Abstract
Background The flowers of core eudicots and monocots are generally determined by the number of floral organs they produce, and their developmental set-up tolerates little change from the bauplan once the floral primordium is initiated. Many species outside the core eudicots and monocots are more plastic in the number of floral organs they produce. For example, the Nymphaeales (water lilies), within the basal angiosperms, arrange their floral organs spirally and show smooth transitions between floral organs, and many Ranunculales (buttercups) produce variable numbers of stamens by adjusting the number of stamen whorls generated from a specialized ring meristem. However, the interactions of regulatory genes governing those processes are unknown.
Scope and Conclusions This review provides an overview of the functional analyses of floral homeotic genes carried out in Ranunculales, summarizing knockdown and mutant phenotypes, and protein interactions to identify similarities and differences within the Ranunculales and in comparison with core eudicots. Floral gene regulatory networks in Ranunculales are identified showing intensive re-wiring amongst the floral homeotic genes to allow some degree of plasticity. The ‘fading-border’ model of floral organ identity evolution is extended by a hypothesis on how developmental plasticity can be achieved by interdependent regulation of floral homeotic genes. One aspect of floral plasticity may be achieved by regulation of the activity of a stamen-generating ring meristem and first ideas on its control are presented. While the amazing conservation of the major floral organ identity programme is being unravelled by analysing floral homeotic gene function and expression, we are only just beginning to understand the evolution of the gene network governing the organ identity genes, e.g. how plasticity can be achieved, and which aspects foster the robustness of the core eudicot floral bauplan.
Keywords: flower development, transcription factors, MADS-box genes, developmental plasticity, Eschscholzia californica, evo-devo, ring meristem, Ranunculales, stamen number variation, floral homeotic genes, Papaveraceae, Ranunculaceae
A primer on the evolution of labile and fixed floral traits between species
Angiosperms produce flowers of amazing diversity in terms of colour, size, shape and merosity. By observing and comparing well-known flowers growing in many gardens and fields, e.g. lily (Lilium marthagon), snapdragon (Antirrhinum majus), poppy (Meconopsis horridula), wheat (Triticum aestivum), water lily (Nymphaea colorata) and roses (Rosa sp.) (Fig. 1) one can already obtain a glimpse of the floral diversity that has evolved since the time of angiosperm origin in the Early Cretaceous. Despite the enormous differences in appearance, all these flowers follow the same blueprint of floral organ type and arrangement, with only a few notable exceptions. Four types of floral organs, the sepals, petals, stamens and carpels, are arranged spirally or in whorls with sepals on the outside safeguarding the more delicate inner floral organs. Next come the petals, which play a major role in the attraction of pollinators, further inward the stamens and innermost the carpels. The array of flowers in Fig. 1 includes a range of only distantly related species such as basal angiosperms (water lily) monocots (lily and wheat), core eudicots (snapdragon and rose) and a basal eudicot (poppy). These six species also produce flowers distinct in their planes of symmetry: zygomorphic flowers (wheat and snapdragon) and radially symmetric flowers (lily, rose and poppy), and also flowers with labile (water lily, rose, and poppy) versus fixed number of floral organs (lily, wheat and snapdragon). This short excursion into fields and gardens indicates that several floral traits have evolved multiple times independently during the evolution of flowering plants and may occur convergently in very divergent groups of plants.
Fig. 1.
Photos of flowers from plants of diverse orders with a simplified phylogeny above, sketching their relationships. From left to right: Nymphaea colorata (water lily, basal angiosperms); Lilium marthagon (lily, monocot); Triticum aestivum (bread wheat, monocot); Meconopsis horridula (prickly blue poppy, basal eudicot); Antirrhinum majus (snapdragon, fabid); Rosa sp. (rose, malvid).
Whenever evolutionary changes lead to different morphologies, they are enabled by the patterning and morphogenesis potential of the species’ genetics. However, these changes are constricted by ecological limitations and functional constraints, which serve as ‘stabilizing’ forces (Endress, 2011). However, in some directions, more flexibility for change is apparent; for example, organ size or floral symmetry seem overly flexible (e.g. in the Ranunculales alone monosymmetric corollas evolved three times independently; Damerval and Nadot, 2007). In contrast, other traits barely change over time, such as thecal anther and ovule organization, such that the ancestral state of bitegmic (surrounded by two integuments) ovules and a thick nucellus (crassinucellar) has been maintained in the ANA grade plants, magnoliids, most monocots, basal eudicots and most core eudicots (Endress, 2011). However, the molecular networks allowing vs. restraining flexibility in flower morphology within and between species are unknown.
Within angiosperms, major evolutionary trends are clear when major plant groups are compared (Table 1), but only a few of them can be tracked genetically, as the molecular mechanisms resulting in morphological changes are, to a large extent, unknown.
Table 1.
Evolutionary stability versus lability in early-branching (ANA grade) angiosperms versus derived angiosperms (core eudicots and monocots)
| Evolutionarily stable early-branching angiosperm groups | Evolutionarily labile in derived groups | Reference(s) |
|---|---|---|
| Polysymmetric flowers | Polysymmetric, mono- and asymmetric flowers | Endress (2001) |
| Unfused floral organs | Fusion and non-fusion of organs | Endress (2010b) |
| Few pollination syndromes (only insects) | Large number of diverse pollination syndromes (insects plus birds, bats and other mammals) | Endress (2010b) |
| Flower colour limited to yellowish and white | Flower colour includes red and blue | Endress (2010b) |
| Protogynous flowers | Protogynous and protandrous flowers | Endress (2010b) |
| Evolutionary labile in early-branching groups | Evolutionary stable in derived groups | |
| Phyllotaxy whorled or spiral | Phyllotaxy whorled in monocots and core eudicots | Endress and Hufford (1989) |
| Anther dehiscence longitudinal, flaps, H-shaped | Anther dehiscence only longitudinal | Endress and Hufford (1989) |
| Floral organ number variable | 3-merous in monocots, 4- and 5-merous in core eudicots | Endress and Hufford (1989) |
| Ovule curvature and integument thickness | Orthotropous ovule curvature (vs. various types of curvature in more basal lineages) and stable integument thickness | Endress and Igersheim (2000), Endress (2010a) |
This review focuses on the molecular regulation of flower development in the early branching eudicot order Ranunculales. This plant group is of particular interest not only for its phylogenetic position but also for its striking diversity of floral architecture. The Ranunculales are characterized by diversity in the number of floral organs and phyllotaxy, including species with open and others with a closed ground plan, wind-pollinated and animal-pollinated species, and include species that developed novel organs such as nectar spurs and staminodia (Endress, 1999; Ronse De Craene et al., 2003; Kramer et al., 2007; Damerval and Nadot, 2007; Soza et al., 2012). The Ranunculales consist of seven families (Fig. 2): the earliest diverging lineage is that of the monogeneric family Eupteleaceae; the Papaveraceae (including Eschscholzia californica and Papaver somniferum) are the second-diverging clade and are sister to the core Ranunculales. These comprise the two sister families Ciceaeasteraceae and Lardizabalaceae, the Menispermaceae and the two sister families Berberidaceae (including Epimedium) and Ranunculaceae (including the genera Aquilegia and Thalictrum) (Wang et al., 2009).
Fig. 2.
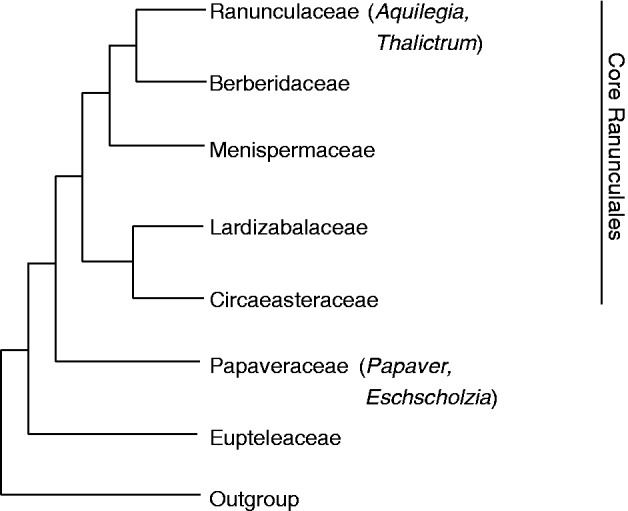
Simplified phylogeny of the Ranunculales with species mentioned here indicated in parentheses. The phylogeny is based on Wang et al. (2009).
A primer on plasticity
Observing and categorizing morphologies in phylogenetically informative species provides the base for generalizing evolutionary trends on a grander scale (Table 1, Fig. 1). This section focuses on within-species variation of development resulting in different morphologies. Phenotypic plasticity is defined as a condition-dependent form of development allowing an organism to transform morphological traits in response to changes in environmental conditions (Debat and David, 2001). In animals, phenotypic plasticity has been described in many classical examples, such as altered gill structure in fish as a response to changes in oxygen level, salinity or temperature (Sollid and Nilsson, 2006). In plants, an obvious case of phenotypic plasticity is skotomorphogenesis of seedlings that grow in near-darkness, and the responsible molecular mechanisms have been elucidated (for a review, see Alabadí and Blázquez, 2009). An intricate network including multiple photoreceptors, phytohormones and transcription factors is involved in the regulation of morphological alterations, such as hypocotyl, petiole and stem elongation, arrest of leaf growth and branching, and promotion of flowering to enable the shade-avoidance growth mode of plants (for a review, see Alabadí and Blázquez, 2009). However, this is an extraordinary case of plasticity, as generally the genetic base of plasticity in plants is unknown.
Developmental plasticity of flowers is more difficult to link to changes in the environment. For example, variation of floral organ numbers has been identified in many taxa, especially in non-core eudicots, such as Ranunculales, and in the core eudicots Caryophyllales, and Oleaceae (Ren et al., 2010; Kitazawa and Fujimoto, 2014), but their ecological significance and molecular regulation remain unclear.
Within the early-diverging eudicots, intra-individual variation was identified in Gunneraceae flowers along the inflorescence axis with various degrees of stamen and petal reduction, with bisexual flowers bearing a perianth being the ancestral condition (Gonzalez and Bello, 2009). Also, instabilities in the number of perianth parts, observed in a few Ranunculales species, such as Anemonella, Trautvetteria, Adonis, Cimicifuga and Ranunculus, is a form of phenotypic plasticity that evolved at least four times independently (Damerval and Nadot, 2007).
Floral homeotic genes in basal eudicots – California poppy as a reference system: (I) the participating genes and their expression
In the core eudicot model species Arabidopsis thaliana (thale cress, Brassicaceae), observation and analysis of floral homeotic mutants led to the classical ‘ABC’ model of flower development which explains how the combinatorial action of genes results in floral organ specification. The A function alone specifies sepal organ identity, A together with the B function specifies petal organ identity, B together with the C function specifies stamen organ identity and C alone specifies carpel identity. APETALA1 (AP1) and APETALA2 (AP2) are required for the A function, PISTILLATA [PI, the orthologue of GLOBOSA (GLO) of A. majus] and APETALA3 [AP3, the orthologue of DEFICIENS (DEF) of A. majus] for the B function and AGAMOUS (AG) for the C function. AP1, PI, AP3 and AG are MADS-box transcription factors, and AP2 is an AP2/EREB family transcription factor (for a recent review, see Irish, 2010, and the references cited therein). Four SEPALLATA1-4 (SEP1-4) genes, also MADS-box gene family members, act together with the other MADS-box genes and their products form higher order complexes which specify organ identity (Theissen and Saedler, 2001). MADS-box proteins bind to DNA with their MADS domains and require the K (for keratin-like) domain which forms three amphipatic helices (K1, K2, and K3) for dimeric interaction and the C domain for forming higher order complexes selectively (Jack, 2001; Lange et al., 2013).
California poppy (E. californica) is a member of the Papaveraceae, an early-diverging lineage within the Ranunculales (Fig. 2) and an emerging model plant, not only for evo-devo studies, but also for research on the biosynthesis and regulation of alkaloid metabolism and invasive plants (Schütz et al., 2014; Anic et al., 2015). Virus-induced-gene-silencing (VIGS) has been used as the method of choice in several studies to knock-down individual genes and observe the resulting phenotype mainly in flowers (Wege et al., 2007; Orashakova et al., 2009; Yellina et al., 2010; Pabón-Mora et al., 2012; Tekleyohans et al., 2013; Lange et al., 2013; Stammler et al., 2013; Fourquin and Ferrándiz, 2014). Taking these studies together, they provide a comparatively detailed picture of the molecular control of flower development in this basal eudicot, which differs in several important aspects from what is known in Arabidopsis, possibly allowing more flexibility in floral architecture.
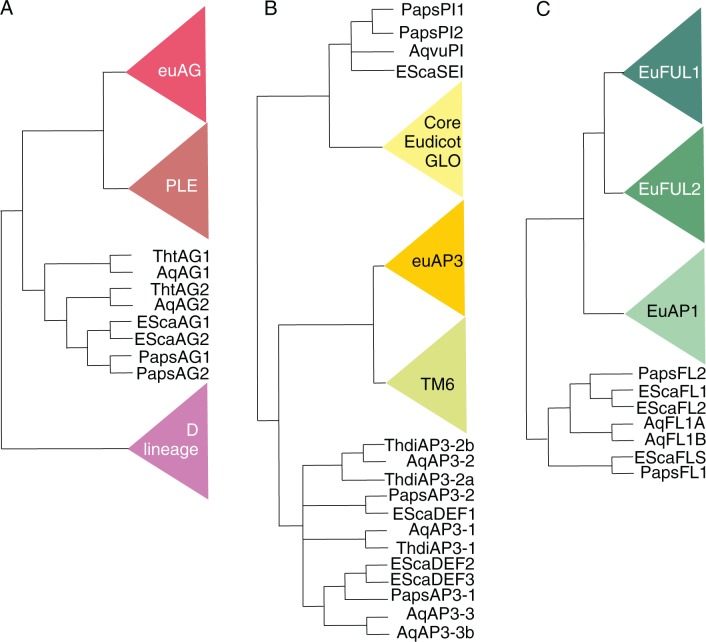
The Ranunculales lineage leading to E. californica experienced a genome duplication independent of that from Arabidopsis (Cui et al., 2006) and another one was detected in Aquilegia formosa (Vanneste et al., 2014) resulting in gene numbers for different developmental gene lineages that deviate from those of Arabidopsis. However, it remains unclear if the two Ranunculales genome duplications occurred in the ancestor of all Ranunculales or are lineage specific. As many MADS-box genes of diverse Ranunculales have been characterized, this section is mainly limited to E. californica genes. Figure 3 and Table 2 provide overviews on phylogeny and expression patterns of additional Ranuculales MADS box genes.
Fig. 3.
Simplified phylogenies of the AP1/FUL (A), AP3/GLO (B) and AG (C) subfamilies of MADS-box genes emphasizing the Ranunculales sequences for which functional analyses have been carried out. The phylogeny is based on Litt and Kramer (2010), Pabón-Mora et al. (2012, 2013a), Lange et al. (2013), and Galimba and Di Stilio (2015).
Table 2.
Simplified overview of the expression patterns of Ranunculales GLO- and DEF-like genes, after Kramer et al. (2007), Di Stilio et al. (2005), Drea et al. (2007), Gonçalves et al. (2013), Sharma and Kramer (2013), Larue et al. (2013) and Lange et al. (2013)
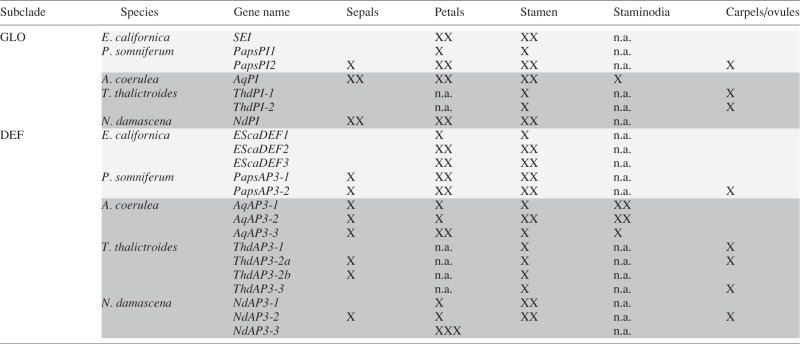 |
XX, strong expression; X, weak expression; Papaveraceae genes are in light grey; Ranunculaceae genes are in dark grey; n.a., absent organ type.
Two homologues of the Arabidopsis C function gene AG, EScaAG1 and EScaAG2, are present in E. californica (Fig. 2A) as well as three genes (EcDEF1, EcDEF2, EcDEF3) that are homologous to the B function gene AP3 of Arabidopsis, and SEIRENE (SEI) is the orthologue of PI from Arabidopsis (Fig. 2B; Zahn et al., 2006; Yellina et al., 2010; Lange et al., 2013).
EScaAG1 and EScaAG2 (Fig. 2A) are mainly expressed in reproductive organs and are most strongly expressed in carpels, fruits and developing buds. EScaAG2 shows its strongest expression in stamens and in the carpels, young fruits and developing buds. It is expressed 40–80 % lower than EScaAG1, suggesting notable differences in the cis-regulatory elements of the two genes. Simultaneous in situ hybridization for EScaAG1 and EScaAG2, which are highly similar in sequence, shows that their expression is restricted to the reproductive organs in developing flowers from their inception and during development. Within the gynoecium, expression remains restricted to the developing ovules. Interestingly, the EScaAG paralogues are not uniformly expressed throughout the multiple stamen whorls of E. californica; rather, their expression in the inner stamen whorls is stronger than in the outer stamen whorls (Yellina et al., 2010; Lange et al., 2013). This expression difference between AG paralogues in terms of transcript abundance, and lower expression of one of the paralogues in outer stamens was also observed in Aquilegia coerulea (Sharma and Kramer, 2013), but as the two respective genes are not orthologues, this feature of the expression pattern may have arisen independently in the two lineages. Alternatively, the ancestor of Ranunculales may have already had differential C gene expression across the stamen primordia and this may have been lost in some lineages.
The E. californica B genes (Fig. 2B) are expressed, as expected, in petals and stamens of flowers at anthesis, but also in this class of MADS-box genes differential expression between the genes is observed. While EScaDEF1 is hardly expressed in petals and stamens at anthesis, EScaDEF3 is most strongly expressed there. In developing buds before anthesis, EscaDEF2 shows low expression levels. The GLO-like gene SEIRENE (SEI) is expressed at roughly uniform levels during bud development and is also found in petals and stamens of flowers at anthesis (Lange et al., 2013).
To identify homologues to the most likely Brassicaceae-specific A function genes, a closer look into the phylogeny of the APETALA1/FRUITFUL (AP1/FUL) lineage of eudicots is necessary (Fig. 2C): in core eudicots, three gene subclades are present, one including AP1 of Arabidopsis, another one including AGL79 and the third one including FUL. However, these gene clades appear to have arisen in two gene duplications shortly before the origin of the core eudicots (Shan et al., 2007). In basal eudicots, a single FUL/AP1/AGL79-like gene, which is equally distant to all three FUL clades, was duplicated and gave rise to the lineage leading to EscaFL1 and EscaFL2 (Shan et al., 2007; Pabón-Mora et al., 2012, 2013a).
Expression of the two FUL-like genes is rather uniform throughout flower development and in all floral organs, fruits and leaves, except that EScaFL1 is expressed at a lower level in petals throughout development. Higher spatial resolution provided by in situ hybridization shows that EScaFL1 and EScaFL2 have essentially the same expression in all floral organs during early flower development, but expression in sepals and petals decreases in later stages. However, expression remains strong in stamens and gynoecium (Pabón-Mora et al., 2012).
Only one FUL-like gene, EsFUL was identified in Epimedium sagittatum and it was shown to be expressed in all floral organs but most strongly in sepals at anthesis and in petals and carpels before anthesis (Sun et al., 2014).
The partially redundantly acting E-function MADS-box genes SEPALLATA1-4 of Arabidopsis are also the product of lineage-specific gene duplications and thus the number of SEP-like genes in E. californica is different. The SEP-like genes duplicated and gave rise to two gene lineages: one includes the Arabidopsis SEP3 (formerly known as AGL9) and the other includes SEP1, 2 and 4 (formerly known as AGL2, 3 and 4). In basal eudicots, this duplication resulted in two genes, one for each lineage, and in E. californica these are called EScaAGL9 (hereafter EScaSEP3) and EScaAGL2. The gene lineage leading to EScaAGL2 and SEP1, SEP2 and SEP4 duplicated at least twice after the lineage leading to E. californica separated from the lineage leading to Arabidopsis (Zahn et al., 2005). Thus, EScaSEP3 is the orthologue of SEP3 and EScaAGL2 is the homologue of and equally distant to the genes SEP1, SEP2 and SEP4 of Arabidopsis (Zahn et al., 2005).
EScaSEP3 is expressed throughout bud development with higher expression in later stages and its expression can be observed throughout petal, stamen and carpel development (Zahn et al., 2005; Lange et al., 2013), but expression data for EScaAGL2 are lacking.
Expression of SEP-like genes was also analysed in E. sagittatum (Sun et al., 2014). Two SEP-like genes, EsAGL2-1 and EsAGL2-2, were identified whose floral organ-specific expression patterns deviate from one another. While EsAGL2-1 is expressed mainly in the petals and in pre-anthesis carpels, EsAGL2-2 is expressed in all floral organs, but more strongly before than at anthesis (Sun et al., 2014).
Based on the expression patterns and duplication history of the MADS-box genes described here, extensive sub- and neofunctionalization can be suggested. A broader analysis comparing Ranunculales genes with each other and in part with model core eudicot genes will be attempted in the following sections.
Floral homeotic genes in basal eudicots – Ranunculales as a reference system: protein interactions
Protein interaction analyses are important tools to elucidate the putative composition of floral homeotic transcription factor complexes. In Ranunculales, these analyses indicate important differences in the composition of floral homeotic complexes between core and basal eudicots, which is in part due to the fact that lineage-specific gene duplication events lead to gene pairs that may have undergone sub- and neofunctionalization.
Interactions between putative floral homeotic proteins of E. californica have been analysed with Y2H and bifluororescence complementation (BiFC), and, for some interactions also with electrophoretic mobility shift assays (EMSAs) (Lange et al., 2013; Table 3). Homodimers of B, C and E class proteins were detected for EScaDEF1, EScaDEF2, EScaDEF3, and SEI, but only the SEI homodimer was shown to bind to CArG boxes. EScaAG1 and EScaAG2 also formed homodimers, but only EScaAG1 homodimers showed CArG box binding. EcSEP3 does not form homodimers in the absence of CArG boxes, but in their presence it does.
Table 3.
Dimeric protein interaction of class B floral homoeotic proteins of Papaveraceae
| Subclade | Species | Protein name | Interacting proteins |
|---|---|---|---|
| GLO | E. californica | SEI | SEI*, EScaDEF1*, EScDEF2*, EScaDEF3 |
| DEF | E. californica | EScaDEF1 | SEI*, EScaDEF1, EScaDEF3, EScaAG1, EScaAG2, EScaSEP3 |
| EScaDEF2 | SEI*, EScaDEF2, EScaDEF3, EScaAG1, EScaAG2, EScaSEP3 | ||
| EScaDEF3 | SEI*, EScaDEF3 | ||
| GLO | P. somniferum | PapsAP3-1 | PapsPI-1 |
| PapsAP3-2 | PapsPI-1 | ||
| DEF | P. somniferum | PapsPI-1 | PapsAP3-1, PapsAP3-2 |
| PapsPI-2 |
Asterisks denote CArG-box binding of the protein dimer (which was tested for only a few combinations).
Heterodimers including B class proteins are formed more promiscuously than in Arabidopsis, where AP3 and PI form heterodimers but neither of them dimerizes with other MADS-box proteins (Table 3). SEI dimerizes with all three DEF-like proteins, but not with EScaSEP3. EscaDEF1 dimerizes with EScaDEF2, EScaAG1, EScaAG2 and EScSEP3. EScaDEF2 dimerizes with EScaDEF3, EScaAG1, EScaAG2 and EScaSEP3, and EScaAG1 interacts with SEP3. Of these heterodimers, only a few were tested for DNA interactions, and the SEI-EScaDEF1 and SEI-EScaDEF2 dimers were able to bind to CArG boxes in a sequence-specific manner.
B protein interactions were tested for other Papaverales species as well (Table 3): in P. somniferum there are two GLO-like and two DEF-like proteins and both, PapsAP3-1 (the orthologue of EScaDEF1) and PapsAP3-2, are able to dimerize with PapsPI-1 but not with PapsPI-2. However, these proteins do not homodimerize in the Y2H system (Drea et al., 2007). Furthermore, as PapsPI-2 does not even homodimerize, its interaction partners remain unclear. PapsPI-1 and PapsPI-2 are paralogous genes and equally distant to SEI, suggesting that at least for PapsPI-2, changes to the protein sequence occurred after the gene duplication that led to this inability to dimerize. However, these results show that the homodimerization capacity of the E. californica GLO- and DEF-like proteins is not conserved throughout the Papaveraceae and may be restricted to the more basal Papaverales such as E. californica.
In Aquilegia vulgaris, all three AP3 homologues are able to form heterodimers with the PI orthologues. However, neither the AP3 homologues nor the PI homologue is able to form homodimers and the AP3 homologues are unable to form heterodimers among each other (Kramer et al., 2007).
Dimeric and trimeric protein interactions were also tested for E. sagittatum proteins showing that EsAG interacts with EsAGL2-1, but not with EsAGL2-2. Also, EsAGL2-1 and EsAGL2-2 are both able to form homodimers and heterodimers with each other. Interactions with DEF- and GLO-like proteins were not tested (Sun et al., 2014). However, higher-order complexes consisting of EsFUL-EsAP3-2-EsPI and EsAGL2-EsAP3-2-EsPI were shown to form in Yeast Three-Hybrid experiments (Sun et al., 2014).
Trimeric interactions in E. californica were tested by yeast three-hybrid (Y3H) and trifluorescence complementation (TriFC) analyses. In both techniques two non-dimerizing tagged proteins and a third, untagged (silent) protein are used. Complexes SEI and EScaAG1 can be formed with EScaDEF2 and EScaDEF3, but not with EScaDEF1. SEI-EScaDEF1-EScaSEP3 and SEI-EScaDEF3-EScaSEP3 can be formed, but not in all combinations, but SEI-EScaDEF2-EScaSEP3 multimerize in all possible TriFC combinations. Complexes including B, C and E proteins that were formed were SEI-EScaAG1-EscaSEP3, DEF1-EScaAG1-EscaSEP3, DEF2-EScaAG1-EScaSEP3 and EScaDEF2-EScaAG2-EScaSEP3 and it was shown that the C-terminal domain of SEI is required for the selectivity of the multimeric complexes (Lange et al., 2013).
These protein interaction analyses show the large number of protein combinations that may form in E. californica plants. Apparently, class B floral homeotic proteins in E. californica are able to form heterodimers with AG orthologues, yet it remains unclear if these dimers also bind to CArG boxes. Interestingly, EScaSEP3 is unable to bind to either of the two AG homologues in Y2H experiments but is able to form homodimers in the presence of CArG boxes in the EMSA analysis. However, the SEP3-like protein of Thalictrum thalictroides dimerizes with the AG homologues (Galimba et al., 2012), indicating that the inability to form EScaSEP3-EScaAG1 or -2 dimers seems to be E. californica-specific. However, experimental evidence for interaction of putative B and E proteins of Amborella trichopoda, the most basal angiosperm, shows that dimeric interactions form between the SEP3 orthologue and orthologues of each of AP3, PI and AG (Melzer et al., 2014). This may indicate that earlier in angiosperm evolution AP3- and PI-like proteins not only formed heterodimers with each other but may also have directly interacted with SEP3 orthologues. Although this capacity was lost in the lineage leading to Arabidopsis where the AP3–PI dimer can interact with the SEP3 dimer, but neither AP3 nor PI can heterodimerize with SEP3 (Melzer and Theissen, 2009), it remained intact within the Ranunculales.
Floral homeotic genes in basal eudicots – Ranunculales as a reference system: (I) knock-down phenotypes
An analysis of knock-down and knock-out mutants of putative A, B and C floral homeotic genes in Ranunculales was carried out in only a few species. Among these are the two Papaveraceae species P. somniferum and E. californica, and the Ranunculaceae A. vulgaris, A. coerulea and T. thalictroides (Drea et al., 2007; Kramer et al., 2007; Yellina et al., 2010; Galimba et al., 2012; Lange et al., 2013; Larue et al., 2013).
FUL knock-down in Ranunculales.
The function of the P. somniferum, E. californica and A. coerulea homologues of the Arabidopsis genes FUL, AGL79, CAL and AP1, for simplicity termed FUL-homologues, was analysed by VIGS, a technique to transiently knock-down target genes by utilizing the cellular machinery to break down viral RNAs (Becker and Lange, 2010). In both the Papaveraceae and the Ranunculaceae species a paralogous gene pair exists (Pabón-Mora et al., 2012, 2013b) and here I focus on the simultaneous knock-down of these two genes. In the Papaveraceae species, the plants with FUL homologue knock-down show a phenotype in both vegetative and reproductive organs in agreement with FUL homologue expression in these organ types. Knocking-down the P. somniferum FUL homologues resulted in a prolonged vegetative phase and misshaped cauline leaves. Untreated plants generally develop a single, terminal flower, but downregulation of the FUL homologs resulted in outgrowth of axillary buds leading to secondary flowers. Also, the sepals of VIGS plants have a leaf-like shape and epidermal structure. Carpels elongate asymmetrically and rupture prematurely, and the fruits show patterning defects with respect to lignification and placentation. The most striking phenotype of FUL homologue downregulation is the partial homeotic conversion of petals into stretches with carpel-like epidermis. However, this is not accompanied by an upregulation of the otherwise weakly expressed AG homologues in the petals, suggesting that FUL homologues do not repress AG homologue expression in P. somniferum (Pabón-Mora et al., 2012). Moreover, the phenotype of partial homeotic conversion into carpel-like organs was limited to only the outer petal whorls, suggesting that the inner and outer petals may be under at least slightly different genetic control, possibly including different responses to gene dosages.
The phenotype of E. californica plants treated with FUL-homologue VIGS was in part similar to what was observed in P. somniferum (Pabón-Mora et al., 2012): increased branching, partial transformation of sepals into leaves and patterning defects in the fruits resulting in premature rupture. However, the FUL homologue VIGS plants do not show differences in leaf shape, and organ identity defects of petals, stamens or carpels were not observed.
Interestingly, the knock-down of the two FUL-like genes of A. coerulea did not affect flower development, such as meristem identity, organ identity or fruit development. Instead, inflorescence height, flower number, axillary meristem activity and, most notably, leaf morphology was modified in the FUL–VIGS-treated plants, suggesting an extension of function towards the regulation of vegetative development in Ranuculaceae (Pabón-Mora et al., 2013b).
These results again raise the central question of the origin and evolution of A-function as we know it from Arabidopsis. Do the FUL-homologues of the common ancestor of Papaveraceae and Brassicaceae have the ability to repress carpel identity genes, which, in the case of Brassicaceae are AG-orthologues and in the case of Papaveraceae these carpel identity genes may be different from or additional to AG-orthologues? And if not the FUL-like genes, which factors confine C expression to whorls 3 and 4?
B gene knock-down in Ranunculales.
Observation of class B floral homeotic mutants and VIGS knock-down lines have demonstrated that class B floral homeotic genes of basal eudicots carry out functions rather similar to their eudicot orthologues. However, there are important differences: gene numbers and protein interaction behaviour are different – the Aquilegia B class genes specify a novel floral organ type and in E. californica B class genes are apparently required for regulation of C-function genes (Kramer et al., 2007; Lange et al., 2013). It is thus worthwhile analysing the phenotypes of B class knock-downs more closely.
Aquilegia vulgaris flowers are special with respect to their organ types. First, the sepals have a petal-like appearance as they are coloured and display conical cells on their adaxial surface, which is typical for petals. Secondly, the flowers contain an extra organ type, called staminodium, between the stamen and the gynoecium whorl (Kramer et al., 2007). These staminodia generally adopt pollination-related functions (Walker-Larsen and Harder, 2000), originate from stamens and, consequently, DEF, GLO and AG homologues are expressed in staminodia (Kramer et al., 2007; Sharma and Kramer, 2013).
Interestingly, none of the B class genes is involved in specifying organ identity of the petal-like sepals, but may play a role later in their development when anthocyanin is accumulated and the papillate cell types differentiate. Silencing of AqvuPI, the only GLO homologue in A. vulgaris, resulted in homeotic conversions of staminodia and stamens into similarly looking carpeloid organs, which fused only partially, and the second whorl petals were converted into sepals (Kramer et al., 2007).
More differentiated phenotypes were observed when the DEF paralogues of A. coerulea were silenced by VIGS: AqAP3-1 VIGS treatment affected staminodia and innermost stamens that were transformed into carpel-like structures, sometimes even bearing ovules. In contrast, AqAP3-2 silencing strictly affected the stamen whorl, in which stamens were not converted into carpels but instead failed to produce anthers, and developed into stunted filaments. AqAP3-1 and AqAP3-2 seem to be partially redundant, as both staminodia and stamens were fully converted into carpel-like organs which even differentiated into style- and ovule-bearing ovaries in double knock-down plants. Also, unlike in the single knock-down lines, petal development was affected (Sharma and Kramer, 2013). Silencing of the petal-specific expressed gene AqAP3-3 consequently resulted in at least partial conversion of petals into sepal-like structures (Sharma and Kramer, 2013). Indeed, the examined AP3-3-like genes of diverse Ranunculales are all expressed in a petal-specific manner and the loss of AP3-3-like genes results in loss of petals (Zhang et al., 2013).
One example of the loss of an AP3-3-like gene was studied in more detail in a naturally occurring homoeotic mutant of Nigella damascena (Gonçalves et al., 2013). This mutant shows a replacement of petals by several sepal-like and chimeric sepal/stamen-like organs. Interestingly, this mutant shows that the AP3-3-like gene whose loss is causative for this phenotype has a function not only in floral organ identity but also in regulating perianth organ number and is in addition required for initiating and maintaining the boundary between petal and stamen organ identity (Gonçalves et al., 2013).
The genus Thalictrum encompasses species with monoecius and dioecius flowers. Thalictrum dioicum develops male and female plants which develop unisexually from inception (Di Stilio et al., 2005). When, in the male plants, any of the two GLO paralogues are silenced, a female flower is formed instead of a male one. In the hermaphroditic species Thalictrum thalictroides, the stamens were converted to carpels while other floral organs remained largely unaffected. These results suggest that the B class gene-based homeosis may account for the change in sexual systems in Thalictrum (Larue et al., 2013).
Papaver somniferum encodes two DEF-like genes (PapsAP3-1 and PapsAP3-2) and two GLO-like genes (PapsPI-1 and PapsPI-2). When the four genes are down-regulated by VIGS, subfunctionalization is evident with distinct phenotypes observed for each silenced gene (Drea et al., 2007). PapsAP3-1 is involved in specifying petal organ identity and has a minor role in conferring stamen identity, and PapsAP3-2 is crucial for specifying stamen organ identity but is of no relevance for petal identity. PapsPI-1 is essential for specifying petal and stamen identity and PapsPI-2 is as well, but to a lesser extent (Drea et al., 2007).
Seirena (sei), a class B floral homeotic mutant, was identified in E. californica from a fast neutron-irradiated mutant population (Lange et al., 2013). The affected gene is the only GLO-like gene encoded in the E. californica genome and a large insertion in the last exon was identified as the causal mutation. This leads to changes in the protein sequence of the C-terminal domain and a premature stop codon, resulting in absence of the conserved PI-motif. The sei mutant shows homeotic conversions of petals into sepals and stamens into single, unfused carpels and, for the outer stamen whorls, also conversions into sepal–carpel-like structures. The three DEF-like genes in E. californica have not been analysed functionally so far but subfunctionalization can be hypothesized, as expression analysis of floral organs at anthesis shows qualitative and quantitative differences between these genes (Lange et al., 2013).
Ranunculales B function genes have experienced a plethora of lineage-specific duplication events, resulting in, for example, two DEF- and two GLO-like genes in P. somniferum or three DEF- and one GLO-like gene in Aquilegia. The DEF genes of P. somniferum are a good example of subfunctionalization as they may have distributed the function of the ancestral gene to the two copies after duplication. However, the GLO-like genes demonstrate how one gene (PapsPI-1) takes over the function of the ancestral gene, while the other one may be prone to non-functionalization. Finally, the GLO-like gene of A. vulgaris is involved in specifying organ identity of staminodia, novel organs most likely not found in the common ancestor of Ranunculales, suggesting that this gene underwent neofunctionalization, a process crucial for the origin of novel structures in evolution.
Thus, the B genes of the Ranunculales may serve as an interesting system to study the processes leading to sub- and neofunctionalization after gene duplications on a molecular level.
C gene knock-down in Ranunculales.
The analysis of putative class C floral homeotic gene functions in Ranunculales has not received as much attention as knock-down of class B genes and only a few examples have been investigated.
In T. thalictoides, a cultivar exhibiting a double-flower phenotype with extra showy petaloid organs called ‘Double White’ or ‘Snowball’ was analysed in detail. Wild-type T. thalictroides flowers lack petals and the sepals take over the function of petals and differentiate into large non-photosynthetic organs. Corroborating the concept of apetaloidy in T. thalictroides, AP3- or PI-like genes are not expressed in the showy sepals until very late in their development (Larue et al., 2013). The double-flowered mutant shows floral homeotic conversions reminiscent of a homeotic C mutant as the reproductive organs are missing. However, unlike in Arabidopsis, where stamens and carpels are converted into petals and sepals, in the T. thalictroides double-flower mutant, the reproductive organs are converted exclusively into sepal-like structures. In addition, the typical over-proliferation of the floral meristem was observed, often causing an extra flower to emerge from the innermost floral whorl (Galimba et al., 2012). The double-flower phenotype is caused by an insertion of an LTR transposon in the fourth exon of the ThtAG1 gene, causing a premature stop codon and a cryptic splice-acceptor site. The resulting protein lacks only a few amino acids but those are located at the end of the K1 and between the K1 and K2 subdomains, a region important for protein dimerization. The mutated protein is unable to interact with ThtSEP3, the T. thalictroides SEP3 orthologue, while the wild-type ThtAG1 heterodimerizes and is thus able to carry out its function in regulating target genes (Galimba et al., 2012).
The T. thalictroides genome codes for one pair of AG paralogues, but lacks representatives of the D-lineage MADS-box genes, such as homologues of STK that participate in specifying the identity of ovule tissue (Galimba and Di Stilio, 2015). The two AG paralogues are also expressed differentially: in flowers at anthesis, ThtAG1 is expressed in stamen, carpels and ovules, but ThtAG2 expression is lacking in sepals and is about three times as high as ThtAG1 expression in ovules. A knock-down of the second AG paralogue, ThtAG2, by VIGS resulted in homeotic transformations of ovules into carpels, suggesting that this gene has acquired the function to specify ovule identity in T. thalictroides (Galimba and Di Stilio, 2015). These results indicate a remarkable flexibility in shifting important gene function between the C and D lineage of MADS-box genes.
When the two AG genes in E. californica are silenced, stamens are converted into petals and the number of the petal-/stamen-like organ is increased when compared to the number of stamens of untreated plants (Yellina et al., 2010). The gynoecia are transformed into sepal-like flat structures, but many of these also show petal characteristics, such as orange coloration and petal surface structure. This suggests that ectopic B function may act in the organs formed instead of gynoecia. Within many of these transformed gynoecia, a second flower develops, with petals, stamens and a gynoecium, indicating that the meristem termination of the central floral meristem is impaired.
Interestingly, the only aspect in which the two genes differ in their silencing phenotype is in the location of the stamens that show conversion into petaloid organs. When EScaAG1 is silenced, the organs of the outer stamen whorls are converted to petaloid organs, when EScaAG2 is silenced, the stamens of the inner whorls are converted, and when both genes are silenced together, the outer and inner stamen whorls are converted into petals, leaving the middle whorls more stamen-like (Yellina et al., 2010).
In plants where the silencing is weaker, the homeotic conversion of stamens into petals is not completed. However, the number of petal-/stamen-like organs remains significantly increased, suggesting a concentration-dependent action of EScaAG proteins. Even when a small amount of EScaAG protein is lacking, the generation of stamen primordia is impaired, but reproductive organ identity remains intact. At a lower EScaAG protein concentration, reproductive organ identity also fails to form properly, and at an even lower concentration, B genes become activated (Yellina et al., 2010).
Generally, the AG-like genes in Ranunculales are required for the specification of stamen and carpel organ identity. However, a unique a shift of functions between C and D class lineage genes can be observed in T. thalictroides, where an AG-like gene specifies ovule identity and the D-lineage is lost. In A. thaliana or Oryza sativa, it is mainly the D-lineage genes that are crucial for ovule identity with only minor involvement of other C-lineage genes (Dreni et al., 2013). In E. californica, a D-lineage gene is present (Zahn et al., 2006) but expression analysis and a description of the knock-down phenotype is lacking.
At present, the interesting phenomenon that a reduction of C function in E. californica does not lead to a conversion of carpels into sepal like but rather into petal-like organs has not been not documented for any other Ranunculales species, as T. thalictroides lacks petals. Thus, it would be interesting to observe if a reduction in C function in other Ranunculales also results in an increase in B function in the floral centre. This would provide a hint towards a regulatory loop between B and C class genes in Ranunculales in general or indicate if this loop is specific to E. californica.
MADSes regulate MADSes – towards understanding the evolution of gene regulatory networks in flower development
VIGS and mutant analyses of plants outside the core eudicot model organisms allow not only the inference of gene functions, but also evaluations of the degree of conservation of functions among orthologous genes of diverse species, and observations on the way related genes subfunctionalize. However, educated guesses on how gene regulatory networks are wired require analyses of a broader spectrum of possibly genetically connected genes in VIGS-treated plants or mutants. This approach then can contribute to our understanding on how genetic networks governing flower development have evolved. Unfortunately, expression analysis without direct evidence for cis-regulatory element binding are limited such that it cannot be determined if the regulation of gene expression is direct or indirect. The genetic interaction is described in the next paragraph without the ability to discriminate whether they are direct or indirect.
As observed for core eudicot B class genes, also the P. somniferum B class genes regulate their own expression: PapsAP3-1 and PapsAP3-2 are required for PapsPI1-1 but not PapsPI-2 expression. When the GLO orthologue of A. vulgaris is silenced, expression of two of the three DEF-like genes, AqvuAP3-2 and AqvuAP3-3, is reduced severely, suggesting a function of GLO in an autoregulatory pathway. However, this autoregulation excludes AqvuAP3-1, which in terms of expression is a rather atypical DEF homologue, as it is also expressed in the petaloid sepals of A. vulgaris (Kramer et al., 2007). In contrast, in A. coerulea, when AqAP3-1 or AqAP3-2 is silenced, expression of both AG paralogues increased significantly in staminodia to the level of carpels of untreated plants. This suggests that in A. coerulea expression of the AG paralogues is negatively regulated by AqAP3-1 and AqAP3-2 (Sharma and Kramer, 2013).
Expression of other putative floral homeotic genes was analysed in wild-type and sei mutant of E. californica. In the sei mutant, expression of all three DEF-like genes was almost completely abolished, during flower development as well as at anthesis, suggesting that the SEI protein is part of a regulatory complex, and probably acts in concert with any EScaDEF proteins, required for activation of expression and maintenance of expression of DEF and the GLO genes (Lange et al., 2013).
The expression of the EScaSEP3 orthologue was unchanged in the sei mutant, but interestingly expression of one of the two AG paralogues was significantly reduced. EScaAG2 is expressed strongly in the inner stamen whorls but to only a lesser extent in the outer stamen and gynoecium. This suggests that SEI, probably together with an EScaDEF gene, is also involved in activating expression of one of the AG paralogues (Lange et al., 2013).
Taken together, this suggests that the autoactivation loop shown for core eudicot DEF- and GLO-like genes was present already in the last common ancestor of core eudicots and basal eudicots. This also seems to be the case when gene duplications lead to multiple copies of DEF- or GLO-like genes (Fig. 3B). With a few notable exceptions, such as PapsPI-2 from P. somniferum, and AqvuAP3-1 from A. vulgaris, the autoregulation mechanism is extended to the multiple copies suggesting that the cis-regulatory elements driving autoregulation have been conserved between core and basal eudicots.
Expression analysis of class C genes in E. californica shows that, as expected, they are expressed in the reproductive organs of the flowers and in carpels; expression of the two is about equal. Putative B class genes are, with the exception of EScaDEF1, not expressed in carpels. However, when EScaAG1 and EScaAG2 are silenced EScaDEF2 and SEI are expressed at a much higher level than in mock treated plants but EScaDEF1 shows no change in expression. In particular, the EScaDEF2 and SEI expression domain now extends into the centre of the flower, inducing even petal-like organs in the place of gynoecia (Yellina et al., 2010). This suggests that EScaDEF2 and SEI are under the transcriptional control of EScaAG1 and/or EScaAG2, but EScaDEF1 is not (Yellina et al., 2010).
While the B gene autoactivation loop was shown for core eudicots and can also be proposed for Ranunculales, tracing the evolutionary history of additional feedback loops is more difficult due to comparatively few data in Ranunculales. Additional regulatory loops can be proposed based mainly on knock-down experiments of B and C class genes of E. californica: SEI, together with at least one EScaDEF gene, activates the expression of an AG-like gene, specificially in the outer stamen whorls. In contrast, AG-like genes repress SEI and EScaDEF expression in the carpel whorl. Interestingly, regulation of AG-like genes by DEF- and GLO-like genes has also been observed in A. coerulea, albeit as repression and not, as seen in E. californica, as activation. Thus, the regulatory dependence on B class genes may have been maintained in the Ranunculales, but lineage-specific activation or repression responses have evolved.
While maintenance of AP3 expression in Arabidopsis is dependent on AG, the presence and expression of the Petunia hybrida B gene TM6 requires the expression of C class genes, and expression of other B genes is independent of the C function (Gómez-Mena et al., 2005; Heijmans et al., 2012). However, C-function genes in core eudicots activate B gene expression while C-function genes in E. californica repress B function activity. Moreover, probably with the help of an unknown cofactor active in the central floral whorl but not in the stamen whorls, C-function genes ensure that B-function activity is excluded from the central floral whorl. In Arabidopsis, SUPERMAN (SUP) represses B gene expression in the fourth whorl (Sakai et al., 1995) but orthologues of SUP were not found in the high-coverage E. californica transcriptome data (K. Pfannebecker, Justus-Liebig-University Gießen, Germany, pers. comm.) and thus another, gynoecium-specific co-factor needs to be hypothesized for B gene repression in the central floral whorl. Taken together, results from P. hybrida, Arabidopsis and E. californica suggest that the binding sites for AG homologues have been maintained in basal eudicots, malvids and fabids, but the complexes incorporating C class proteins may have changed: from repressive in basal eudicots to activating in some, but not all, TM6- and euAP3-like genes in core eudicots.
Increase of stamen number allows escape from constraints by canalized architecture – some insights into the genetic regulation of developmental flexibility
Highly organized flower morphogenesis, as found in Arabidopsis, where the organs initiate in a fixed sequence and are all arranged in a fixed pattern to each other leaves little room for flexibility. Variation in stamen numbers is often found in Ranunculales, other basal eudicots and basal angiosperms, but to a lesser extent in core eudicots (Endress, 2006; Damerval and Nadot, 2007). Interestingly, stamen number in the ancestral Ranunculales was less than 10 but numerous stamens originated at least seven times independently throughout the Ranunculales (Damerval and Nadot, 2007), suggesting that this additional flexibility in the floral bauplan proved successful in this plant order. However, the ecological function of this variable stamen number remains unclear. One aspect that has received little attention so far is that several Ranunculales representatives do not produce nectar, such that from the Ranunculales species analysed by Wang et al. (2009), more than half (53 of 99) did not develop nectar petals. For those species, pollen is the only reward for animal-pollinated species (only few Ranunculales species are wind pollinated; Endress, 2002). This may lead to significant loss in pollen grains and thus lowers the total number of male gametes available for cross-fertilization (Vallejo-Marín et al., 2009).
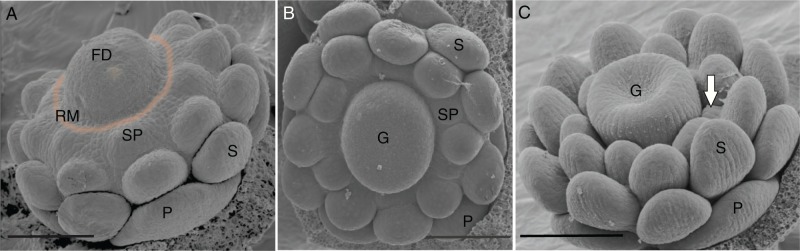
In several Ranunculaes, especially in the Papaveraceae, such as E. californica, stamen primordia form centripetally from an androecial ring meristem that surrounds the gynoecium (Becker et al., 2005; Endress, 2011). This additional ring meristem provides a means to decouple the time of stamen initiation such that the last stamens are initiated after the carpels are far advanced in their development. This is caused by the ring meristem delimiting early from the zone of the floral apex in late stage 4 (Fig. 4A) and maintaining its activity throughout stage 5 and 6 (Fig. 4B and C; Endress, 2006). When the earliest formed stamen primordia already change their shape to accommodate microsporangia formation, new stamen primordia are still formed from the ring meristem (Fig. 4C). While no information is available on the ecological reasons or the molecular mechanism to terminate the action of the ring meristem, analysis of the E. californica transcription factors suggests that there are quantitative and qualitative genetic differences in meristem activity regulation between the ring meristem and the central floral meristem.
Fig. 4.
The ring meristem of E. californica forming stamen primordia. (A) Bud of a late stage 4 flower in which the floral dome separates from the ring meristem (indicated by a blurred orange line). (B) Stage 5 bud showing the continuous formation of stamen primordia while the gynoecium has already initiated. (C) Stage 6 bud showing the formation of new stamen primordia (white arrow) at the time when the early stamens already form a flat surface for microsporangia initiation. The sepals were removed in all images, and staging was done according to Becker et al. (2005). FD, floral dome; G, gynoecium; P, petal; RM, ring meristem; S, stamen; SP, stamen primordium. Scale bar in A = 86 μm, in B = 100 μm, in C = 120 μm.
Action of the two EScaAG genes is required for both the ring and the central floral meristems to terminate. However, a mild reduction in EScaAG transcripts by VIGS, not affecting organ identity or central floral meristem activity, is already sufficient to lead to a prolonged activity of the ring meristem, resulting in a significant increase in the number of stamens. Only when silencing of the EScaAG genes is strong enough to affect organ identity does the central floral meristem fail to terminate. This suggests a quantitative difference in the regulation of the two meristem types, such that the termination of the ring meristem requires a higher concentration of EScaAG proteins than the termination of the central floral meristem and reproductive organ identity (Yellina et al., 2010). As the B function genes enhance EScaAG gene expression, even small stochastic changes in B or C gene expression may thus lead to changes in stamen number, but targeted gene regulation is equally well conceivable.
Additional VIGS experiments support the hypothesis that the ring and central floral meristem activities in E. californica are regulated differently: when the YABBY transcription factor EcCRC is down-regulated, the central floral meristem fails to terminate and forms gynoecia within gynoecia, reminiscent of a Russian doll, a process that is stopped only by space constraints. However, in EcCRC VIGS-treated plants, stamen number does not deviate from mock-treated plants, suggesting that EcCRC is required for terminating the central floral meristem, but has no role in ring meristem regulation (Orashakova et al., 2009).
Silencing STM-like KNOX transcription factors resulted in a more general phenotype: when the silencing was very successful, the flower produced no stamens or carpels, probably because the floral meristem ceases activity before the ring meristem can be formed or the gynoecium initiated. In weakly silenced plants, which started to initiate the gynoecium that then aborted early in development, the number of stamens was also significantly lower. This indicates that EcSTM gene activity affects both types of meristems within the E. californica flower (Stammler et al., 2013).
The gene regulatory network controlling floral meristem activity of Arabidopsis includes a plethora of interacting transcription factors (Sun and Ito, 2015, and references therein), and the genes shown to be involved in regulating E. californica’s meristems are orthologues of the Arabidopsis floral meristem regulators. However, as with most other core eudicots, Arabidopsis has a highly robust floral architecture, and is thus not suitable to study aspects of stamen number variation. However, homologues of other Arabidopsis floral meristem regulators are probably involved in regulating the ring meristem activity and may thus provide suitable candidates for further investigation. As few steps have been undertaken to date to study the molecular regulation of floral plasticity in E. californica, the interesting questions remain unanswered, for example: what is the ecological relevance of this type of developmental flexibility? What is the cue triggering change in ring meristem activity? How are the two meristem types partitioned? How do the surrounding organs expand to provide the space for the multiplying and expanding stamen primordia? What is the molecular difference in gene regulatory networks between the two meristem types? Moreover, as stamen-producing ring meristems have evolved several times independently, what are the morphogenetic and molecular differences between them?
A hypothesis for the evolutionary trend from plasticity to robustness in floral development
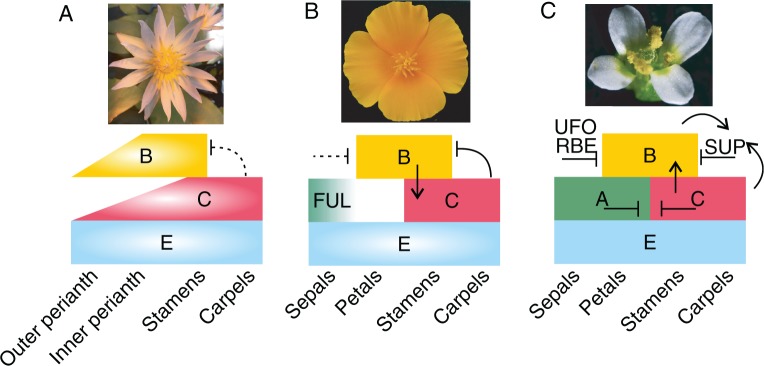
Basal angiosperms, the only survivors of ancient plants lineages, are a species-poor group of plants that represent less than 3 % of all angiosperms and include taxa such as A. trichopoda, the water lilies (Nymphaeaceae) or star anise (Illicium verum). Although their species number is small, their diversity in floral structure and organization is extreme (Endress, 2006; Soltis et al., 2007). The Nymphaeaceae, as an example, develop an undifferentiated perianth, and spiral phyllotaxy and a smooth transition between stamens and the perianth can be observed (Fig. 5A). Expression analysis of homologues of core eudicot floral homeotic genes has been carried out in this and some other basal angiosperm species and it turns out that their domains of expression are not as clearly defined as has been shown for Arabidopsis for example. AG homologues may be expressed in reproductive organs but also in perianth organs and AP3 and PI homologues may also be expressed throughout the flower (Kim et al., 2005; Soltis et al., 2007). The ‘fading border’ model has been proposed which correlates the more diffuse expression pattern of putative floral homeotic genes with the gradual transitions of floral organs in basal angiosperms. According to this model, the gradual transitions between neighbouring organs results from a gradient in floral homoeotic gene expression (Buzgo et al., 2004). This transition can be observed between the perianth organs and the stamens in many species, for example within the Nymphaeaceae, in which several species and horticultural cultivars show this morphology. However, experimental evidence for the ‘fading border’ model is, except for MADS gene expression in early-branching angiosperms (Kim et al., 2005), very weak.
Fig. 5.
Schematic representation of the ABCE model of flower development in angiosperm lineages and the genetic interaction of the floral homeotic genes in (A) basal angiosperms (ANA grade) with Nymphaea colorata (Nymphaeales) as a representative, (B) basal eudicots with E. californica as a representative and (C) core eudicots, malvids, with A. thaliana as a representative. Boxes with a light centre represent putative gene functions based solely on expression data. Boxes with a lighter side represent changes in organ identity upon gene knock-down different from what is predicted from the ‘classical’ ABC model. Arrows indicate direct or indirect activation based on gene expression experiments in mutants or knock-down lines. Lines ending in horizontal bars symbolize direct or indirect repression of at least some gene function in the respective classes based on gene expression experiments in mutants or knock-down lines. Dashed lines represent hypothetical interaction. SUP stands for the gene SUPERMAN, which is known to repress the Arabidopsis B class genes in the fourth whorl (Sakai et al., 1995). The drawings are based on several sources (Kim et al., 2005; Krizek and Fletcher, 2005; Zahn et al., 2005; Yellina et al., 2010; Pabón-Mora et al., 2012; Heijmans et al., 2012; Galimba and Di Stilio, 2015).
Interestingly, a smooth transition between stamens and the gynoecium is much rarer and although AP3- and PI-like genes extend their domain of expression into the floral centre in basal angiosperms they may not have a function in organ identity in the flower centre. The presence of a highly conserved genetic factor needs to be proposed that inhibits the expression of AP3- and PI-like genes with a homeotic gene function from the floral centre to ensure a clear distinction between the stamen and the carpel whorls (Fig. 5A).
In E. californica it was observed that the B function gene expression extends into the gynoecium whorl when the C function is silenced, suggesting that in wild-type plants, B class gene expression is repressed, at least in part by C function in the central part of the flower. Apparently, another gynoecium- or floral centre-specific factor is required to act together with the C function genes for this negative regulation as otherwise the B class genes would also be repressed in the regions where stamen primordia arise (Fig. 5B; Yellina et al., 2010). If this molecular mechanism of B class gene repression were to be found already in the most recent common ancestor of angiosperms, it could provide an explanation for the discrete appearance of the stamen and carpel whorls in flowers of the vast majority of plants, even those with otherwise smoothly transitioning organs (Fig. 5A). Possibly, only late in evolution, within the core eudicots, SUPERMAN (SUP) homologues limit B expression independently of the C function from expanding into the central floral whorl to demarcate the boundary between whorl 3 and 4. AG, PI and AP3 expression is required for SUP expression at the stamen/carpel boundary, suggesting a feedback loop to maintain this demarcation (Fig. 5C; Sakai et al., 1995; Nakagawa et al., 2004).
Another such factor may limit C function expression extension in the first and second whorl of flowers, which show a clear morphological separation of petals and stamens, as do most monocots, basal eudicots and core eudicots do. In A. thaliana, the A function genes directly and indirectly repress AG (Gregis et al., 2006), but a classical A function is lacking in plants outside the Brassicaceae and the genes homologous to AP1 and AP2 have a function in floral meristem regulation (Litt, 2007). Two more genes, RABBIT EARS (RBE) and UNUSUAL FLORAL ORGANS (UFO), promote petal development by inhibiting AG expression in the second whorl (Fig. 5C; Durfee et al., 2003; Krizek et al., 2006). However, a genetic factor repressing AG function analogous to the A function genes/RBE/UFO in Arabidopsis nonetheless needs to be postulated for the non-Brassicaceae angiosperms. The regulation of genes directing flowering has been elucidated to a large extent for Arabidopsis, but as a core eudicot, Arabidopsis and its Brassicaceae relatives are unsuited to serve as models for, for example, developmental plasticity and many other morphological traits found in diverse species outside the Brassicaceae. Ranunculales are a morphologically diverse plant order and lack the α and β genome and segmental duplications specific to the lineage leading to Brassicaceae and may therefore regulate their reproductive development with a more diverse set of genes only loosely related to their A. thaliana homologues. They are certainly more difficult experimental systems than Arabidopsis, as species such as the poppies are obligate outcrossers, and the columbines include novel organ types. However, with their phylogenetic position as early-branching eudicots, the Ranunculales floral developmental programme may represent a more generally applicable system.
ACKNOWLEDGEMENTS
I thank Dietmar Haffer and Holger Laake of the Botanical Garden Gießen for growing most of the plants I took photographs from, and David Smyth in whose lab the Eschscholzia scanning electron micrographs were taken. Further, I thank the current and previous members of the lab for fruitful discussions on this topic. Work in the Becker lab is mainly funded by the German Research Foundation (BE 2547/3-1, 3-2, 6-1, 6-2, 7-2), and the Justus-Liebig-University Gießen.
LITERATURE CITED
- Alabadí D, Blázquez MA. 2009. Molecular interactions between light and hormone signaling to control plant growth. Plant Molecular Biology 69: 409–417. [DOI] [PubMed] [Google Scholar]
- Anic V, Henríquez CA, Abades SR, Bustamante RO. 2015. Number of conspecifics and reproduction in the invasive plant Eschscholzia californica (Papaveraceae): is there a pollinator-mediated allee effect? Plant Biology 17: 720–727. [DOI] [PubMed] [Google Scholar]
- Becker A, Lange M. 2010. VIGS – genomics goes functional. Trends in Plant Science 15: 1–4. [DOI] [PubMed] [Google Scholar]
- Becker A, Gleissberg S, Smyth DR. 2005. Floral and vegetative morphogenesis in California poppy (Eschscholzia californica Cham.). International Journal of Plant Science 166: 537–555. [Google Scholar]
- Buzgo M, Soltis PS, Soltis DE. 2004. Floral developmental morphology of Amborella trichopoda (Amborellaceae). International Journal of Plant Science 165: 925–947. [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Research 16: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, Nadot S. 2007. Evolution of perianth and stamen characteristics with respect to floral symmetry in Ranunculales. Annals of Botany 100: 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debat V, David P. 2001. Mapping phenotypes: canalization, plasticity and developmental stability. Trends in Ecology and Evolution 16: 555–561. [Google Scholar]
- Di Stilio VS, Kramer EM, Baum DA. 2005. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae) – a new model for the study of dioecy. The Plant Journal 41: 755–766. [DOI] [PubMed] [Google Scholar]
- Drea S, Hileman LC, Martino G de, Irish VF. 2007. Functional analyses of genetic pathways controlling petal specification in poppy. Development 134: 4157–4166. [DOI] [PubMed] [Google Scholar]
- Dreni L, Osnato M, Kater MM. 2013. The ins and outs of the rice AGAMOUS subfamily. Molecular Plant 6: 650–664. [DOI] [PubMed] [Google Scholar]
- Durfee T, Roe JL, Sessions RA, et al. 2003. The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 100: 8571–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 1999. Symmetry in flowers: diversity and evolution. International Journal of Plant Science 160: S3. [DOI] [PubMed] [Google Scholar]
- Endress PK. 2001. Evolution of floral symmetry. Current Opinion in Plant Biology 4: 86–91. [DOI] [PubMed] [Google Scholar]
- Endress PK. 2002. Morphology and angiosperm systematics in the molecular era. Botanical Review 68: 545–570. [Google Scholar]
- Endress PK. 2006. Angiosperm floral evolution: morphological developmental framework In: Soltis DE, Lebens-Mack JH, Soltis PS, eds. Developmental genetics of the flower. Amsterdam: Elsevier, 1–61. [Google Scholar]
- Endress PK. 2010a. Flower structure and trends of evolution in eudicots and their major subclades. Annals of the Missouri Botanical Garden 97: 541–583. [Google Scholar]
- Endress PK. 2010b. The evolution of floral biology in basal angiosperms. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 365: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 2011. Evolutionary diversification of the flowers in angiosperms. American Journal of Botany 98: 370–396. [DOI] [PubMed] [Google Scholar]
- Endress PK, Hufford LD. 1989. The diversity of stamen structures and dehiscence patterns among Magnoliidae. Botanical Journal of the Linnean Society 100: 45–85. [Google Scholar]
- Endress PK, Igersheim A. 2000. Gynoecium structure and evolution in basal angiosperms. International Journal of Plant Sciences 161: S211–S223. [DOI] [PubMed] [Google Scholar]
- Fourquin C, Ferrándiz C. 2014. The essential role of NGATHA genes in style and stigma specification is widely conserved across eudicots. The New Phytologist 202: 1001–1013. [DOI] [PubMed] [Google Scholar]
- Galimba KD, Di Stilio VS. 2015. Sub-functionalization to ovule development following duplication of a floral organ identity gene. Developmental Biology 405: 158–172. [DOI] [PubMed] [Google Scholar]
- Galimba KD, Tolkin TR, Sullivan AM, Melzer R, Theißen G, Di Stilio VS. 2012. Loss of deeply conserved C-class floral homeotic gene function and C- and E-class protein interaction in a double-flowered ranunculid mutant. Proceedings of the National Academy of Sciences of the United States of America 109: E2267–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Mena C, Folter S de, Costa MM, Angenent GC, Sablowski R. 2005. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438. [DOI] [PubMed] [Google Scholar]
- Gonçalves B, Nougué O, Jabbour F, et al. 2013. An APETALA3 homolog controls both petal identity and floral meristem patternin in Nigella damscena L. (Ranunculaceae). The Plant Journal 76: 223–235. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Bello AM. 2009. Intra-individual variation of flowers in Gunnera subgenus Panke (Gunneraceae) and proposed apomorphies for Gunnerales. Botanical Journal of the Linnean Society 160: 262–283. [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2006. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. The Plant Cell 18: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans K, Ament K, Rijpkema AS, et al. 2012. Redefining C and D in the petunia ABC. The Plant Cell 24: 2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF. 2010. The flowering of Arabidopsis flower development. The Plant Journal 61: 1014–1028. [DOI] [PubMed] [Google Scholar]
- Jack T. 2001. Plant development going MADS. Plant Molecular Biology 46: 515–520. [DOI] [PubMed] [Google Scholar]
- Kim S, Koh J, Yoo M, et al. 2005. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. The Plant Journal 43: 724–744. [DOI] [PubMed] [Google Scholar]
- Kitazawa MS, Fujimoto K. 2014. A developmental basis for stochasticity in floral organ numbers. Frontiers in Plant Science 5, doi:10.3389/fpls.2014.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM. 2007. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. The Plant Cell 19: 750–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. 2005. Molecular mechanisms of flower development: an armchair guide. Nature Reviews Genetics 6: 688–698. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Lewis MW, Fletcher JC. 2006. RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. The Plant Journal 45: 369–383. [DOI] [PubMed] [Google Scholar]
- Lange M, Orashakova S, Lange S, et al. 2013. The seirena B class floral homeotic mutant of California Poppy (Eschscholzia californica) reveals a function of the enigmatic PI motif in the formation of specific multimeric MADS domain protein complexes. The Plant Cell 25: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue NC, Sullivan AM, Di Stilio VS. 2013. Functional recapitulation of transitions in sexual systems by homeosis during the evolution of dioecy in Thalictrum. Frontiers in Plant Science 4: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A. 2007. An evaluation of A‐function: evidence from the APETALA1 and APETALA2 gene lineages. International Journal of Plant Science 168: 73–91. [Google Scholar]
- Litt A, Kramer EM. 2010. The ABC model and the diversification of floral organ identity. Seminars in Cell & Developmental Biology 21: 129–137. [DOI] [PubMed] [Google Scholar]
- Melzer R, Theissen G. 2009. Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Research 37: 2723–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R, Härter A, Rümpler F, et al. 2014. DEF- and GLO-like proteins may have lost most of their interaction partners during angiosperm evolution. Annals of Botany 114: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Ferrario S, Angenent GC, Kobayashi A, Takatsuji H. 2004. The petunia ortholog of Arabidopsis SUPERMAN plays a distinct role in floral organ morphogenesis. The Plant Cell 16: 920–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orashakova S, Lange M, Lange S, Wege S, Becker A. 2009. The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. The Plant Journal 58: 682–693. [DOI] [PubMed] [Google Scholar]
- Pabón-Mora N, Ambrose BA, Litt A. 2012. Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiology 158: 1685–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N, Hidalgo O, Gleissberg S, Litt A. 2013a. Assessing duplication and loss of APETALA1/FRUITFULL homologs in Ranunculales. Frontiers in Plant Science 4: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N, Sharma B, Holappa LD, Kramer EM, Litt A. 2013b. The Aquilegia FRUITFULL-like genes play key roles in leaf morphogenesis and inflorescence development. The Plant Journal 74: 197–212. [DOI] [PubMed] [Google Scholar]
- Ren Y, Chang H, Endress PK. 2010. Floral development in Anemoneae (Ranunculaceae). Botanical Journal of the Linnean Society 162: 77–100. [Google Scholar]
- Ronse De Craene Louis P, Soltis PS, Soltis DE. 2003. Evolution of floral structures in basal angiosperms. International Journal of Plant Science 164: S329. [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. 1995. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203. [DOI] [PubMed] [Google Scholar]
- Schütz I, Moritz GB, Roos W. 2014. Alkaloid metabolism in thrips–Papaveraceae interaction: recognition and mutual response. Journal of Plant Physiology 171: 119–126. [DOI] [PubMed] [Google Scholar]
- Shan H, Zhang N, Liu C, et al. 2007. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Molecular Phylogenetics and Evolution 44: 26–41. [DOI] [PubMed] [Google Scholar]
- Sharma B, Kramer E. 2013. Sub- and neo-functionalization of APETALA3 paralogs have contributed to the evolution of novel floral organ identity in Aquilegia (columbine, Ranunculaceae). The New Phytologist 197: 949–957. [DOI] [PubMed] [Google Scholar]
- Sollid J, Nilsson GE. 2006. Plasticity of respiratory structures - Adaptive remodeling of fish gills induced by ambient oxygen and temperature. Respiratory Physiology and Neurobiology 154: 241–251. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Chanderbali AS, Kim S, Buzgo M, Soltis PS. 2007. The ABC model and its applicability to basal angiosperms. Annals of Botany 100: 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soza VL, Brunet J, Liston A, Smith PS, Di Stilio VS. 2012. Phylogenetic insights into the correlates of dioecy in meadow-rues (Thalictrum, Ranunculaceae). Molecular Phylogenetics and Evolution 63: 180–192. [DOI] [PubMed] [Google Scholar]
- Stammler A, Meyer SS, Plant AR, Townsley BT, Becker A, Gleissberg S. 2013. Duplicated STM-like KNOX I genes act in floral meristem activity in Eschscholzia californica (Papaveraceae). Development Genes and Evolution 223: 289–301. [DOI] [PubMed] [Google Scholar]
- Sun B, Ito T. 2015. Regulation of floral stem cell termination in Arabidopsis. Frontiers in Plant Science 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Huang W, Li Z, et al. 2014. Functional and evolutionary analysis of the AP1/SEP/AGL6 superclade of MADS-box genes in the basal eudicot Epimedium sagittatum. Annals of Botany 113: 653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekleyohans DG, Lange S, Becker A. 2013. Virus-induced gene silencing of the alkaloid-producing basal eudicot model plant Eschscholzia californica (California Poppy). Methods in Molecular Biology 975: 83–98. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. 2001. Plant biology. Floral quartets. Nature 409: 469–471. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marín M, Manson JS, Thomson JD, Barrett SCH. 2009. Division of labour within flowers: heteranthery, a floral strategy to reconcile contrasting pollen fates. Journal of Evolutionary Biology 22: 828–839. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y. 2014. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Research 24: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Larsen J, Harder LD. 2000. The evolution of staminodes in angiosperms: patterns of stamen reduction, loss, and functional re-invention. American Journal of Botany 87: 1367–1384. [PubMed] [Google Scholar]
- Wang W, Lu A, Ren Y, Endress ME, Chen Z. 2009. Phylogeny and classification of Ranunculales: evidence from four molecular loci and morphological data. Perspectives in Plant Ecology, Evolution and Systematics 11: 81–110. [Google Scholar]
- Wege S, Scholz A, Gleissberg S, Becker A. 2007. Highly efficient virus-induced gene silencing (VIGS) in California poppy (Eschscholzia californica): an evaluation of VIGS as a strategy to obtain functional data from non-model plants. Annals of Botany 100: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellina AL, Orashakova S, Lange S, Erdmann R, Leebens-Mack J, Becker A. 2010. Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica). EvoDevo 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, et al. 2005. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack JH, Arrington JM, et al. 2006. Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evolution & Development 8: 30–45. [DOI] [PubMed] [Google Scholar]
- Zhang R, Guo C, Zhang W, et al. 2013. Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proceedings of the National Academy of Sciences of the United States of America 110: 5074–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]