Abstract
Clustered regularly interspersed short palindromic repeats (CRISPR) interference (CRISPRi) is a powerful technology for sequence-specifically repressing gene expression in bacterial cells. CRISPRi requires only a single protein and a custom-designed guide RNA for specific gene targeting. In Escherichia coli, CRISPRi repression efficiency is high (~300-fold), and there are no observable off-target effects. The method can be scaled up as a general strategy for the repression of many genes simultaneously using multiple designed guide RNAs. Here we provide a protocol for efficient guide RNA design, cloning, and assay of the CRISPRi system in E. coli. In principle, this protocol can be used to construct CRISPRi systems for gene repression in other species of bacteria.
Keywords: CRISPRi, dCas9, sgRNA, Escherichia coli
1 Introduction
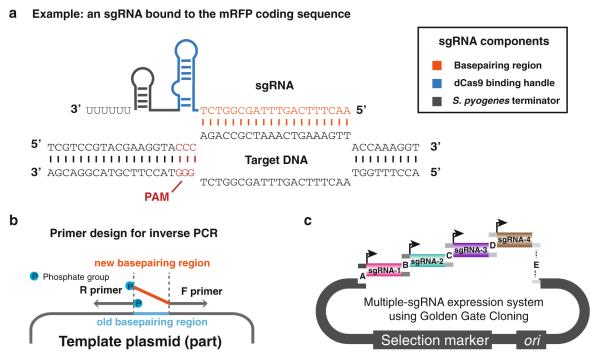
About 40 % of bacteria and 90 % of archaea possess an endogenous clustered regularly interspersed short palindromic repeats (CRISPR) system for defense against foreign DNA elements, such as viruses, bacteriophages, and plasmids [1, 2]. CRISPR utilizes CRISPR-associated (Cas) proteins and noncoding CRISPR (cr) RNA elements to confer genetic immunity [1–4]. In type II CRISPR systems, a single endonuclease protein Cas9, in complex with the mature form of crRNA-transacting (tracr) RNA complex, binds specifically to the DNA target by sequence complementarity and induces a double-stranded break. Jinek et al. have shown that a designed chimeric single-guide (sg) RNA derived from Streptococcus pyogenes could solely direct the DNA targeting and cleavage activities of Cas9 [5]. Thus the system allows easy programming of DNA target specificity, offering a modular, efficient, and multiplexable tool for genome editing [6–8]. In addition to using CRISPR for gene editing, we recently developed a new technology, CRISPR interference (CRISPRi) [9], that repurposes the CRISPR system for use in transcription regulation. In CRISPRi, a catalytically dead variant of Cas9 (dCas9) without endonucleolytic activity is coupled with designed small guide RNAs (sgRNAs) complementary to a desired DNA target. The dCas9-sgRNA complex acts to sterically hinder transcription of the targeted DNA, causing gene repression. Because repression by CRISPRi depends on base pairing between a short segment of the sgRNA and the DNA target, new DNA targets can be specified simply by altering the sgRNA sequence. Furthermore, repression by CRISPR is titratable and reversible through use of inducible promoters for both Cas9 and sgRNA expression. The current sgRNA design is a 102-nt chimeric noncoding RNA consisting of a 20-nt base pairing region, followed by a 42-nt Cas9-binding “handle,” and a 40-nt transcription terminator (Fig. 1a). The 20-nt base pairing region can be freely modified, allowing broad flexibility in targeting genes for repression. Here, we provide guidance on sgRNA design and plasmid construction for repression of specific genes of interest [10, 11]. We also provide a protocol for the modular assembly of multiple sgRNA expression cassettes into a single vector using Golden Gate Cloning [12]. Finally, we provide methods for quantifying CRISPRi repression.
Fig. 1.
Design and cloning of sgRNAs for targeted gene repression. (a) Schematic of sgRNA binding to target DNA. The sgRNA consists of a 20-nt base-pairing region (orange), a 42-nt dCas9 binding handle (blue), and a 40-nt S. pyogenes transcription terminator (grey). The PAM is shown in red. Depicted is DNA from the coding sequence of RFP. (b) Schematic of generating vectors with new sgRNA sequences using inverse PCR. 5′ phosphorylation of oligos is indicated by blue circles. (c) Schematic of cloning multiple sgRNAs into a single vector using Golden Gate cloning
2 Materials
2.1 Materials for sgRNA Design
The genome sequence of the target bacterial strain (e.g., a FASTA reference genome).
The 102-nt sgRNA reference sequence (see below).
(Optional) An RNA secondary structure prediction algorithm such as the Vienna software suite for RNA folding analysis.
2.2 Materials for Single-sgRNA Cloning
E. coli sgRNA expression plasmid with ampicillin resistance (Addgene ID no. 44251). The sgRNA is expressed from a strong constitutive promoter J23119 (http://parts.igem.org/Part:BBa_J23119).
Designed forward sgRNA primers (e.g., 5′-N20 GTTTTAGA GCTAGAAATAGCAAGTTAAAATAAGGC-3′ for Addgene ID no. 44251, N20 is the sgRNA base pairing region).
Universal reverse primer (e.g., 5′-ACTAGT ATTATACCTAGGACTGAGCTAGC-3′ for Addgene ID no. 44251).
10 mM ATP (NEB).
T4 Polynucleotide Kinase Kit (NEB).
2× Phusion PCR Master Mix (NEB).
QiaQuick PCR Purification Kit (Qiagen).
DpnI Kit (NEB).
QiaQuick Gel Extraction Kit (Qiagen).
Quick Ligase Kit (NEB).
One Shot TOP10 chemically competent E. coli cells (Life Technologies).
LB plates with 100 μg/mL ampicillin.
Ampicillin (Sigma).
TYGPN broth (can be purchased from Amresco) (see Note 1).
Breatheable Sealing Films (e.g., AeraSeal Breatheable Sealing Films; Excel Scientific).
QiaPrep 96 Turbo Miniprep Kit (Qiagen).
QiaVac 96 vacuum manifold (Qiagen).
2.3 Cloning Multi-sgRNA Constructs for Multiplex CRISPRi
T4 DNA Ligase Kit, 400,000 units/mL (NEB).
BsaI (NEB).
2× Taq Master Mix (NEB).
2.4 Materials for CRISPRi Repression Assays
E. coli test strain (e.g., K12-strain MG1655).
E. coli dCas9 expression plasmid with chloramphenicol resistance (Addgene ID no. 44249). The dcas9 gene is under the control of an anhydrotetracycline-inducible promoter PLTetO-1.
LB plates with 100 μg/mL ampicillin and 20 μg/mL chloramphenicol.
Chloramphenicol (Sigma).
Anhydrotetracycline (Clontech).
RNeasy RNA Purification Kit (Qiagen).
DNA-free Kit (Ambion).
Superscript III First Strand Synthesis System (Life Technologies).
Random Hexamers (Life Technologies).
qPCR primers.
Brilliant II SYBR Green Master Mix (Agilent).
3 Methods
3.1 sgRNA Design
The CRISPRi system requires two components, the dCas9 protein and a sequence-specific sgRNA. The key to successful gene repression is to custom design an appropriate sgRNA for the gene target of interest. The main body of the sgRNA, the Cas9 handle and transcriptional terminator, remains constant for different DNA targets. The region that must be designed specifically for each DNA target is a 20-nt base pairing region of the sgRNA (see Note 2).
Example: Design an sgRNA target to repress mRFP [13] expressed in E. coli
There are four primary constraints to consider in designing the sgRNA base pairing region:
-
Choice of target site: For better repression efficiency, the base pairing region should bind to the non-template DNA strand of the coding region for the gene to be repressed. Targeting the template DNA strand of the coding sequence is generally ineffective, at most leading to mild repression (~50 %) [9]. Choosing a target closer to the 5′ end of the gene generally results in greater repression efficiency.
Example: See Fig. 2 for initial sequence of mRFP.
PAM adjacency: In addition to the 20-bp base pairing region, the Cas9-sgRNA complex requires a juxtaposed proto-spacer adjacent motif (PAM) to bind DNA [5, 14, 15], which is specific to the Cas9 being used [16, 17]. In the case of S. pyogenes Cas9, the motif is NGG or NAG (N as any nucleotide) [6]. Thus, the targetable sites are restricted to 20-nt regions 5′ to NGG in the genome (Fig. 1a).
Example: To target a particular site on the template DNA strand, search for N20-N(G/A)G, and N20 is the sequence of the base pairing region of sgRNA; to target a site on the non-template strand, search for C(C/T)N-N20, and the reverse complementary sequence of N20 is the sequence of the sgRNA base pairing region. For example, to target the non-template DNA strand of the mRFP sequence listed above, one target site is AGACCGCTAACTGAAAGTT (in bold italic) on the non-template strand, with a PAM (CCC, in bold underline). The sgRNA base pairing sequence is thus the reverse complementary, i.e., AACTTTCAGTTTAGCGGTCT.
-
Genomic specificity: The dCas9-sgRNA complex will bind to any sufficiently similar DNA sequences that are adjacent to PAM sites. To identify such sequences, run BLAST (blastn; default settings) [18] searches with each designed sgRNA base pairing region against the complete genome of the organism in which the system will be used. Potential off-target DNA-binding sites with partial complementarity to the sgRNA should be evaluated for degree of homology; nucleotides closer to the PAM have a greater impact on relative binding affinity. One or two mismatches, particularly in the PAM-adjacent 12-nt portion of the target (the “seed” region), are sufficient to reduce binding efficiency by an order of magnitude or more [9].
Example: BLAST [19] the above sgRNA base pairing region (AACTTTCAGTTTAGCGGTCT) against the E. coli genome. There should be no exact 20-nt matches with adjacent PAM sites.
Folding quality (optional): Generate the full sgRNA sequence by appending the target DNA sequence (replacing T with U) to the 5′ end of the sgRNA sequence (5′ 20-nt base pairing reg ion + GUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGG CACCGAGUCGGUGCUUUUUUU -3′). Using a folding algorithm such as the Vienna suite [20], compare the predicted structure of the sgRNA containing the new 20-nt base pairing region to the structure of a functionally validated sgRNA. If the dCas9 binding handle is disrupted the sgRNA will bind poorly to dCas9, and repression efficiency will be reduced.
Fig. 2.
An example for designing an sgRNA that targets the mRFP gene. The PAM sequence (CCC) is underlined and bold. The base pairing sequence is in bold italics. The bottom DNA strand is the DNA transcription template strand
3.2 Single-sgRNA Cloning Using Inverse PCR
Primers for the target-specific inverse PCR (iPCR; described in [21] and Fig. 1b) are paired with a universal reverse primer to generate new sgRNA expression vectors. These vectors are transformed into a cloning strain of E. coli and then grown to a sufficient quantity for plasmid purification. The forward and reverse primers are homologous to the template vector, with the forward primer containing a 20-nt base pairing region at the 5′ end unique to each sgRNA (Fig. 1b). The forward primer anneals to the part of the vector that encodes the dcas9-binding handle, immediately downstream of the 20-nt base pairing region. The following protocol addresses parallel or pooled cloning of many vectors, each of which expresses a single sgRNA (see Note 3).
iPCR can be performed using individual primer pairs (i.e., one sgRNA product per reaction) or by pooling sets of forward primers that contain different sgRNA sequences with a universal reverse primer (i.e., numerous sgRNA products per reaction). Pooling can reduce labor and reagent consumption, but may cause bias (e.g., differential amplification of specific sgRNA plasmids) or other problems (e.g., mispriming) in the PCR reaction. To form pools, combine multiple forward primers in equal portion so that the total concentration of the primer pool is the concentration listed below. (The concentration of any individual forward primer will be 1/N times the total concentration, where N is the number of primers in the pool. The reverse primer should be used at the same concentration, as listed.) For construction of large sgRNA libraries (>96), we typically start with multiple iterations of a pooled approach followed by individual cloning of sgRNA plasmids to complete the library. For small libraries (<96), individual cloning of the entire library may be more efficient.
-
Inverse PCR requires primer phosphorylation (required for the subsequent blunt-end ligation step) prior to PCR, which can be done as a pool or as multiple sub-pools (which may reduce PCR bias). To prepare a 100-μM primer pool, mix 1 μL of individual 100 μM primers. To phosphorylate primers:
100 μM Forward primer (or primer pool) 1 μL 100 μM Universal reverse primer 1 μL 10× PNK buffer 1 μL 10 mM ATP 1 μL T4 PNK 0.5 μL ddH2O 6.5 μL Total 10 μL Incubate at 37 °C for 30 min, and then heat-inactivate the reaction at 65 °C for 20 min.
- Amplify new sgRNA vectors using iPCR:
Phosphorylation reaction 10 μL 100 ng/μl E. coli sgRNA plasmid template 0.5 μL 2× Phusion PCR Master Mix 12.5 μL ddH2O 2 μL Total 25 μL - Perform iPCR using the following thermal cycler program:
Step Temperature (°C) Duration Initial denaturation 98 30 s 25 cycles 98 10 s 62 30 s 72 1 min Final extension 72 5 min If more than one pool is used for iPCR, combine pools for the remaining steps.
Purify PCR products using the QiaQuick PCR Purification Kit (Qiagen) following the manufacturer’s instructions. Elute using 30 μL of EB.
-
Treat the purified PCR products with DpnI to degrade the PCR template plasmid.
Purified PCR products 10 μL DpnI restriction endonuclease 1.25 μL 10× CutSmart Buffer 2.5 μL ddH2O 11.25 μL Total 25 μL Incubate at 37 °C for 1 h
Run 2 μL of the DpnI-digested PCR product on a 1 % agarose gel, and then stain with 1 μg/mL ethidium bromide for 20 min. If using the E. coli sgRNA plasmid as a template (Addgene ID no. 44251), the iPCR product should be ~2,500 bp in size. Wrong PCR products may interfere with subsequent ligation or may form a circularized plasmid that can transform E. coli, which greatly reduces the cloning efficiency. To remove wrong PCR products, purify the ~2,500 bp band from a 1 % agarose gel using the QiaQuick Gel Extraction Kit (Qiagen) following the manufacturer’s instructions. Elute using 30 μL of EB.
-
Circularize the PCR products by ligating the ends:
DpnI-treated PCR product 9 μL 2× Quick Ligase Buffer 10 μL Quick Ligase 1 μL Total 20 μL Incubate at 25 °C for 5 min.
Transform 10 μL of the ligation reaction into One Shot TOP10 chemically competent E. coli cells following the manufacturer’s instructions.
Spread onto LB agar plates containing ampicillin (100 μg/mL) and incubate overnight at 37 °C.
Prepare 96-well 2 mL deep-well blocks with 1.7 mL per well of TYGPN liquid medium + 100 μg/mL ampicillin.
Use pipette tips to inoculate individual wells of a 96-well deep-well block with single colonies.
Cover the plate with an AeraSeal breatheable film. Incubate overnight at 37 °C without shaking.
Pellet cells by centrifugation at 2,000 × g on a tabletop centrifuge for 10 min.
Aspirate medium off of the cell pellets.
Prep plasmid DNA using a QIAprep 96 Turbo miniprep kit and a QIAVac 96-well vacuum manifold following the manufacturer’s instructions. Elute in 100 μL EB. Alternatively, minipreps can be performed on individual cell pellets using the QIAprep Spin Miniprep kit or an equivalent kit from another manufacturer.
3.3 Cloning Multi-sgRNA Constructs for Multiplex CRISPRi
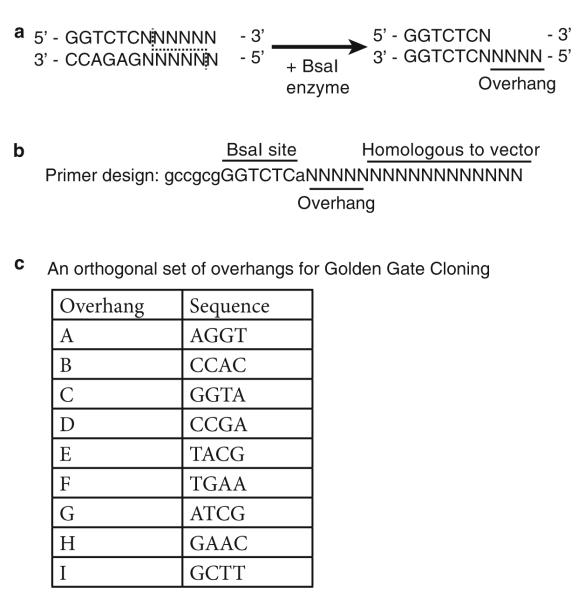
This protocol describes construction of a single vector that contains multiple sgRNAs for targeting two or more genes in E. coli using Golden Gate cloning [12], which facilitates ligation of up to ten DNA fragments in one reaction. Golden Gate cloning employs BsaI, a type IIS restriction enzyme that recognizes a sequence 5′ of the actual cut site, allowing creation of arbitrary 4-nt overhangs (Fig. 3a). Ligation of non-palindromic 4-nt overhangs allows efficient assembly of sgRNAs in a defined order (Fig. 1c). We use primers with added BsaI sites and overhangs to amplify sgRNA expression cassettes using PCR (Fig. 3b). BsaI recognition sites are removed from the sgRNA insert during digestion and thus are not present in the final plasmid. Each sgRNA expression fragment contains a constitutive promoter (promoter J23119; http://parts.igem.org/Part:BBa_J23119) that drives expression of a single sgRNA. The Golden Gate cloning procedure described below streamlines creation of multi-sgRNA vectors for multiplex CRISPRi (Fig. 1c) (see Note 4):
Fig. 3.
Primer design and overhang sequences for Golden Gate cloning. (a) BsaI is a type IIS restriction enzyme that cuts outside of the recognition site. (b) The primers (both forward and reverse) contain a BsaI site, a unique 4-nt overhang, and homologous sequence binding to the PCR vector. (c) An orthogonal set of overhangs for Golden Gate cloning. These orthogonal overhangs (A–I) prevent erroneous ligation during the ligation step
Design primers to amplify the sgRNA expression fragments with added BsaI sites and custom overhang sequences (Fig. 3c).
- PCR amplify fragments using NEB Phusion Polymerase Master Mix, with 0.5 μL of 100 μM primers:
sgRNA expression vector 0.5 μL Forward primer (100 μM) 0.5 μL Reverse primer (100 μM) 0.5 μL 2× Phusion PCR Master Mix 25 μL ddH2O 23 μL Total 50 μL Run the PCR products on 1 % agarose gel to verify that the PCR is successful and specific (no secondary bands).
PCR purification using standard protocol using, e.g., QiaQuick PCR Purification Kit.
Quantify the concentration of each purified PCR product using a Nanodrop.
- Set up the Golden Gate reaction in a PCR reaction tube:
DNA fragments 25 ng each Vector DNA 75 ng T4 DNA Ligase 15 units BsaI 10 units 10× T4 DNA Ligase Buffer 1.5 μL ddH2O Up to 15 μL - In a thermocycler, perform the program:
Step Temperature (°C) Duration (min) 50 cycles of digestion
and ligation37 10 16 10 Final digestion 50 10 Heat inactivation 80 20 Add 10 units of DpnI and incubate at 37 °C for 1 h to remove the template vector.
Directly transform ligation reactions into One Shot TOP10 chemically competent E. coli cells following the manufacturer’s instructions.
Spread the cells on LB plates with 100 μg/mL ampicillin. Grow cells overnight at 37 °C.
Inoculate and miniprep cells as described in Subheading 3.2, steps 13–17.
- While incubating the cells, perform colony PCR to validate clones prior to miniprep. Use Primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) to design PCR primers; the two primers are designed to flank the sgRNA expression cassette and generate approximately 1-kb PCR product. Primers should have annealing temperatures of 60 °C. Set up the following PCR reaction:
Cell culture 1 μL Forward colony PCR primer, 10 μM 0.5 μL Reverse colony PCR primer, 10 μM 0.5 μL Taq Master Mix, 2× 12.5 μL ddH2O 10.5 μL Total 25 μL - Perform colony PCR using the following program:
Step Temperature (°C) Duration Initial denaturation 95 30 s 30 cycles 95 20 s 55 30 s 68 1 min per kb Final extension 68 5 min Hold 10 Select positive clones for DNA miniprep.
3.4 CRISPRi Gene Repression Assay Using Quantitative PCR
If the target gene encodes a fluorescent protein, transcriptional repression using CRISPRi can be assayed using methods such as flow cytometry [9]. More generally, transcriptional repression of endogenous genes can be assayed using quantitative PCR (qPCR) (see Note 5).
Co-transform the cloned sgRNA vector with an inducible dCas9-expressing vector (e.g., Addgene cat. no. 44249) into desired E. coli strain (e.g., MG1655) for targeted gene expression.
Grow overnight without inducers at 37 °C.
Inoculate single colonies into a 2-mL deep-well 96-well plate. Each well should contain 200 μL of LB with ampicillin (100 μg/mL) and chloramphenicol (20 μg/mL) but without inducers.
Grow overnight at 37 °C at 900 RPM shaking speed in a deep-well plate shaker.
Dilute overnight cultures 1:1,000 into 3 mL LB in 18 mL glass culture tubes, supplemented with 1 μM anhydrotetracycline to induce dCas9 expression. Grow cultures for 6 h at 37 °C in a culture tube rotator.
When cells reach late-exponential phase, mix 600 μL of culture with 600 μL of −20 °C methanol in a microcentrifuge tube, and then pellet cells at maximum speed (e.g., 18,000 × g) for 1 min.
Pipette off the supernatant and then extract the RNA using Qiagen RNeasy RNA purification kit following the manufacturer’s instructions.
Following RNA isolation, remove genomic DNA using Ambion DNA-Free kit following the manufacturer’s instructions.
Arbitrarily choose six DNA-free RNA samples and add 1 μL of 100 mg/mL RNase A. These will be used to ensure that there is no contaminating DNA in the samples when the qPCR is run.
Synthesize cDNA using Invitrogen SuperScript III First-Strand Synthesis System with random hexamers following the manufacturer’s instructions. Make working stocks by diluting the cDNA 1:10.
Using Primer3 software [22], design primers with annealing temperatures of approximately 60 °C flanking a 200-bp region in the middle of the target gene. Design primers to amplify a nontargeted control gene in the genome (a gene unlikely to be perturbed by the knockdown, such as the rpoD gene that encodes the primary σ factor in E. coli) as control for qPCR reactions.
-
Set up the qPCR reaction:
Agilent Brilliant II SYBR Green qPCR 2× Master Mix 12.5 μL Each qPCR primer, 5 μM 3 μL Diluted cDNA 1 μL Nuclease-free water Up to 25 μL Include three biological replicates for each gene target.
Using a real-time thermal cycler (e.g., Stratagene Mx3005P qPCR System), perform the qPCR reaction.
Calculate the relative gene expression using the cycle threshold values (Ct). First, subtract the experimental strain’s control gene’s average Ct from the experimental strain’s target gene’s average Ct (difference called A for simplicity). Then subtract the control strain’s control gene’s average Ct from the control strain’s target gene’s average Ct (difference called B for simplicity). Finally, subtract B from A and use this difference as an exponent for 2, i.e., 2(A-B) to calculate gene expression. Errors are calculated with the formula for propagation of error [23].
Acknowledgements
We thank the Lei Qi lab, Carol Gross lab, and Wendell Lim lab for their support. J.S.H. acknowledges the support from Biophysics Graduate Program at UCSF. Spencer Wong acknowledges the support from Summer Research Training Program (SRTP) at UCSF. This work was supported by NIH P50 (grant GM081879, L.S.Q.), NIH Director’s Early Independence Award (grant OD017887, L.S.Q.), and a Ruth L. Kirschstein National Research Service Award (F32GM108222-01, J.M.P.).
Notes
TYGPN medium:
For 1 L: 750 mL ddH2O, 5 g Na2HPO4, 10 g KNO3, 20 g tryptone, 10 g yeast extract, 10.7 mL 75 % glycerol
Be sure to dissolve the salts thoroughly first and then mix in the remainder of the reagents. Autoclave for 45 min.
The final repression effect of a given sgRNA may depend on multiple factors, including sequence composition of the base-pairing region, off-target binding, target location relative to the transcription start site, and possibly even local chromosome architecture [9].
Protocol 3.2 should be preferentially performed in a single day, or samples should be stored at −20 °C and used soon after to avoid loss of phosphate groups prior to the ligation step.
For protocol 3.3, we found that DNA containing eight or more tandem sgRNA cassettes does not efficiently amplify by PCR. If desired constructs consist of eight or more sgRNA cassettes, it may be necessary to use two compatible plasmids for co-expression of multiple sgRNAs.
Repressing the first gene in an operon likely has a similar repressive effect on all downstream genes in the same operon.
References
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S-H. Inverse polymerase chain reaction. Mol Biotechnol. 1994;2:15–22. doi: 10.1007/BF02789286. [DOI] [PubMed] [Google Scholar]
- 11.Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc. 2011;6:242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- 12.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden Gate shuffling: a one-pot DNA Shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 16.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 17.Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.BLAST: Basic Local Alignment Search Tool. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 20.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA web-suite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Clifford AA. Multivariate error analysis: a handbook of error propagation and calculation in many-parameter systems. Wiley; New York: 1973. ISBN 0470160551. [Google Scholar]