loci of flower morphology in Arabidopsis thaliana. Genetics 156: 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. 1998. Heterochrony and allometry: the analysis of evolutionary change in ontogeny. Biological Reviews 73: 79–123. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. 2011. MorphoJ: an integrated software package for geometric morphometrics. Molecular Ecology Resources 11: 353–357. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Ekau W. 1996. A combined morphometric and phylogenetic analysis of an ecomorphological trend: pelagization in Antarctic fishes (Perciformes: Nototheniidae). Biological Journal of the Linnean Society 59: 143–177. [Google Scholar]
- Klingenberg CP, Duttke S, Whelan S, Kim M. 2012. Developmental plasticity, morphological variation and evolvability: a multilevel analysis of morphometric integration in the shape of compound leaves. Journal of Evolutionary Biology 25: 115–129. [DOI] [PubMed] [Google Scholar]
- Lenser T, Theißen G, Dittrich P. 2009. Developmental robustness by obligate interaction of class B floral homeotic genes and proteins. PloS Computational Biology 5: e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu Y, Zheng L, et al. 2012. The plant-specific G proteincsubunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytologist 194: 690–703. [DOI] [PubMed] [Google Scholar]
- Maie T, Schoenfuss HL, Blob RW. 2007. Ontogenetic scaling of body proportions in waterfall-climbing gobiid fishes from Hawai’i and Dominica: implications for locomotor function. Copeia 3: 755–764. [Google Scholar]
- McGuire JA, Witt CC, Remsen Jr. JV, et al. 2014. Molecular phylogenetics and the diversification of hummingbirds. Current Biology 24: 910–916. [DOI] [PubMed] [Google Scholar]
- Miller CE, Basu C, Fritsch G, Hildebrandt T, Hutchinson JR. 2008. Ontogenetic scaling of foot musculoskeletal anatomy in elephants. Journal of the Royal Society Interface 5: 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro L. 1999. Multivariate regression models and geometric morphometrics: the search of causal factors in the analysis of shape. Systematic Biology 48: 192–199. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. 2015. vegan: Community Ecology Package. R package version 2.2–1. http://CRAN.R-project.org/package=vegan. [Google Scholar]
- Ollerton J, Killick A, Lamborn E, Watts S, Whiston M. 2007. Multiple meanings and modes: on the many ways to be a generalist flower. Taxon 56: 717–728. [Google Scholar]
- Olson ME, Aguirre-Hernández RA, Rossel JA. 2009. Universal foliage-stem scaling across environments and species in dicot trees: plasticity, biomechanics and Corner’s Rules. Ecology Letters 12: 210–219. [DOI] [PubMed] [Google Scholar]
- Pélabon C, Armbruster WS, Hansen TF. 2011. Experimental evidence for the Berg hypothesis: vegetative traits are more sensitive than pollination traits to environmental variation. Functional Ecology 25: 247–257. [Google Scholar]
- Pérez-Barrales R, Arroyo J, Armbruster SW. 2007. Differences in pollinator faunas may generate geographic differences in floral morphology and integration in Narcissus papyraceus (Amaryllidaceae). Oikos 116: 1904–1918. [Google Scholar]
- Rausher MD. 2008. Evolutionary transitions in floral colour. International Journal of Plant Sciences 169: 7–21. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–233. [Google Scholar]
- Rice SH. 1990. A geometric model for the evolution of development. Journal of Theoretical Biology 143: 319–342 [Google Scholar]
- Rohlf FJ. 2009. TPS series software. Available at: http://life.bio.sunysb.edu/ee/rohlf/software.html. [Google Scholar]
- Rosas-Guerrero V, Quesada M, Armbruster WS, Pérez-Barrales R, DeWitt Smith S. 2010. Influence of pollinator speciation and breeding system in floral integration and phenotypic variation. Evolution 65: 350–364. [DOI] [PubMed] [Google Scholar]
- Rosas-Guerrero V, Aguilar R, Martén-Rodríguez S, et al. 2014. A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecology Letters 17: 388–400. [DOI] [PubMed] [Google Scholar]
- Schenk JJ, Hufford L. 2010. Effects of substitution models on divergence time estimates: Simulations and an empirical study of model uncertainty using Cornales. Systematic Botany 35: 578–592. [Google Scholar]
- Schluter D. 2000. The ecology of adaptive radiation. Oxford: Oxford University Press. [Google Scholar]
- Shoval O, Shefter H, Shinar G, et al. 2012. Evolutionary trade-offs, Pareto optimality, and the geometry of phenotype space. Science 336: 1157–1160. [DOI] [PubMed] [Google Scholar]
- Sidlauskas B. 2008. Continuous and arrested morphological diversification in sister clades of characiform fishes: a phylomorphospace approach. Evolution 62: 3135–3156. [DOI] [PubMed] [Google Scholar]
- Silva MFS, de Andrade IM, Mayo SJ. 2012. Geometric morphometrics of leaf blade shape in Montrichardia linifera (Araceae) populations from the Rio Parnaíba delta, north-east Brazil. Botanical Journal of the Linnean Society 170: 554–572. [Google Scholar]
- Simpson GG. 1953. The major features of evolution. New York: Columbia University Press. [Google Scholar]
- Stebbins GL. 1970. Adaptive radiation of reproductive characteristics in angiosperms. Annual Review of Ecology and Systematics 1: 307–326. [Google Scholar]
- Stebbins GL. 1974. Flowering plants: evolution above the species level. Cambridge, MA: Harvard University Press. [Google Scholar]
- van der Niet T, Johnson SD. 2012. Phylogenetic evidence for pollinator driven diversification of angiosperms. Trends in Ecology and Evolution 27: 353–361. [DOI] [PubMed] [Google Scholar]
- Viscosi V, Antonecchia G, Lepais O, Fortini P, Gerber S, Loy A. 2012. Leaf shape and size differentiation in white oaks: assessment of allometric relationships among three sympatric species and their hybrids. International Journal of Plant Sciences 173: 875–884. [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of adquired characters. Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Weigend M. 2004. Four new species of Nasa ser. Alatae (Loasaceae) in the Amotape-Huancabamba Zone of Peru. Novon 14: 134–146. [Google Scholar]
- Weigend M, Ackermann M, Henning T. 2010. Reloading the revolver- male fitness as a simple explanation for complex reward partitioning in Nasa macrothyrsa (Loasaceae, Cornales). Botanical Journal of the Linnean Society 100: 124–131. [Google Scholar]
- West-Eberhard MJ. 2005. Developmental plasticity and the origin of species differences. Proceedings of the National Academy of Sciences 102: 6543–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall JB, Hodges SA. 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 447: 706–710. [DOI] [PubMed] [Google Scholar]
- Williams GC. 1966. Natural selection, the costs of reproduction and the refinement of Lack’s principle. American Naturalist 100: 687–690. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.