Abstract
NSD1 and EZH2 are SET domain-containing histone methyltransferases that play key roles in the regulation of transcription through histone modification and chromatin modelling: NSD1 preferentially methylates lysine residue 36 of histone 3 (H3K36) and is primarily associated with active transcription, whilst EZH2 shows specificity for lysine residue 27 (H3K27) and is associated with transcriptional repression. Somatic dysregulation of NSD1 and EZH2 have been associated with tumorigenesis. NSD1, as a fusion transcript with NUP98, plays a key role in leukemogenesis, particularly childhood acute myeloid leukemia. EZH2 is a major proto-oncogene and mono- and biallelic activating and inactivating somatic mutations occur as early events in the development of tumors, particularly poor prognosis hematopoietic malignancies. Constitutional NSD1 and EZH2 mutations cause Sotos and Weaver syndromes respectively, overgrowth syndromes with considerable phenotypic overlap. NSD1 mutations that cause Sotos syndrome are loss-of-function, primarily truncating mutations or missense mutations at key residues in functional domains. EZH2 mutations that cause Weaver syndrome are primarily missense variants and the rare truncating mutations reported to date are in the last exon, suggesting that simple haploinsufficiency is unlikely to be generating the overgrowth phenotype although the exact mechanism has not yet been determined. Many additional questions about the molecular and clinical features of NSD1 and EZH2 remain unanswered. However, studies are underway to address these and, as more cases are ascertained and technology improves, it is hoped that these will, in time, be answered.
Keywords: NSD1, EZH2, histone methyltransferases, Sotos, Weaver
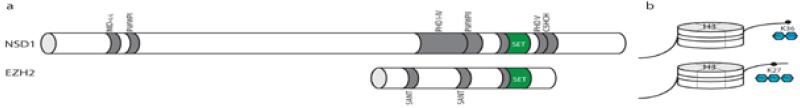
NSD1 (Nuclear receptor-binding SET domain-containing protein 1) and EZH2 (Enhancer of Zeste, drosophila, homolog 2) both encode SET domain-containing histone methyltransferases and their germline abrogation results in the childhood overgrowth syndromes, Sotos syndrome and Weaver syndrome respectively (MIM 117550 and MIM 614421). NSD1 is located at chromosome 5q35.3 and contains ten conserved domains in addition to the SET (Su(Var)3-9, Enhancer of zeste, Trithorax) and preceding SAC (SET associated cysteine rich) domains: two distinct nuclear interacting domains (NID+L and NID−L); two PWWP domains (Pro-Try-Try-Pro motif); five zinc finger, PHD, domains and one C5HCH (cysteine/histidine rich) domain. EZH2, at chromosome location 7q36.1, has two SANT (Swi3, Ada2, N-cor TFIIIB) domains in addition to the SET and SAC domains (figure 1a).
Figure 1. Domain structure of NSD1 and EZH2 SET domain-containing histone methyltransferases.
a) NSD1 is a 2696 amino acid protein with ten conserved domains in addition to the SET and preceding SAC domains whilst EZH2 is a 746 amino acid protein with two SANT domains in addition to the SET and SAC. b) NSD1 is a histone methyltransferase which preferentially mono or di- methylates lysine residue 36 of histone 3 whereas EZH2 can catalyse the transfer of up to three methyl groups to lysine residue 27 of histone 3. NID, Nuclear interacting domain; PWWP, Proline, tryptophan, tryptophan, proline; PHD, plant homeodomain; SANT, Swi3, Ada2, N-cor TFIIIB.
There is considerable phenotypic overlap between Sotos and Weaver syndromes with both characterized by pre and post natal overgrowth, a variable intellectual disability and similar facial appearance. In addition somatic disruption of both NSD1 and EZH2 has been implicated in multiple tumor types, particularly hematological malignancies. It is interesting and noteworthy that these two histone methyltransferases should have similar, dual roles in tumorigenesis and human development and the current review will explore the similarities and key differences between these two proto-oncogenes.
NSD1 and EZH2 are histone methyltransferases but with differing histone/lysine specificities
Histone methyltransferases play a critical role in the epigenetic modification of histones thereby determining chromatin compaction and transcriptional activity. Most histone methyltransferases contain a conserved SET domain that catalyses the transfer of methyl groups to specific lysine and arginine residues of histone and non-histone proteins (Rea et al 2000). By comparing homology of the SET domains, four subfamilies of histone methyltransferases have been defined; SUV39, SET1, SET2 and RIZ (Kouzarides 2002). NSD1 and EZH2 belong to the SET2 and SET1 subfamilies respectively and each is associated with specific post-translational histone modifications or “marks”: NSD1 preferentially catalyses the transfer of up to two methyl residues to lysine residue 36 of histone 3 (H3K36) and may additionally have specificity for lysine residue 20 of histone 4 (H4K20, Qiao et al 2011; Rayasam et al 2003). In contrast, EZH2, (when associated with EED (Embryonic Ectoderm Development protein, mouse, homolog of) and SUZ12 (Suppressor of Zeste 12, drosophila, homolog of) to form the core components of the polycomb repressor complex 2 (PCR2)), catalyses the tri-methylation of lysine residue 27 of histone 3 (H3K27me3, figure 1b, Cao et al 2002). The histone modifications catalyzed by EZH2 have been associated with transcriptional repression whereas those catalyzed by NSD1 have primarily been associated with activation but can be associated with repression depending on the cellular context (Cao et al 2002; Huang et al 1998; Wagner et al).
NSD1 and EZH2 are proto-oncogenes
NSD1 and EZH2 are proto-oncogenes with somatic mutations identified in multiple tumor types. A recurrent cryptic translocation t(5;11)(q35.3;p15.5) involving NSD1 has been identified in approximately 5% of childhood acute myeloid leukemia (Cerveira et al 2003). The translocation fuses NSD1 to nucleoporin 98 (NUP98), a component of the nuclear core complex, and the resultant NUP98-NSD1 fusion protein plays a key role in leukemogenesis through H3K36 methylation and subsequent HOX-A gene activation (Wang et al 2007). Although there are conflicting reports, it is believed that the alternative NSD1-NUP98 transcript does not have a biologically relevant role in tumorigenesis (Cerveira et al 2003). In addition, somatic epigenetic silencing of NSD1, through promoter hypermethylation, has been associated with neuroblastoma and gliomas and somatic NSD1 mutations have been associated with carcinoma of the upper airway digestive tract (Berdasco et al 2009, Catalogue of Somatic Mutations in Cancer).
Somatic disruption of EZH2 is a key event in the development of many tumors, particularly hematopoietic malignancies (Chase et al 2011). EZH2 is noteworthy in that both activating and inactivating mutations have been associated with tumorigenesis. A recurrent monoallelic, gain-of-function alteration, affecting the 646 tyrosine residue and associated with enhanced di- and trimethylation of H3K27, has been identified in approximately 7% of follicular lymphomas and 22% of diffuse large cell B-cell lymphomas of germinal center origin (Morin et al 2010; Sneeringer et al 2010). In contrast, both monoallelic and biallelic inactivating mutations, distributed throughout the gene, have been identified in poor prognosis myeloproliferative neoplasms and myelodysplastic syndromes (Ernst et al 2010). This association of both activating and inactivating mutations with tumorigenesis has been interpreted as suggesting that a critical dosage level of EZH2 is required for normal stem cell homeostasis and that hematological malignancies develop where there is disruption of this normal balance (Sauvageau et al 2010).
Germline disruption of NSD1 and EZH2 function cause overgrowth
Germline, monoallelic disruption of both NSD1 and EZH2 cause the overgrowth syndromes, Sotos and Weaver syndromes respectively, but the spectra of mutations associated with the genes differs (Kurotaki et al 2002; Gibson et al 2011; Tatton-Brown et al 2011). NSD1 causes Sotos syndrome through germline haploinsufficiency and, amongst the non-Japanese population, intragenic loss-of-function mutations, primarily truncating mutations, account for over 80% of NSD1 mutation-positive individuals (Cecconi et al 2005; Rio et al 2003; Tatton-Brown et al 2005b; Turkmen et al 2003; Waggoner et al 2005). In the Japanese, whole NSD1 gene deletions are the primary cause of Sotos syndrome (Kamimura et al 2003). This difference in microdeletion frequency between individuals of Japanese and non-Japanese descent has been attributed to the genomic architecture of the 5q35.3 region where three distinct low copy repeat elements flank NSD1. Two of these low copy repeats are in the same orientation whilst the third, located between these same orientation elements but telomeric to NSD1, is inversely orientated (Tatton-Brown et al 2005a). An inversion polymorphism, between the inversely orientated low copy repeats, is quite common in Japanese individuals and likely predisposes to deletions mediated by non-allelic homologous recombination (Tatton-Brown et al 2005a). The EZH2 mutational spectrum in Weaver syndrome is markedly different: >90% of mutations are missense and the rare truncating mutations reported to date are in the last exon and are therefore unlikely to initiate nonsense-mediated RNA decay. This suggests that simple haploinsufficiency is unlikely to be the primary pathogenic mechanism of germline EZH2 mutations (Tatton-Brown et al 2011). Given these differences, it is interesting that, although mouse models null for both genes die in early embryonic development, the NSD1 heterozygous mutant embryo is viable and fertile whereas the EZH2 heterozygous mutants die during the transition from pre- to post-implantation (O’Carroll et al 2001; Rayasam et al 2003).
The key clinical criteria associated with germline NSD1 and EZH2 alterations are overgrowth; a characteristic facial appearance and intellectual disability
There is considerable overlap between the NSD1 and EZH2-associated phenotypes as exemplified by our initial paper on NSD1 and Sotos syndrome that, erroneously, reported that Sotos and Weaver syndromes were allelic conditions (Douglas et al 2003). Subsequent studies by ourselves and others, have shown that NSD1 alterations are specific to Sotos syndrome and are identified in over 90% of individuals with a clinical diagnosis of Sotos syndrome (Tatton-Brown et al 2005b). Although EZH2 alterations do not appear to be as sensitive or specific for Weaver syndrome (i.e some individuals with EZH2 alterations do not have classic Weaver syndrome whilst others with a clinical diagnosis of Weaver do not have a germline EZH2 alteration) for the purposes of this review we have used Sotos syndrome to describe individuals with an NSD1 mutation and Weaver syndrome to describe individuals with an EZH2 mutation.
Facially, children with disruption of both NSD1 and EZH2 can have a high, broad forehead and prominent chin (figure 2). However, in classic Sotos syndrome, the palpebral fissures are usually down slanting, there is frontotemporal hair sparsity, the face is long and thin and, although the eyes can appear hyperteloric, this is generally because of an associated bitemporal narrowing rather than a true hypertelorism (figure 2a). In classic Weaver syndrome, young children are retrognathic and have large, fleshy ears whilst both children and adults with classic Weaver syndrome are hyperteloric and the eyes are almond shaped (figure 2b).
Figure 2. Facial appearance of children and adults with a)NSD1 mutations and b) EZH2 mutations.
Although germline disruption of both NSD1 and EZH2 results in overgrowth, the associated growth profiles differ. Children with EZH2 mutations are consistently tall, with heights up to eight standard deviations above the mean, but macrocephaly is not a consistent finding (~50% of EZH2 mutation-positive individuals are not macrocephalic, Tatton-Brown, Rahman submitted). In contrast, not all NSD1 mutation-positive individuals are tall and some have isolated macrocephaly (~20%) whilst a small proportion (~10%) are neither tall nor macrocephalic (Tatton-Brown et al 2005b)
Most individuals with NSD1 or EZH2 mutations have an intellectual disability, although the degree is variable for both genes. Amongst the NSD1 mutation-positive individuals, intellectual disability ranges from mild through to severe with a moderate disability most frequently reported (~45% of NSD1 mutation-positive individuals have a moderate disability compared with 30% and 20% with mild and severe intellectual disabilities respectively, Tatton-Brown et al 2005b). In contrast, a mild disability is most frequently reported in individuals with EZH2 mutations (~45%) with a moderate and severe disability reported in the minority (~30% and ~5% respectively). The proportion of individuals with no intellectual disability is correspondingly different, present in 2% of NSD1 mutation-positive individuals and ~20% of EZH2 mutation-positive individuals (Tatton-Brown et al, 2005, Tatton-Brown, Rahman, submitted).
Advanced bone age is reported in individuals with both NSD1 and EZH2 mutations. However, whilst the bone age is consistently advanced in EZH2 mutation-positive individuals, it has been shown to be advanced in only 80% of NSD1 mutation-positive individuals (Tatton-Brown et al 2005b), Tatton-Brown, Rahman submitted).
The Sotos/Weaver syndrome phenotypic overlap can make it challenging, even for the experienced clinician, to clinically distinguish the two conditions. However, congenital cardiac disease, renal anomalies and seizures are more frequently reported in individuals with NSD1 than EZH2 alterations. In contrast, a connective tissue phenotype with soft, loose skin, umbilical hernia and thin, deep-set nails are more commonly described in the EZH2 mutation-positive group. Other potentially distinguishing features described amongst individuals with EZH2 alterations include a deep hoarse voice and camptodactyly of the fingers and/or toes evolving into boutonniere deformities in adulthood (Tatton-Brown, Rahman submitted).
There is evidence of an increased risk of certain tumors in Sotos and Weaver syndromes, but the absolute risk of cancer is small, in the order of 3% and 5% respectively (Tatton-Brown et al, 2005; Tatton-Brown, Rahman, submitted). The spectra of tumors may differ between the conditions, for example sacrococcygeal teratoma has been reported in three individuals with Sotos syndrome but no individual with Weaver syndrome, although neuroblastoma has been reported, very rarely, in both conditions. As the risk of tumors is small and there are currently no effective screening modalities for the tumor types observed in either condition, screening is not recommended. More appropriate is clinical vigilance and thorough investigation of any possible tumor related signs and symptoms.
Conclusions and future directions
The current review has highlighted key similarities and differences between the two histone methyltransferases, NSD1 and EZH2, and their associated overgrowth syndromes, Sotos and Weaver syndromes. However, there is still much to understand about the molecular and clinical features of these two overgrowth genes.
Although NSD1 loss of function mutations cause Sotos syndrome, it is currently not known how the germline, predominantly missense, EZH2 mutations cause Weaver syndrome. Given that the few truncations to have been identified all target the final exon, and are therefore unlikely to be initiating nonsense-mediated RNA decay, haploinsufficiency seems an unlikely mechanism. It is also interesting that three identical mutations, Gly159Arg, Arg684Cys and Tyr733ter, have been identified, somatically, in myeloid malignancies and, constitutionally, in eight unrelated individuals with Weaver syndrome (Chase et al 2011; Ernst et al 2010; Nikoloski et al 2010; Tatton-Brown et al 2011). None of the Weaver individuals with these mutations have developed malignancies to date. The reason for the divergent phenotypes associated with these identical germline and somatic mutations is currently unclear but may be related to the age of onset of myeloid malignancies, which tend to occur in later life, compared to the young age of the current EZH2 mutation-positive cohort. Alternatively it may reflect additional or different mechanisms whereby the somatic mutations are causing tumors and the germline mutations are disrupting normal human development and growth.
The NSD1-associated phenotype is well characterized with many hundreds of reported cases. However, we do not yet understand what factors determine the variability of the Sotos syndrome phenotype. This variability is exemplified by unrelated individuals with the same, recurrent mutation but differing degrees of intellectual disability and frequency of associated medical issues such as cardiac and renal anomalies, seizures and scoliosis. In addition, we still do not understand why there are so few familial Sotos syndrome cases and what factors are reducing vertical transmission of mutations.
The number of known individuals with constitutional EZH2 mutations is small. This is primarily because EZH2 was only identified as the cause of Weaver syndrome in 2011. It may also be because Weaver syndrome is rarer than Sotos syndrome and/or because Weaver syndrome is more likely to evade diagnosis because individuals are more often mildly affected. The paucity of cases limits current knowledge of the associated clinical features and there is also likely to have been a bias towards more severely affected individuals having been tested thus far, which might result in inflation of the frequencies of associated clinical features.
As genetic testing becomes more accessible it is likely that many more individuals with EZH2 and NSD1 mutations are identified enabling (further) clarification of the associated phenotypes. In addition, long-term prospective studies will address associated tumor risks and the evolution of both the NSD1-associated and EZH2-associated phenotypes. Currently, functional work is underway to investigate how somatic EZH2 mutations are causing malignancies and constitutional mutations are causing Weaver syndrome. Finally, as technology improves, so will our understanding of the other factors which determine phenotype and, in the future, we may be able to not only make a diagnosis of Sotos or Weaver syndrome, but be able to offer prognostic information about the nature and severity of associated clinical sequelae.
Biographies
Bios of Authors
Dr Kate Tatton-Brown is a Clinical Geneticist at the Institute of Cancer Research (ICR), St George’s University of London and the Royal Marsden Hospital. She has a strong research interest and clinical experience in childhood overgrowth syndromes and has published widely on Sotos syndrome, the 15q overgrowth syndrome and, more recently, on Weaver syndrome.
Professor Nazneen Rahman is Professor of Human Genetics at the Institute of Cancer Research (ICR). She is both a clinician and world leading researcher in the field of genetic susceptibility. Her research work has been directed towards the mapping and identification of adult and paediatric cancer predisposition genes and childhood overgrowth genes. She has extensive publications and during her 13 years as a clinical academic researcher has mapped/ identified 14 disease genes.
Footnotes
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.gov/OMIM/ (for Sotos syndrome and Weaver syndrome)
Catalogue of Somatic Mutations in Cancer (COSMIC), http://www.sanger.ac.uk/genetics/CGP/cosmic/
References
- Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R, Alaminos M, Cheung NK, Rahman N, Esteller M. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci U S A. 2009;106(51):21830–5. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cecconi M, Forzano F, Milani D, Cavani S, Baldo C, Selicorni A, Pantaleoni C, Silengo M, Ferrero GB, Scarano G, Della Monica M, Fischetto R, Grammatico P, Majore S, Zampino G, Memo L, Cordisco EL, Neri G, Pierluigi M, Bricarelli FD, Grasso M, Faravelli F. Mutation analysis of the NSD1 gene in a group of 59 patients with congenital overgrowth. Am J Med Genet A. 2005;134(3):247–53. doi: 10.1002/ajmg.a.30492. [DOI] [PubMed] [Google Scholar]
- Cerveira N, Correia C, Doria S, Bizarro S, Rocha P, Gomes P, Torres L, Norton L, Borges BS, Castedo S, Teixeira MR. Frequency of NUP98-NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia. 2003;17(11):2244–7. doi: 10.1038/sj.leu.2403104. [DOI] [PubMed] [Google Scholar]
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17(9):2613–8. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- Douglas J, Hanks S, Temple IK, Davies S, Murray A, Upadhyaya M, Tomkins S, Hughes HE, Cole TR, Rahman N. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am J Hum Genet. 2003;72(1):132–43. doi: 10.1086/345647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, Mungall AJ, Eydoux P, Babul-Hirji R, An J, Marra MA, Chitayat D, Boycott KM, Weaver DD, Jones SJ. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2011;90(1):110–8. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998;17(12):3398–412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura J, Endo Y, Kurotaki N, Kinoshita A, Miyake N, Shimokawa O, Harada N, Visser R, Ohashi H, Miyakawa K, Gerritsen J, Innes AM, Lagace L, Frydman M, Okamoto N, Puttinger R, Raskin S, Resic B, Culic V, Yoshiura K, Ohta T, Kishino T, Ishikawa M, Niikawa N, Matsumoto N. Identification of eight novel NSD1 mutations in Sotos syndrome. J Med Genet. 2003;40(11):e126. doi: 10.1136/jmg.40.11.e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12(2):198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita Ha HA, Kinoshita A, Mizuguchi T, Yoshiura Ki K, Ohta T, Kishino T, Fukushima Y, Niikawa N, Matsumoto N. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet. 2002;30(4):365–6. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42(8):665–7. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330–6. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Li Y, Chen Z, Wang M, Reinberg D, Xu RM. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem. 2011;286(10):8361–8. doi: 10.1074/jbc.M110.204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003;22(12):3153–63. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rio M, Clech L, Amiel J, Faivre L, Lyonnet S, Le Merrer M, Odent S, Lacombe D, Edery P, Brauner R, Raoul O, Gosset P, Prieur M, Vekemans M, Munnich A, Colleaux L, Cormier-Daire V. Spectrum of NSD1 mutations in Sotos and Weaver syndromes. J Med Genet. 2003;40(6):436–40. doi: 10.1136/jmg.40.6.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107(49):20980–5. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Douglas J, Coleman K, Baujat G, Chandler K, Clarke A, Collins A, Davies S, Faravelli F, Firth H, Garrett C, Hughes H, Kerr B, Liebelt J, Reardon W, Schaefer GB, Splitt M, Temple IK, Waggoner D, Weaver DD, Wilson L, Cole T, Cormier-Daire V, Irrthum A, Rahman N. Multiple mechanisms are implicated in the generation of 5q35 microdeletions in Sotos syndrome. J Med Genet. 2005a;42(4):307–13. doi: 10.1136/jmg.2004.027755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Douglas J, Coleman K, Baujat G, Cole TR, Das S, Horn D, Hughes HE, Temple IK, Faravelli F, Waggoner D, Turkmen S, Cormier-Daire V, Irrthum A, Rahman N. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005b;77(2):193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte Sdel V, Ramsay E, Snape K, Murray A, Perdeaux ER, Seal S, Loveday C, Banka S, Clericuzio C, Flinter F, Magee A, McConnell V, Patton M, Raith W, Rankin J, Splitt M, Strenger V, Taylor C, Wheeler P, Temple KI, Cole T, Douglas J, Rahman N. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2(12):1127–33. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkmen S, Gillessen-Kaesbach G, Meinecke P, Albrecht B, Neumann LM, Hesse V, Palanduz S, Balg S, Majewski F, Fuchs S, Zschieschang P, Greiwe M, Mennicke K, Kreuz FR, Dehmel HJ, Rodeck B, Kunze J, Tinschert S, Mundlos S, Horn D. Mutations in NSD1 are responsible for Sotos syndrome, but are not a frequent finding in other overgrowth phenotypes. Eur J Hum Genet. 2003;11(11):858–65. doi: 10.1038/sj.ejhg.5201050. [DOI] [PubMed] [Google Scholar]
- Waggoner DJ, Raca G, Welch K, Dempsey M, Anderes E, Ostrovnaya I, Alkhateeb A, Kamimura J, Matsumoto N, Schaeffer GB, Martin CL, Das S. NSD1 analysis for Sotos syndrome: insights and perspectives from the clinical laboratory. Genet Med. 2005;7(8):524–33. doi: 10.1097/01.GIM.0000178503.15559.d3. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–26. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9(7):804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]