Abstract
Diverse microarray and sequencing technologies have been widely used to characterize molecular changes in malignant epithelial cells in salivary neoplasms. Such gene expression studies to identify markers and targets in tumor cells are, however, compromised by the cellular heterogeneity of these tumors and by the difficulties to accrue matching controls representing normal salivary glands. Seventeen samples of primary salivary epithelial–myoepithelial carcinoma along with tissue from six normal major salivary glands were microdissected from paraffin-embedded tissue. Pools of RNA from highly enriched preparations of these cell types were subjected to expression profiling using a whole-transcriptome shotgun sequencing experiment. In parallel, extracted genomic DNA was used for the 50 gene hotspot panel sequenome. KRAS mutations in three patients (18%), NRAS mutations in one patient (6%), but no HRAS, MET, PIK3CA, or BRAF mutations. Using strict and conservative criteria, 220 differentially expressed transcripts were found, with 36% up- and 64% downregulated. The transcripts were annotated using NCBI Entrez Gene, and computationally analyzed with the Ingenuity Pathway Analysis program. From these significantly changed expressions, the analysis identified 26 cancer-related transcripts and 16 transcripts related to mitochondrial dysfunction overlapping with three cancer-related genes. These 220 differentially expressed genes including micro-RNAs provide here a sufficiently large set to specifically define epithelial–myoepithelial carcinoma and to identify novel and potentially important targets for diagnosis, prognosis, and therapy of this cancer.
INTRODUCTION
First described by Donath et al. in 1972 (Donath et al. 1972), epithelial-myoepithelial carcinoma (EMC) of the salivary gland is a rare tumor, comprising about 0.4–1% of salivary gland tumors. Typically, EMC has been described as a low-grade neoplasm that enlarges slowly (Donath et al. 1972; Brocheriou et al. 1991; Seethala et al. 2007). The tumors are usually firm and well circumscribed, with little to no invasion to nearby structures. Histologically, the neoplasm consists of both epithelial and myoepithelial cells arranged in tubules, trabeculae, small islands or sheets. Although typical EMC has a high rate of recurrence (35–40%), mortality is low (Tralongo and Daniele 1998; Seethala et al. 2007). Adverse features that have been shown to confer a worse prognosis include invasion, metastasis, necrosis, and anaplasia. Tumors with anaplasia have been labeled as either dedifferentiated EMC or EMC with high-grade transformation (Baker et al. 2013). In contrast, the molecular profile of EMC has not been well studied, although an association with Harvey rat sarcoma viral oncogene homolog (HRAS) mutations has been noted in recent studies (Prior et al. 2012; Cros et al. 2013; Chiosea et al. 2014).
Advances in DNA sequencing, called next-generation sequencing (NGS), have allowed massive parallel throughput, and data volumes that eclipse the nucleic acid information content possible with other technologies, making feasible extensive genome analysis of groups of individuals, including analysis of sequence differences, polymorphisms, mutations, copy number variations, epigenetic variations, and transcript abundance. Biomarker discovery is an attractive potential application of this new technology.
Aberrant transcript expression includes changes in expression levels, isoforms, and polymorphisms, which are commonly observed in cancer; these aberrations could alter biological pathways and disease phenotypes. NGS of RNA (RNA-Seq) has become a powerful tool for studying the comprehensive transcriptome (Wang et al. 2009; Wolf 2013).
The RNA-Seq method described here enables transcriptome-wide cancer biomarker discovery with archival fixed paraffin-embedded (FFPE) tissue. The FFPE material is linked to mature clinical records in hospital pathology archives. This material can be used for tumor gene expression profiling and, therefore, may enable rapid clinical biomarker discovery in studies that are statistically well-powered (Li et al. 2010; Li and Dewey 2011; Rehrauer et al. 2013; Long et al. 2014).
To determine the gene expression levels in salivary EMC tissues, we used high-throughput mRNA sequencing (RNA-Seq) to characterize the differences and similarities of transcriptome expression in tissues from patients with salivary EMC and healthy salivary tissues, in a retrospective study. We also investigated the molecular profile of tumor tissues in the same cohort using a platform that probed 190 potentially targetable common oncogenic mutations.
MATERIALS AND METHODS
Patients
Patients were identified by a head and neck pathologist (IF) from pathology archives of the Portuguese Oncology Hospital, with institutional review board approval.
Tissue Specimens
Primary salivary EMC FFPE tumor specimens were available from 17 patients with clinical outcome data available. Six nonmatching normal salivary FFPE tissues samples were analyzed in parallel.
Mutation Analysis
Genomic DNA was isolated from 5-µm-thick paraffin sections using the Epicentre Master pure DNA and RNA isolation kit (Illumina, San Diego, CA) according to the manufacturer’s protocol following deparaffinization and proteinase K treatment. The Sequenom MALDI-TOF Mass-ARRAY platform was used to profile 190 common oncogenic point mutations in 50 genes. One microgram of genomic DNA per sample was submitted to the Sequencing and Microarray Facility at our institution. Each specimen was tested in duplicate for every mutation in the Sequenom panel.
RNA-Seq Sample Preparation and Sequencing
Tissue was scraped from FFPE tissue slides for 6 normal and 17 EMC samples, and total RNA was extracted using an RNeasy Universal kit (Qiagen, Gaithersburg, MD). Briefly, FFPE tissue was deparaffinized, proteinase K treated, and genomic DNA removed for total RNA extraction. Quality and quantity of total RNA were verified spectro-photometrically (NanoDrop 100 spectrometer, ThermoScientific, Wilmington, DE) and electro-phoretically (Bioanalyzer 2100, Agilent Technologies, Palo Alto, CA).
RNA-Seq was performed by EA-Quintiles (EA-Quintiles, Durham, NC) using the library kit Illumina TrueSeq RNA Gold (Illumina, San Diego, CA). Briefly, total RNA was depleted of ribosomal RNA with the Ribo-Zero method. The residual RNA was fragmented, reverse transcribed using random hexamer priming into double-stranded RNA/DNA hybrids for first strand synthesis. For second strand synthesis, double-stranded cDNA was created incorporating dUTP, rendering the second strand an inefficient template for a subsequent PCR step, which in turn leads to strand selection. End repair, A-tailing, sequencing-adapter ligation, and polymerase chain reaction (PCR) amplification to generate short double-stranded cDNA fragments followed, products of which were then used to create a transcriptome cDNA library for RNA-Seq. Sequencing was performed on an Illumina HiSeq 2000 (Illumina, San Diego, CA) sequencer with paired end reads of 50 base pairs of RNA insert. The actual total cluster varied from 40.5 to 79 million, with most samples between 50 and 60 million clusters (100–120 millions if counting each paired read separately) depending on the quality of the reads.
RNA-Seq Analysis for Differentially Expressed Genes and Isoforms
The RNA-Seq and its results analysis were performed by EA-Quintiles using their software pipeline mRNAv7-RNA seq by Expectation-Maximization (RSEM) (EA-Quintiles, Durham, NC). In detail, expressed transcripts were normalized with upper-quartile normalization, which sets the upper-quartile value of each transcript in each sample to be 1,000. The data were then quantified with the RSEMv1.2.0 program (RNA-Seq data analysis by Expectation-Maximization from EA-Quintiles, Durham, NC) optimized for Illumina 50 × 50 paired-end reads (Illumina, San Diego, CA) using the reference gene transcriptome and annotations from UCSC (hg19; Genome Reference Consortium GRCh37 at http://genome.uscs.edu). UCSC Human RNA reference data, especially lncRNA reference data, were supplemented with data available from ENSEMBL (http://www.ensembl.org/index.html). Annotation from RefSeq (http://www.ncbi.nlm.nih.gov/refseq/) was also provided.
The transcript counts for gene expression levels were calculated, and the relative transcript abundance was determined using normalized counts. Using this approach, the expression levels were measured for 27976 UCSC KnownGene and 77099 UCSC KnownGene isoforms uniquely aligned based on RNA sequencing reads.
Raw data were extracted as normalized count values across all samples, and samples with zero values across more than 50% of the genes were excluded.
The statistical significance of the fold change in expression was determined using paired t-tests with the null hypothesis being that no difference existed between the two values.
The false discovery rate (FDR) was controlled by adjusting the P-values using the Benjamini– Hochberg algorithm and after exclusion of ribosomal and control transcripts the results were assigned to 24567 gene transcripts and to 62700 isoform transcripts, each including non-coding RNA and representing the human transcriptome.
From the total of 24567 gene transcripts only 220 genes were selected, with the most statistical evidence of being differentially expressed at FDRs < 3.8 E −3 (based on P-values<3.4 E −5) for usage in pathway analysis and screening for biomarkers.
All data analyses and visualization of differentially expressed genes were conducted using the R2.15.1 package (www.r-project.org).
Cluster Analysis and Heatmaps
To show the dissimilarity between all the samples a dendrogram cluster analysis was performed on all log transformed genes, not just the differentially expressed ones, using the “matrix-cluster-sample-analysis” correlation procedure (EA-Quintiles, Durham, NC).
Two heatmaps with cluster analyses were calculated from the top 100 differential expressed genes and the top 100 differential expressed isoforms based on their log2 expression values. The similarity calculations were done as average linkage clustering, based on the Euclidean distance of the samples comparing the top 100 gene/isoform expressions with each other (EA-Quintiles, Durham, NC).
Immunohistochemical Analysis
Immunohistochemical analysis was performed with antibodies against human CST2, PDK4, PLCG1, FGFR1 (all from LifeSpan Biosciences, Seattle, WA) and BPIFA4P (BASE) (Abnova, Taipei). All slides were analyzed by the same pathologist (DB). For each protein, the results were categorized as positive (>10% of the area in the entire tissue specimen was positive) or negative (<10% of the entire tissue specimen was stained).
Pathway Analysis
The extracellular matrix (ECM) pathway network was created using the Ingenuity Pathway Analysis (IPA) program from Ingenuity Systems (http://www.ingenuity.com/).
RESULTS
Clinical and Histologic Data
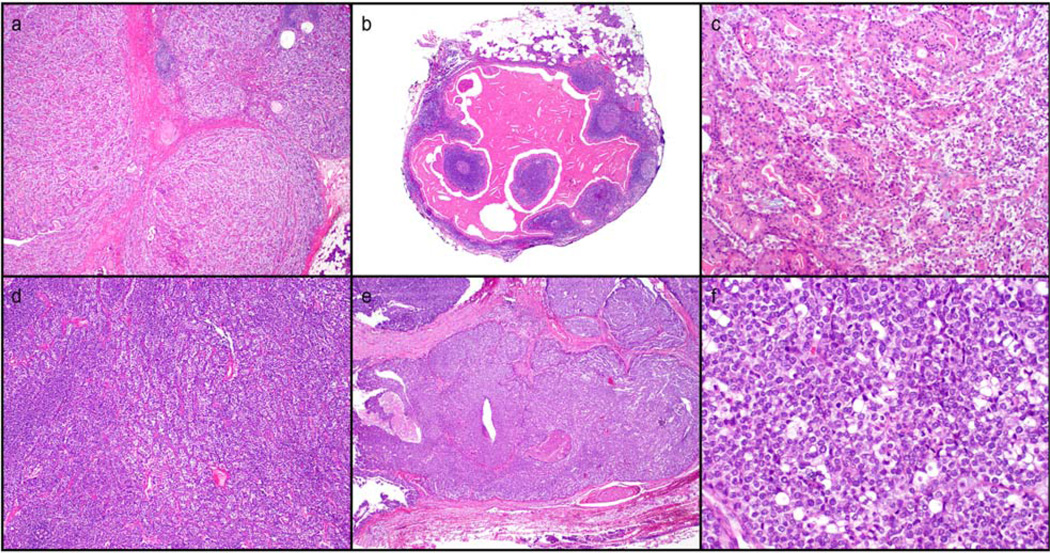
The diagnosis of EMC was established individually by two experienced head and neck patholo-gists (IF, DB). Clinical characteristics of the 17 EMC patients are given in Table 1. Age at diagnosis ranged between 42 and 85 years. Most patients were male (59%). The parotid gland was the most frequent primary site (n = 10) Morphologic subtypes of EMC are illustrated in Figure 1 (lower grade histology in patient 15-a,b,c; higher grade histology in patient 11-d,e,f).
TABLE 1.
Clinical Characteristics of the 17 EMC Patients, and the Mutation Status Detected by Sequenome (the Arrow Pointing to MET Indicates Polymorphism)
| Age | Sex | Location | Size (cm) |
Resection | pN | RT | FU (months) |
FU (status) |
Mutation status | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 47 | m | Parotid | – | – | – | – | 0 | No follow-up | Negative | |
| P2 | 61 | f | Parotid | 0.9 | Complete | 0 | No | 122 | A&W | Currently in treatment for multiple myeloma |
Negative |
| P3 | 76 | f | Parotid | 1.0 | Incomplete | 0 | Yes | 78 | A&W | KRAS_G12DAV_G35ACT | |
| P4 | 65 | m | Maxillary sinus | 2.0 | Complete | 0 | Yes | 28 | A&W | Negative | |
| P5 | 52 | f | Nasal cavity | 4.0 | Incomplete | 0 | No | 7 | AWD | 1st rec 2002 | Negative |
| P6 | 72 | m | Palate | 5.0 | Incomplete | 0 | No | 78 | A&W | Negative | |
| P7 | 51 | f | Palate | – | – | – | – | 167 | DOD | 1st rec 1997; 2nd rec 1998; CNS mets 2002; DOD 2005 |
Negative |
| P8 | 85 | f | Palate | 1.5 | Incomplete | 0 | No | 25 | DOC | KRAS_G12DAV_G35ACT | |
| P9 | 59 | m | Parotid | 7.5 | Incomplete | 0 | Yes | 138 | A&W | MET_N375S_A1124G (polymorphism) |
|
| P10 | 61 | m | Parotid | – | – | – | – | 80 | DOD | 1st rec 1986; ln mets 1986; 2nd rec1989; skin mets 1991; DOD 1992 |
Negative |
| P11 | 70 | m | Parotid | 4.0 | Incomplete | 0 | Yes | 24 | DOD | Lung mets 2001; DOD 2001 |
KRAS_G12DAV_G35ACT |
| P12 | 42 | m | Submandib | 3.0 | Complete | 0 | Yes | 147 | A&W | Negative | |
| P13 | 69 | f | Trachea | 3.5 | Incomplete | 0 | Yes | 101 | A&W | Negative | |
| P14 | 84 | m | Parotid | 1.1 | Incomplete | 0 | na | 11 | REC | 1st rec 2007 | Negative |
| P15 | 84 | f | Parotid | 0.9 | Complete | 0 | na | 18 | A&W | Negative | |
| P16 | 78 | m | parotid | 4.0 | Complete | 0 | Yes | 83 | DOD | Negative | |
| P17 | 70 | m | Parotid | 3.0 | Incomplete | 0 | No | 39 | A&W | NRAS_G12DAV_G35ACT |
A&W = alive and well; AWD = alive with disease; DOD = died of disease; DOC = died of other cause; REC = recurrence; na = not available
Figure 1.
Morphologic subtypes of EMC. a, b, c-P15 (patient 15), classic morphology of EMC and incidental Warthins tumor; d, e, f-P13 (patient 13), conventional and higher grade EMC with basaloid, comedonecrosis, and sebaceous elements. Hematoxilin and Eosin. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Mutations
Three patients (18%) had a KRAS mutation and 1 patient (6%) had an NRAS mutation (Table 1). No patient had an HRAS, MET, PIK3CA, or BRAF mutation. We did not detect any trends between the presence of an oncogenic mutation or specific oncogenic mutation and AJCC T category or patient outcome.
Genome-Wide RNA Sequencing Analysis
RNA-Seq results were generated for all 17 EMC patient samples and 6 normal salivary tissue samples.
Statistical correlation analysis of the resulting data allowed continuing with a comparative analysis of 6 normal samples versus 17 neoplastic samples. Using strict and conservative criteria, we identified 220 significantly differentially expressed transcripts (at FDR<3.8 E −3 based on P-values<3.4 E −5), of which 36% were upregulated and 64% downregulated. The transcripts were annotated using the UCSC Genome Browser (http://genome.uscs.edu), and the NCBI Entrez Gene data base (http://www.ncbi.nlm.nih.gov/gene) and further analyzed using the IPA program (http://www.ingenuity.com/).
From these differentially expressed transcripts, the IPA identified 26 cancer-related transcripts (ACAT1, ARPC1A, ATP5J, CANX, DDX39B, DDOST, EXT1, FGFR1, GOLPH3, MAGT1, MAPK8IP3, MAT2A, MMADHC, P4HB, PDK4, RHOA, SCARB2, SDHB, SDHD, SEC63, SSR1, SSR3, TM9SF2, TMED2, TRAM1, ZMPSTE24) and 16 transcripts related to mitochondrial dysfunction overlapping with three cancer genes (ATP5A1, ATP5F1, ATP5J, COX7B, COX7C, MT-ND5, NCSTN, NDUFA4, NDUFA5, NDU-FAB1, NDUFB11, NDUFV2, PDHA1, PRDX3, SDHB, SDHD).
For other strongly differentially expressed transcripts of microRNAs and certain genes the biological functions and their role in this cancer are still not clear.
For example, two micro RNAs were detected as significantly strongly upregulated, MIR663A (miR-663) at a log2-fold change of 2.8 and MIR3648 (miR-3648) at the high log2-fold change of 4.4. miR-663 is known to be associated with cellular senescence, immunity, and cancer (Yi et al. 2012). miR-663 can function as tumor suppressor, for example, downregulated in gastric cancer (Pan et al. 2010) as well as an oncogene, for example, silencing p21 to promote G1/S transition for proliferation and tumorigenesis of nasopharyngeal carcinoma (Yi et al. 2012), and as oncogene for chemotherapy resistance in other kinds of cancer from prostate, ovarian to breast cancer (Hu et al. 2013; Jiao et al. 2014; Kim et al. 2014). The role that miR-3648 may play in cancer is yet unknown. miR-3648 was described in solid tumors (Meiri et al. 2010), and it may be processed differently to suppress the expression of different target genes (Marco et al. 2012). This opens up the possibility of miR-3648 exerting different functions in different kinds of cancer, or exerting different functions in the same tumor tissue thus causing, or contributing to tumor heterogeneity and plasticity.
Supporting Information with detailed gene annotations are available.
Figures 2 and 3 show panels based on the top 100 most strongly differentially expressed genes and gene isoforms in the form of heatmaps and dendrograms. The similarities were calculated as average linkage clustering, based on the Euclidean distance of the samples comparing the top 100 gene/isoform expressions with each other.
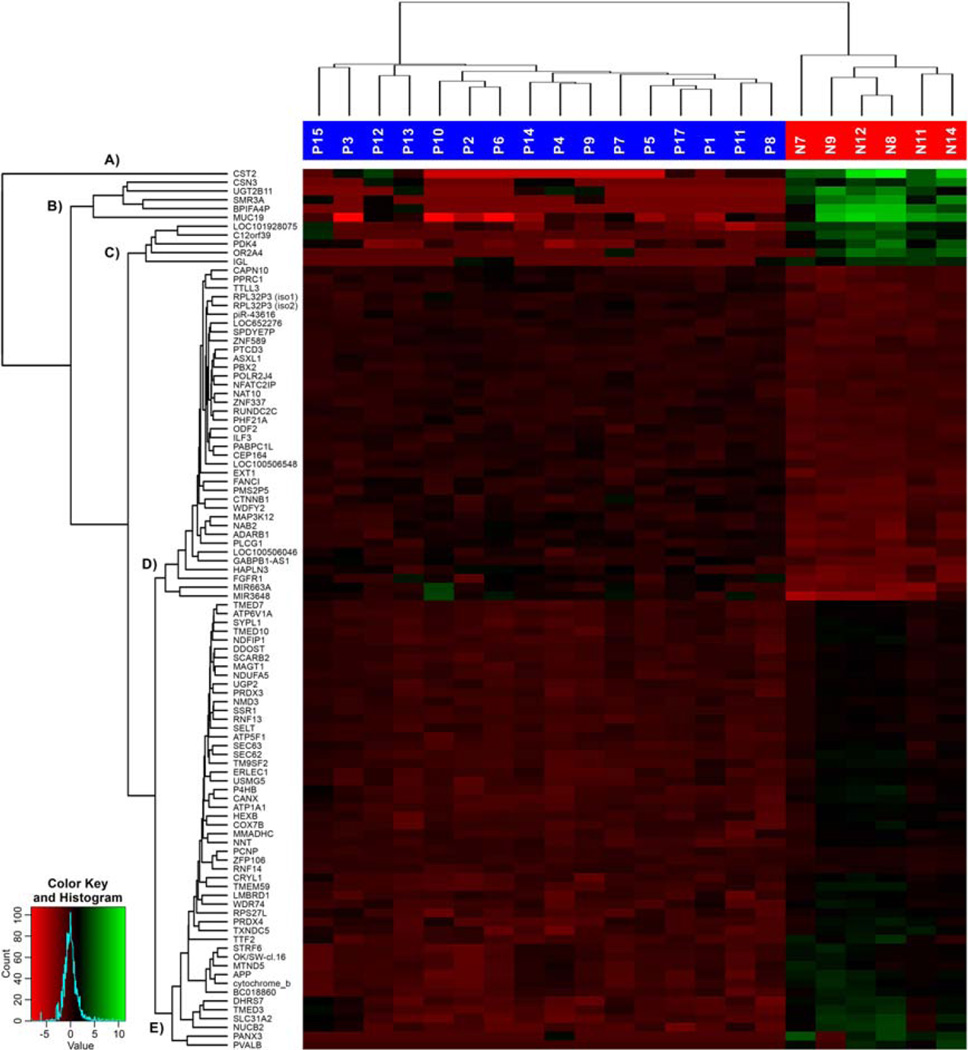
Figure 2.
Heatmap of top 100 highest differentially expressed genes with sample and gene dendrograms, showing five major gene cluster groups labeled A to E. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
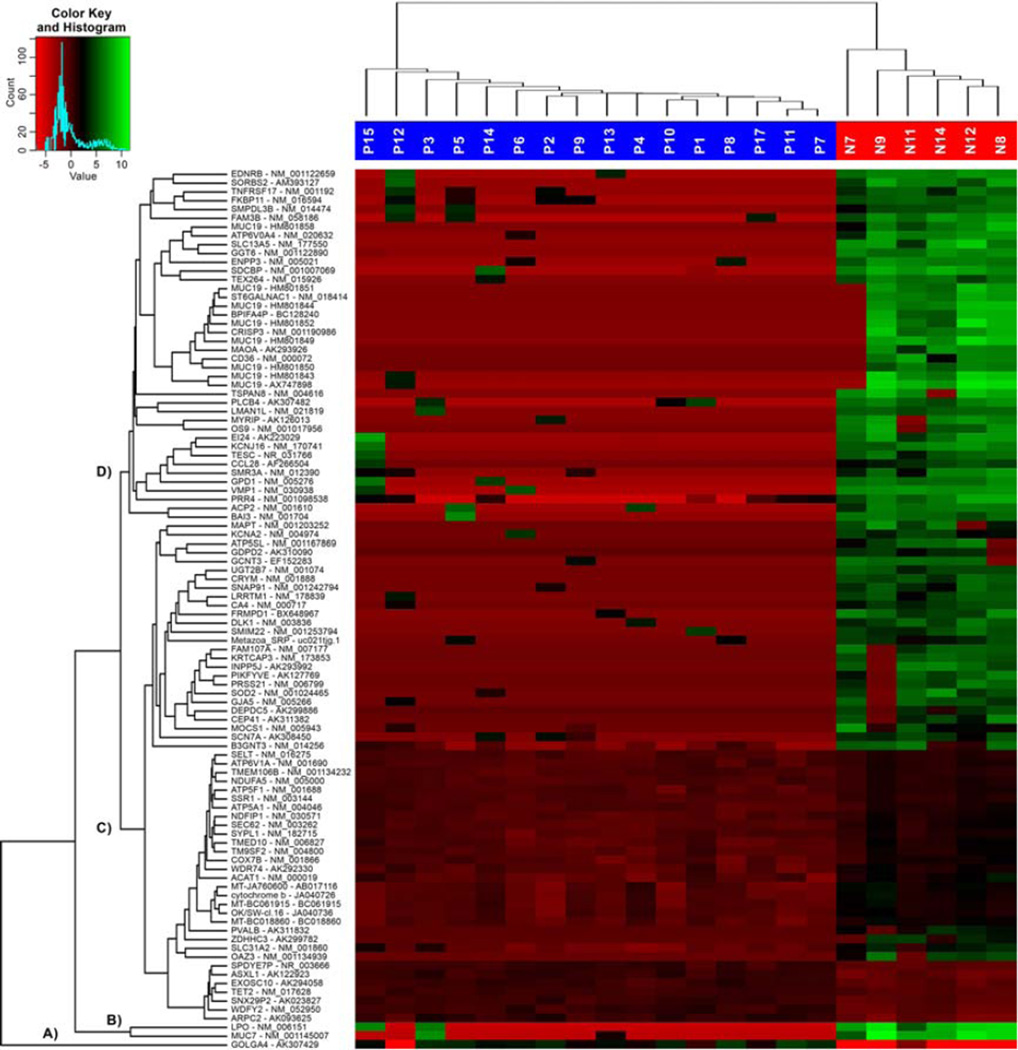
Figure 3.
Heatmap of top 100 highest differentially transcribed gene isoforms with sample and gene isoform dendrograms, showing four major gene cluster groups labeled A–D. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
A heatmap of the top 100 most strongly differentially expressed genes, with sample and gene-dendrograms are illustrated in Figure 2. Their gene hierarchical clustering dendrogram shows five major gene groups: group (A) only including one individual gene (CST2), and four groups (B– E) of gene clusters of different sizes.
Figure 3 depicts the top 100 most strongly differentially transcribed gene isoforms presented as a heat-map, with sample and gene-isoform dendrograms. The isoform hierarchical clustering dendrogram shows four major transcribed isoform groups: (A) one single transcribed isoform (GOLGA4), (B) two separately transcribed isoforms (MUC7, LPO), (C) one iso-form cluster with 4 subgroups, and (D) one isoform cluster with 5 subgroups with high similarities.
The differences in the detection of the top 100 most strongly differentially expressed genes and the top 100 differentially expressed gene isoforms are based on the following facts. The RSEM method uses an isoform abundance method which models the read generation from the isoforms and estimates the isoform abundance based on the observed reads. This limits isoform detection to alignment to known isoforms and does not discover new isoforms or isoform switching (Rehrauer et al. 2013).
For many genes, the RSEM isoform analysis produces strongly differentially expressed gene isoforms since it separates the highly transcribed isoforms from the low transcribed isoforms of the same gene, whereas the RSEM gene analysis uses all the expression values of highly and low expressed isoforms of one gene to calculate its gene expression value.
Therefore, the isoform analysis detects certain genes which might not show up in the gene analysis because in the gene analysis the high expression of one gene isoform can be strongly decreased by the low expressions of multiple isoforms of the same gene.
Biomarker Analysis
To validate the gene expression patterns detected by RNA-seq analysis, protein expression of five selected genes was characterized by immunohistochemistry (IHC).
The five genes were selected as representatives of four of the five major gene groups found in the cluster analysis of the top 100 most strongly differentially expressed genes (Fig. 2).
Their selection additionally depended on the availability of gene annotations or literature corroborating a potential involvement or causality in EMC (see table “Prot-Cluster 1–4, Genes selected for Test” in Supporting Information), and on the availability of high-quality monoclonal antibodies suited for IHC of FFPE tissue slides.
The five antibodies were selected to detect the protein expression of CST2, BPIFA4P (BASE), PDK4, PLCG1, and FGFR1. The log2-fold changes of the corresponding genes were: −11.6, −9.8, −4.6, 3.4, and 3.3.
CST2 expression was negative in 12 of 16 tumors (one sample was not evaluable because it did not withstand processing) but positive in all 6 normal salivary parenchyma samples (confined to the ducts, while acini were devoid of expression). PDK4 expression was positive in three samples within both epithelial and myoepithelial cells, while variable weak to negative in the remainder. Normal salivary parenchyma expression pattern was similar to CST2. Eight of fourteen tumors were positive within epithelial/ductal component for BASE (and weak to negative expression of myoepithelial outer cells); similar expression levels and findings were seen with PLCG1 (11 of 15 tumors positivity mainly confined to ductal cells). FGFR1 was found to be expressed in 12 of 14 tumors while weaker detection seen in normal salivary tissue (Fig. 4). Detailed analysis of tissue samples and cell-type immunoreactivity with various biomarkers is shown in Table 2 (E-epithelial cells, M-myoepithelial cells).
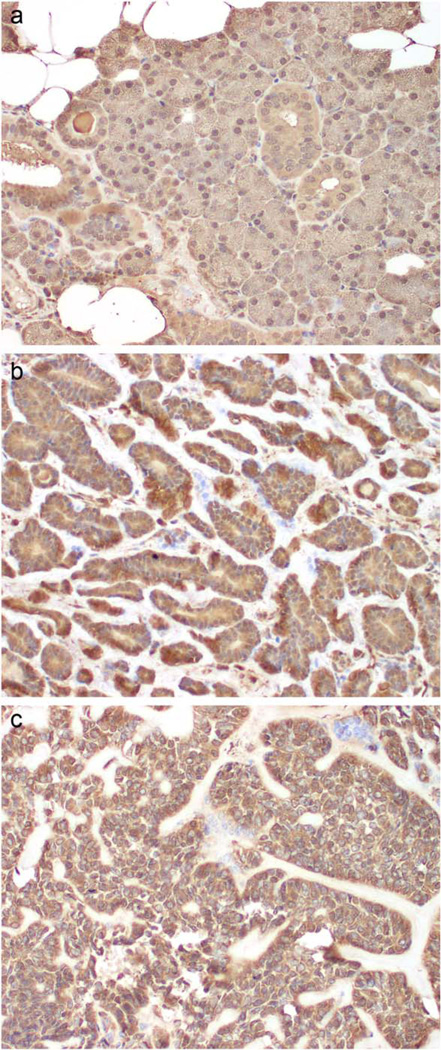
Figure 4.
Expression of FGFR1 in tumor tissue from EMC patients. IHC was performed as described for tumor tissue (b, c) and normal salivary tissue (a). Brown staining indicates immunoreactivity (weak in normal, strong expression in tumor). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
TABLE 2.
Detailed Analysis of Tissue Samples and Cell-Type Immunoreactivity with Various Biomarkers (E-Epithelial Cells, M-Myoepithelial Cells)
| CST2 | FGFR1 | PDK4 | PLCG1 | BASE | |
|---|---|---|---|---|---|
| P1 | Positive (E, M) | Positive (E, M) | positive (E, M) | Positive (E, M) | Positive (E, M) |
| P2 | Negative | Positive (E, M) | positive (E) | Positive (E, M) | Positive (E) |
| P3 | n/a | n/a | n/a | n/a | n/a |
| P4 | Negative | Positive (E, M) | Positive (E) | Positive (E, M) | Positive (E, M) |
| P5 | Negative | Positive (E, M) | Positive (E, M) | Positive (E, M) | Positive (E) |
| P6 | Negative | n/a | n/a | Positive (E, M) | Positive (E) |
| P7 | Negative | Positive (E, M) | Positive (E) | Positive (E, M) | Positive (E) |
| P8 | Negative | Positive (E, M) | Positive (E, M) | Positive (E, M) | Negative |
| P9 | Negative | Positive (E, M) | Positive (E, M) | Positive (E, M) | Positive (E, M) |
| P10 | Positive (E, M) | ||||
| P11 | Negative | Positive (M) | Positive (E, M) | Positive (M) | Negative |
| P12 | Positive | Positive (E, M) | Positive (E) | Positive (E, M) | Positive (E, M) |
| P13 | Negative | Positive (E, M) | Positive (E) | Positive (E, M) | Negative |
| P14 | Negative | Positive (E, M) | Positive (E) | Positive (E, M) | Positive (E, M) |
| P15 | Negative | Positive (E, M) | Positive (E) | Positive (E, M) | Positive (E) |
| P16 | Negative | Positive (E, M) | Positive (E) | Positive (E, M) | Positive (E, M) |
| P17 | Positive | Positive (E, M) | Positive (E) | Positive (E, M) | Positive (E) |
| N1 | Positive | Positive | Positive | Positive | Positive |
| N2 | Positive | Positive | Positive | Positive | Positive |
| N3 | Positive | Positive | Positive | Positive | Positive |
| N4 | Positive | Positive | Positive | Positive | Positive |
Pathway Analysis
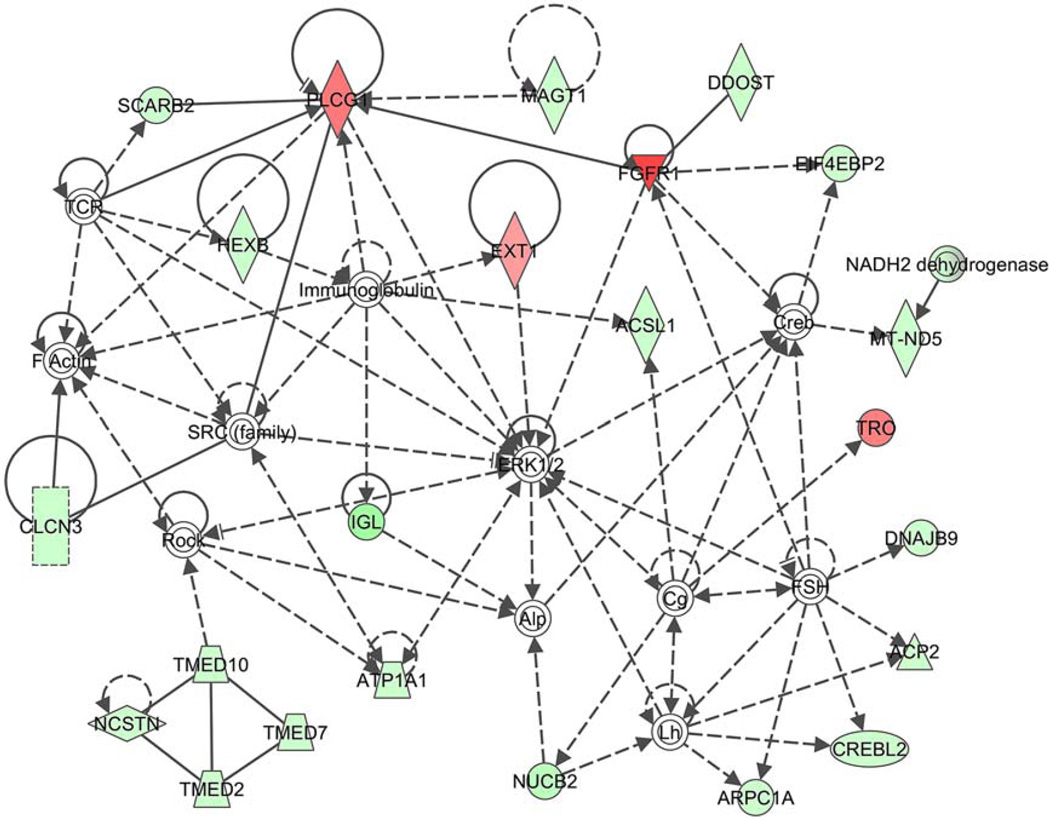
Figure 5 depicts the pathway network created by the IPA program of differentially expressed RNAs related to cell morphology, nervous system development and function, and lipid metabolism. FGFR1 was consistently upregulated in neoplastic tissue.
Figure 5.
Pathway network created by the IPA program, depicts genes of differentially expressed RNA, related to cell morphology, nervous system development and function, and lipid metabolism. FGFR1 shows consistent upregulation in neoplastic tissue (red triangle). Green, downregulated; red, upregulated. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Sample Cluster Analysis
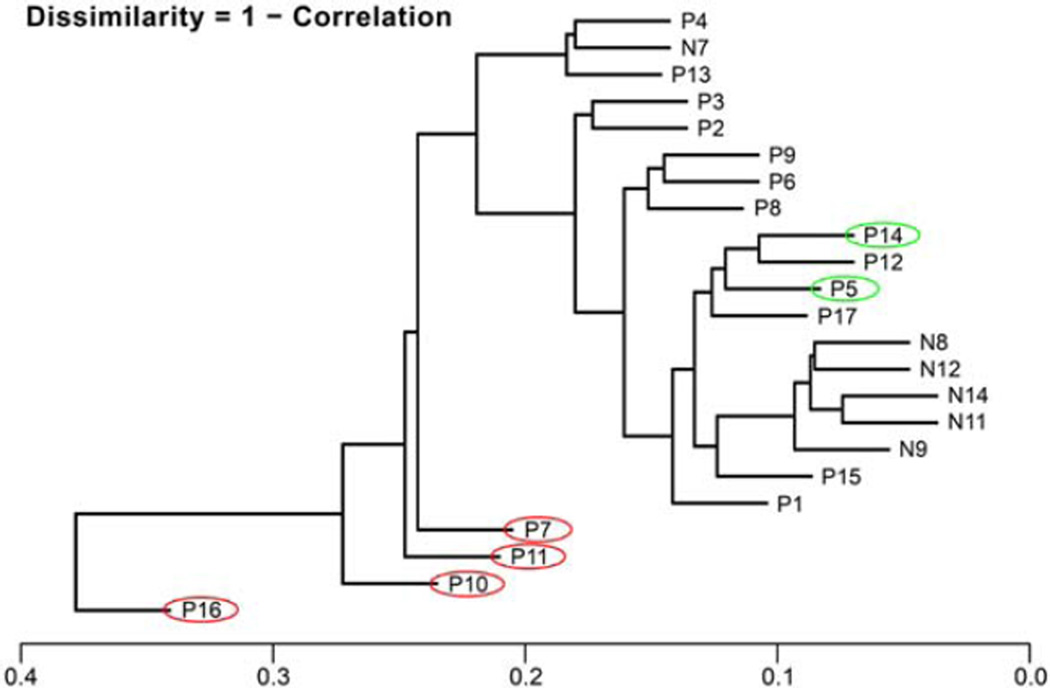
The cluster dendrogram in Figure 6 shows the dissimilarities between neoplastic and normal samples. It is calculated as Dissimilarities 5 1– Correlation from neoplastic samples versus normal samples, based on the log values of all expressed RNA. Clustering of samples P7, P10, P11, P16 with clinical follow up reported as dead of disease (labeled with red circles); P5 and P14 both alive with recurrences (labeled with green circles). Normal salivary tissue clusters as N11, N14, N8, N12, N9, while N7 is an outlier, grouped with P4 and P13.
Figure 6.
The cluster dendrogram shows the dissimilarities between neoplastic samples compared to normal samples (red circles indicate patients DOD, and green circles indicate patients with recurrent disease and alive). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
We have demonstrated that RNA-Seq analysis of FFPE salivary tissue is feasible and can provide insights into salivary cancer progression. There is a great need for robust biomarkers to predict which tumors are more likely to result in specific clinical outcomes to optimize treatment decisions. Biomarker studies in salivary cancer in general (and in particular in EMC) have faced a challenge in which the number of specimens available, associated with very long-term follow-up (>8 years) is extremely limited.
To address this need, we used the Sequenome platform to profile 190 common oncogenic point mutations in 50 genes and identified KRAS mutations in 3/17 and NRAS mutations in one case, with the possibility of targeting EGFR-RAS-RAF signaling cascade. Our findings are at variance with a recent study with 15 salivary EMC samples, where the HRAS exon 3 codon 61 mutation was the main mutation 29%, 4 of 14 tumors. In this group, there were no significant differences in mutation-positive and mutation-negative cases by age, gender, or clinical outcome. A high-throughput molecular screening of 107 malignant salivary tumors reports HRAS mutations in 7% (7/107), particularly tumors with myoepithelial component. RNA-seq as a platform of analysis has a better sensitivity and dynamic range over traditional microarrays and additionally enables identification of novel transcripts as well as more detailed studies of biological pathways in EMC.
Of the 220 biomarker genes, at least 26 have been previously associated with some type of cancer (Supporting Information Table 3). There are several biologic functions that can be used to functionally categorize these biomarker genes (Supporting Information Table 3).
CST2 (cystatin 2) gene encodes a secreted thiol protease inhibitor found at high levels in saliva, tears, seminal fluid, and plasma. Cystatin SA1 and SA2 adhere to human fibroblasts, which results in thyrosine phosphorylation and upregulation of the released IL-6 mediated by the enhancement of NF-kappa B activity. Thus CST2 probably functions as a cytokine activator causing an IL-6 pro-inflammatory response (Kato et al. 2004).
BPIFA4P (BPI fold containing family A, member 4 pseudogene) also known as breast cancer and salivary gland expression (BASE) gene is repressed by estrogen via estrogen receptor alpha (ESR), and it may function as a breast cancer marker (Egland et al. 2003; Bretschneider et al. 2008). Tissue distribution of BASE and its expression in cancer cell lines is described (Egland et al. 2003).
PDK4 (pyruvate dehydrogenase kinase isozyme 4) gene is a member of the PDK/BCKDK protein kinase family and encodes a mitochondrial protein with a histidine kinase domain. This protein is located in the matrix of the mitochondria and inhibits the pyruvate dehydrogenase complex by phosphorylating one of its subunits, thereby contributing to the regulation of glucose metabolism. PDK4 is downregulated in colonic adenocarcinoma (Blouin et al. 2011).
PLCG1 (phospholipase C gamma 1 catalyses the hydrolysis of phosphatidylinositol 4,5-biphosphate into inositol 1,4,5-triphosphate (IP3) and diacylglycerol. This reaction uses calcium as a cofactor and plays an important role in the intra-cellular transduction of receptor-mediated tyrosine kinase activators. High expression of PLCG1, and its activated form, is associated with a worse clinical outcome in terms of incidence of distant metastases in breast cancer. PLCG1 expression was increased in colon cancer cells (Reid et al. 2009) and described in the interplay of HER2/HER3/PI3K and EGFR/HER2/PLCG1 signaling in breast cancer cell migration and dissemination (Uhlmann et al. 2010; Balz et al. 2012).
FGFR1 (fibroblast growth factor receptor 1), the protein encoded by this gene is a member of the fibroblast growth factor receptor family, where the amino acid sequence is highly conserved between members and throughout evolution. The extracellular portion of FGFR1 protein interacts with fibroblast growth factors (FGFs), setting in motion a cascade of downstream signals, ultimately influencing mitogenesis and differentiation. FGFR1 gene amplification is a common genetic event occurring at a frequency of 16% in lung squamous cell cancer; lymph node metastases derived from FGFR1 amplified lung squamous cancer also exhibit FGFR1 amplification (Azuma et al. 2014). Novel FGFR inhibitor ponatinib suppresses the growth of nonsmall cell lung cancer overexpressing FGFR1. A high frequency of FGFR1 gene amplification in oral tongue squamous cell carcinoma and association with clinical features and patient outcome was shown (Freier et al. 2007). Also, amplification/upregulation of FGFR1 is associated with gastric cancer (Murase et al. 2014).
An upregulation of FGFR1 in EMC was found in our RNA-Seq experiment and at the protein level by IHC. Additionally, our mutation study shows that there were no somatic mutations of the FGFR1 gene in EMC.
Based on these results, we assume that the increased FGFR1 expression either is caused by an increased transcriptional activation, activating epigenetic changes, like CpG-demethylation, or by post-transcriptional activation, for example, by a decrease in a microRNA silencing of FGFR1.
Although our study was done on a small cohort of patients, the dendrogram of our sample cluster analysis exhibits correlation from neoplastic samples versus normal samples. Based on the log values of all expressed RNA (Fig. 6), patients with similar clinical outcome grouped together.
FFPE specimens with long-term follow-up clinical data are much more abundant than frozen tissue, and sequence analysis of this resource opens up this large resource for further analysis. To date, very few studies have reported RNA-seq analysis of FFPE tissues with the notable exception of breast and prostate cancer (Horvath et al. 2013; Long et al. 2014). Development of biomarkers from FFPE tissues also increases the likelihood that they can be translated into clinically useful biomarkers with direct application into traditional clinical practice that may not have access to frozen specimens. However, challenges remain in translating an RNA-seq biomarker test to the clinic without additional validation using alternative platforms. Therefore, further validation using TaqMan, NanoString, Digital droplet PCR, Bio-Mark PCR, or gene-focused RNA-seq methods such as PCR-amplicon and capture-sequencing in a Clinical Laboratory Improvement Amendments laboratory environment is important before developing a biomarker panel into a clinical test. This offers double advantage as simplify the assay and provide independent validation that data from RNAseq of FFPE samples are sufficiently robust across technologies.
Supplementary Material
Acknowledgments
Supported by: MD Anderson Cancer Center start-up funds (DB).
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Azuma K, Kawahara A, Sonoda K, Nakashima K, Tashiro K, Watari K, Izumi H, Kage M, Kuwano M, Ono M, Hoshino T. FGFR1 activation is an escape mechanism in human lung cancer cells resistant to afatinib, a pan-EGFR family kinase inhibitor. Oncotarget. 2014;5:5908–5919. doi: 10.18632/oncotarget.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AR, Ohanessian SE, Adil E, Crist HS, Goldenberg D, Mani H. Dedifferentiated epithelial-myoepithelial carcinoma: Analysis of a rare entity based on a case report and literature review. Int J Surg Pathol. 2013;21:514–519. doi: 10.1177/1066896912468153. [DOI] [PubMed] [Google Scholar]
- Balz LM, Bartkowiak K, Andreas A, Pantel K, Niggemann B, Zanker KS, Brandt BH, Dittmar T. The interplay of HER2/HER3/PI3K and EGFR/HER2/PLC-gamma1 signalling in breast cancer cell migration and dissemination. J Pathol. 2012;227:234–244. doi: 10.1002/path.3991. [DOI] [PubMed] [Google Scholar]
- Blouin JM, Penot G, Collinet M, Nacfer M, Forest C, Laurent-Puig P, Coumoul X, Barouki R, Benelli C, Bortoli S. Butyrate elicits a metabolic switch in human colon cancer cells by targeting the pyruvate dehydrogenase complex. Int J Cancer. 2011;128:2591–2601. doi: 10.1002/ijc.25599. [DOI] [PubMed] [Google Scholar]
- Bretschneider N, Brand H, Miller N, Lowery AJ, Kerin MJ, Gannon F, Denger S. Estrogen induces repression of the breast cancer and salivary gland expression gene in an estrogen receptor alpha-dependent manner. Cancer Res. 2008;68:106–114. doi: 10.1158/0008-5472.CAN-07-5647. [DOI] [PubMed] [Google Scholar]
- Brocheriou C, Auriol M, de Roquancourt A, Gaulard P, Fornes P. [Epithelial-myoepithelial carcinoma of the salivary glands. Study of 15 cases and review of the literature] Ann Pathol. 1991;11:316–325. [PubMed] [Google Scholar]
- Chiosea SI, Miller M, Seethala RR. HRAS mutations in epithelial-myoepithelial carcinoma. Head Neck Pathol. 2014;8:146–150. doi: 10.1007/s12105-013-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros J, Sbidian E, Hans S, Roussel H, Scotte F, Tartour E, Brasnu D, Laurent-Puig P, Bruneval P, Blons H, Badoual C. Expression and mutational status of treatment-relevant targets and key oncogenes in 123 malignant salivary gland tumours. Ann Oncol. 2013;24:2624–2629. doi: 10.1093/annonc/mdt338. [DOI] [PubMed] [Google Scholar]
- Donath K, Seifert G, Schmitz R. [Diagnosis and ultrastruc-ture of the tubular carcinoma of salivary gland ducts. Epithelial-myoepithelial carcinoma of the intercalated ducts] Virchows Arch A Pathol Pathol Anat. 1972;356:16–31. [PubMed] [Google Scholar]
- Egland KA, Vincent JJ, Strausberg R, Lee B, Pastan I. Discovery of the breast cancer gene BASE using a molecular approach to enrich for genes encoding membrane and secreted proteins. Proc Natl Acad Sci USA. 2003;100:1099–1104. doi: 10.1073/pnas.0337425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier K, Schwaenen C, Sticht C, Flechtenmacher C, Muhling J, Hofele C, Radlwimmer B, Lichter P, Joos S. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC) Oral Oncol. 2007;43:60–66. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Horvath A, Pakala SB, Mudvari P, Reddy SD, Ohshiro K, Casimiro S, Pires R, Fuqua SA, Toi M, Costa L, Nair SS, Sukumar S, Kumar R. Novel insights into breast cancer genetic variance through RNA sequencing. Sci Rep. 2013;3:2256. doi: 10.1038/srep02256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, Yao H, Song E, Chen Y, Wang M, Lin L. The overexpression of hypomethylated miR-663 induces chemotherapy resistance in human breast cancer cells by targeting heparin sulfate proteoglycan 2 (HSPG2) J Biol Chem. 2013;288:10973–10985. doi: 10.1074/jbc.M112.434340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, Deng Z, Xu C, Yu Y, Li Y, Yang C, Chen J, Liu Z, Huang G, Li LC, Sun Y. miR-663 induces castration-resistant prostate cancer transformation and predicts clinical recurrence. J Cell Physiol. 2014;229:834–844. doi: 10.1002/jcp.24510. [DOI] [PubMed] [Google Scholar]
- Kato T, Ito T, Imatani T, Minaguchi K, Saitoh E, Okuda K. Cystatin SA, a cysteine proteinase inhibitor, induces interferon-gamma expression in CD4-positive T cells. Biol Chem. 2004;385:419–422. doi: 10.1515/BC.2004.047. [DOI] [PubMed] [Google Scholar]
- Kim YW, Kim EY, Jeon D, Liu JL, Kim HS, Choi JW, Ahn WS. Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug Des Dev Ther. 2014;8:293–314. doi: 10.2147/DDDT.S51969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Xu J, Osunkoya AO, Sannigrahi S, Johnson BA, Zhou W, Gillespie T, Park JY, Nam RK, Sugar L, Stanimirovic A, Seth AK, Petros JA, Moreno CS. Global transcriptome analysis of formalin-fixed prostate cancer specimens identifies bio-markers of disease recurrence. Cancer Res. 2014;74:3228–3237. doi: 10.1158/0008-5472.CAN-13-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Macpherson JI, Ronshaugen M, Griffiths-Jones S. MicroRNAs from the same precursor have different targeting properties. Silence. 2012;3:8. doi: 10.1186/1758-907X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri E, Levy A, Benjamin H, Ben-David M, Cohen L, Dov A, Dromi N, Elyakim E, Yerushalmi N, Zion O, Lithwich-Yanai G, Sitbon E. Discovery of microRNAs and other small RNAs in solid tumors. Nucl Acids Res. 2010;38:6234–6246. doi: 10.1093/nar/gkq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase H, Inokuchi M, Takagi Y, Kato K, Kojima K, Sugihara K. Prognostic significance of the co-overexpression of fibro-blast growth factor receptors 1, 2 and 4 in gastric cancer. Mol Clin Oncol. 2014;2:509–517. doi: 10.3892/mco.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L, Sun L, Liu J, Yang Z, Ran Y. Tumor-suppressive mir-663 gene induces mitotic catastrophe growth arrest in human gastric cancer cells. Oncol Rep. 2010;24:105–112. doi: 10.3892/or_00000834. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrauer H, Opitz L, Tan G, Sieverling L, Schlapbach R. Blind spots of quantitative RNA-seq: The limits for assessing abundance, differential expression, and isoform switching. BMC Bioinf. 2013;14:370. doi: 10.1186/1471-2105-14-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JF, Gariboldi M, Sokolova V, Capobianco P, Lampis A, Perrone F, Signoroni S, Costa A, Leo E, Pilotti S, Pierotti MA. Integrative approach for prioritizing cancer genes in sporadic colon cancer. Genes Chromosomes Cancer. 2009;48:953–962. doi: 10.1002/gcc.20697. [DOI] [PubMed] [Google Scholar]
- Seethala RR, Barnes EL, Hunt JL. Epithelial-myoepithelial carcinoma: A review of the clinicopathologic spectrum and immunophenotypic characteristics in 61 tumors of the salivary glands and upper aerodigestive tract. Am J Surg Pathol. 2007;31:44–57. doi: 10.1097/01.pas.0000213314.74423.d8. [DOI] [PubMed] [Google Scholar]
- Tralongo V, Daniele E. Epithelial-myoepithelial carcinoma of the salivary glands: a review of literature. Anticancer Res. 1998;18:603–608. [PubMed] [Google Scholar]
- Uhlmann S, Zhang JD, Schwager A, Mannsperger H, Riazalhosseini Y, Burmester S, Ward A, Korf U, Wiemann S, Sahin O. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29:4297–4306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB. Principles of transcriptome analysis and gene expression quantification: An RNA-seq tutorial. Mol Ecol Resour. 2013;13:559–572. doi: 10.1111/1755-0998.12109. [DOI] [PubMed] [Google Scholar]
- Yi C, Wang Q, Wang L, Huang Y, Li L, Liu L, Zhou X, Xie G, Kang T, Wang H, Zeng M, Ma J, Zeng Y, Yun JP. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31:4421–4433. doi: 10.1038/onc.2011.629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.