Abstract
Importance
Germline mutations in BRCA1 and BRCA2 are relatively common in women with ovarian, fallopian tube, and peritoneal carcinoma (OC) causing a greatly increased lifetime risk of these cancers, but the frequency and relevance of inherited mutations in other genes is less well characterized.
Objective
To determine the frequency and importance of germline mutations in cancer-associated genes in OC.
Design
Subjects were ascertained from two phase III clinical trials in newly diagnosed advanced stage OC (GOG 218 and GOG 262), and a university-based gynecologic oncology tissue bank. Germline DNA was sequenced from women with OC using the targeted capture and multiplex sequencing assay BROCA.
Setting
Referral centers participating in NRG Oncology studies, and a University-based gynecologic oncology practice (UW).
Participants
The study population was 1915 women with OC with available germline DNA, unselected for age or family history, enrolled at the time of OC diagnosis (GOG 218, N=788; GOG 262, N=557; UW, N=570).
Main Outcomes and Measures
Mutation frequencies in OC were compared to the NHLBI GO Exome Sequencing Project (ESP) and the Exome Aggregation Consortium (ExAC). Clinical characteristics and survival were assessed by mutation status.
Results
Of 1915 subjects, 280 (15%) had mutations in BRCA1 (182), or BRCA2 (98) and 8 (0.4%) had mutations in DNA mismatch repair (MMR) genes. Mutations in BRIP1 (26), RAD51C (11), RAD51D (11), PALB2 (12) and BARD1 (4), were significantly more common in OC patients than in the ESP or ExAC, and in total were present in 3.3% of patients. Race, histologic subtype, and disease site were not predictive of mutation frequency. Mutation status affected survival, in particular for BRCA2 mutation carriers with HR 0.60 (95% CI 0.45 – 0.79, p<0.001) for progression-free survival, and HR 0.39 (95% CI 0.25 – 0.60, p<0.001) for overall survival in the GOG patients.
Conclusions and Relevance
In total, 347/1915 (18%) OC patients carried pathogenic germline mutations in genes associated with OC risk. PALB2 and BARD1 are suspected OC genes and together with established OC genes (BRCA1, BRCA2, BRIP1, RAD51C, RAD51D, MSH2, MLH1, PMS2, and MSH6) bring the total number of genes suspected to cause hereditary OC to 11.
INTRODUCTION
Ovarian carcinoma (OC) remains the deadliest gynecologic malignancy.1 Identifying genetic predisposition offers opportunities for cancer prevention. According to previous studies, 13–18% of OC is associated with germline mutations in BRCA1 and BRCA2.2–4 BRCA1 and BRCA2 mutations confer a lifetime risk of developing OC of approximately 20% (BRCA2) to 50% (BRCA1)5 and risk-reducing salpingo-oophorectomy has been shown to significantly reduce the risk of OC and all-cause mortality.6,7 Other genes from the BRCA-Fanconi anemia pathway such as BRIP1, RAD51C, and RAD51D have been implicated in hereditary OC.8–13 We previously reported the frequency of these mutations in a small (n=360) set of unselected patients with OC.4 Several publications on a recent large series of unselected OC reported lower rates of mutations in all OC genes, perhaps secondary to different sequencing methods.12–14 Mutations in other genes within this pathway (PALB2, BARD1, NBN, and CHEK2, amongst others) have been identified in OC patients, but it is unknown if these mutations confer an elevated risk of OC.4
We sought to determine the frequency of mutations in known or suspected OC susceptibility genes in a large unselected group of patients with ovarian, peritoneal, and fallopian tube carcinoma, using a comprehensive targeted sequencing method.
METHODS
Patients with primary ovarian, peritoneal, and fallopian tube carcinoma (collectively OC), were identified from three sources: 1) patients undergoing primary treatment at the University of Washington (UW) Medical Center, 2) patients consented for translational research with available DNA from Gynecologic Oncology Group (GOG) protocol 218, and 3) GOG protocol 262. Patients were enrolled at diagnosis and were not selected for age or family history. Pathology was centrally reviewed by gynecologic pathologists, and unsure cases were resolved by consensus. GOG-218 and GOG-262 were large randomized phase III trials for primary advanced stage OC. A subset of the UW patients have been previously described.4 All patients provided written informed consent on protocols approved by an Institutional Review Board.
Germline DNA extracted from blood was sequenced using BROCA, a targeted capture, massively parallel sequencing test developed at the University of Washington.15 For this study, we sequenced ATM, BARD1, BRCA1, BRCA2, BRIP1, CHEK2, FAM175A, FANCP, MLH1, MSH2, MSH6, MRE11A, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C, RAD51D, and TP53. Sequencing reads were aligned to the human reference genome (hg19). Variants were identified using GATK37 and Pindel after indel realignment and base quality recalibration. Variants including copy number variations (CNVs) were detected as previously described.15–17 Missense mutations were only included if proven to be damaging (e.g., BRCA1 C61G18). TP53 missense mutations were classified as deleterious based on available functional data as per the International Agency for Research on Cancer.19
Two publically available, overlapping, online exome sequencing datasets were utilized to estimate population mutation frequencies, the European American (EA) dataset from the NHLBI Exome Sequencing Project (ESP)20, and the Exome Aggregation Consortium (ExAC).21 To account for inaccuracy in indel calling in the ESP, we visually inspected the read data for all coding indel calls in the ESP dataset for the genes of interest. Low quality (< 17) and low coverage (< 5) calls were excluded. The EA group was used as most OC subjects were Caucasian and the total ESP population over-represents African Americans. ExAC mutation frequencies were weighted to match the racial distribution in OC cases. As these exome databases do not include CNVs, CNVs were removed from the OC frequencies for this comparison. Splice mutations were also excluded from OC and population mutation frequencies as they could not all be verified to be damaging.
Clinical information was collected as per the GOG 218 and 262 protocols, or extracted from the medical record for UW patients. Race and ethnicity (self-reported in the GOG patients, and as noted in the medical record for UW patients) were assessed to determine if they impacted mutation rates.
Contingency tables were analyzed with χ2 or Fisher’s exact test. In GOG patients, progression free survival (PFS) and overall survival (OS) were defined as the time period between enrollment and progression22 or death, respectively. Proportional hazards models were used to provide estimates of relative hazards adjusted for clinical characteristics (further description, as well as the log files of the statistical analyses are available in the supplemental materials). Wald’s test was used to assess the null hypotheses of equal hazards. All p-values are two-sided.
RESULTS
Description of study population
Clinical characteristics are provided in Table 1. The median age at diagnosis was 60 (range 28 – 91) in the UW patients and 61 (range 23 – 87) in the GOG patients. Differences between the GOG patients versus UW patients included an increased fraction of black women in the GOG (4.3% versus 1.4%, p=0.009, Fisher’s exact) and higher proportions in the UW series of fallopian tube carcinomas (13.3% versus 5.7%, p<0.001), stage I and II disease (14.6% versus 0% as GOG trials were restricted to advanced stage), and non-serous carcinomas (29.9% versus 13.1%, p<0.001).
Table 1.
Clinical Characteristics of OC Patients
| Characteristic | GOG 218 and 262 |
UW | |
|---|---|---|---|
| N | 1345 | 570 | |
| Age | <40 | 35 (2.6%) | 16 (2.8%) |
| 40 – 49 | 176 (13.1%) | 90 (15.8%) | |
| 50 – 59 | 426 (31.7%) | 165 (28.9%) | |
| 60 – 69 | 451 (33.5%) | 172 (30.2%) | |
| 70 – 79 | 229 (17.0%) | 92 (16.1%) | |
| ≥80 | 28 (2.1%) | 35 (6.1%) | |
| Race/Ethnicity | Non-Hispanic White | 1176 (87.4%) | 505 (88.6%) |
| Hispanic | 53 (4.3%) | 16 (2.8%) | |
| Non-Hispanic Black | 58 (4.3%) | 8 (1.4%) | |
| Asian/Pacific Islander | 27 (2.0%) | 11 (1.9%) | |
| Other or Unknown | 31 (2.3%) | 30 (5.3%) | |
| Disease Site | Ovary | 1076 (80.0%) | 426 (74.7%) |
| Peritoneal | 192 (14.3%) | 57 (10.0%) | |
| Fallopian Tube | 77 (5.7%) | 76 (13.3%) | |
| Ovary/Endometrial | 0 | 11 (1.9%) | |
| Stagea | Stage I | 0 | 41 (7.2%) |
| Stage II | 0 | 42 (7.4%) | |
| Stage III | 977 (72.6%) | 380 (66.7%) | |
| Stage IV | 365 (27.1%) | 107 (18.8%) | |
| Histology | High-Grade Serous | 1,118 (83.1%) | 380 (66.7%) |
| Low-Grade Serous | 51 (3.8%) | 19 (3.3%) | |
| Carcinoma, NOS | 84 (6.2%) | 81 (14.2%) | |
| Low-Grade Endometrioid | 4 (0.3%) | 9 (1.6%) | |
| High-Grade Endometrioid | 38 (2.8%) | 26 (4.6%) | |
| Clear Cell | 30 (2.2%) | 28 (4.9%) | |
| Carcinosarcoma | 3 (0.2%) | 19 (3.3%) | |
| Mucinous | 9 (0.7%) | 7 (1.2%) | |
| Transitional Cell | 8 (0.6%) | 1 (0.2%) |
Stage was not available for 3 GOG patients.
Frequency of mutations in BRCA1, BRCA2, and mismatch-repair genes
Of 1,915 patients, 182 (9.5%) had mutations in BRCA1 and 98 (5.1%) had mutations in BRCA2. Of BRCA1 and BRCA2 mutations, 38/280 (13.6%) were in Ashkenazi Jewish founder mutations, BRCA1 185delAG (c.68_69delAG) (18) and 5382insC (c.5266dupC) (14) and BRCA2 6174delT (c.5946delT) (6). 16/182 (8.8%) BRCA1 mutations were large genomic duplications or deletions, also called copy number variants (CNVs). 8/1915 (0.4%) carried mutations in genes involved in mismatch repair (MMR), including 4 in PMS2, 3 in MSH6, and 1 in MLH1. There were no mutations found in MSH2. Mutation frequencies for BRCA1, BRCA2, and MMR did not differ by ascertainment. Median coverage was 219 fold.
Mutations in other genes and comparison with population frequency
Mutations in BRCA1, BRCA2, BRIP1, PALB2, RAD51C, RAD51D, and BARD1 were all significantly more common in women with OC compared to the ESP or ExAC (Table 2) and using these data, we categorized these genes together with the MMR genes as the “OC-associated genes”. Other putative cancer-associated genes such as CHEK2, NBN, RAD50, FAM175A, and MRE11A were not more frequently mutated in women with OC. Deleterious mutations in ATM and TP53 were more frequent in OC patients when compared to ExAC, but this was not significant when compared to the ESP. Odds ratios for OC are presented in Table 2. There were no mutations found in PTEN or FANCP.
Table 2.
Mutation Frequencies in Ovarian Cancer Cases Compared with Population
| Gene | Mut in Cases, N=1915 (Freq) |
Mut minus CNVs/ splice N=1915 (Freq) |
Mut in Adjusted ESP EA N=4300 (Freq) |
OR (95% CI) of Cases vs. ESP EA |
P Value |
Mut in ExACa N=36,276 (Freq) |
OR (95% CI) of Cases vs. EXAC |
P Value |
|---|---|---|---|---|---|---|---|---|
| Genes more frequently mutated in ovarian cancer | ||||||||
| BRCA1 | 182 (0.0950) | 160 (0.0836) | 8 (0.0019) | 48.9 (24.0 – 100) | <0.001 | 114 (0.0031) | 29.0 (22.7 – 37.1) | <0.001 |
| BRCA2 | 98 (0.0512) | 95 (0.0496) | 16 (0.0037) | 14.0 (8.2 – 23.8) | <0.001 | 149 (0.0041) | 12.7 (9.7 – 16.4) | <0.001 |
| BRIP1 | 26 (0.0136) | 20 (0.0104) | 5 (0.0012) | 9.1 (3.4 – 24.2) | <0.001 | 60 (0.0017) | 6.4 (3.8 – 10.6) | <0.001 |
| PALB2 | 12 (0.0062) | 9 (0.0047) | 2 (0.0005) | 10.2 (2.2 – 47.0) | <0.001 | 39 (0.0011) | 4.4 (2.1 – 9.1) | <0.001 |
| RAD51C | 11 (0.0057) | 7 (0.0037) | 1 (0.0002) | 15.8 (1.9 – 128) | 0.002 | 39 (0.0011) | 3.4 (1.5 – 7.6) | 0.005 |
| RAD51D | 11 (0.0057) | 8 (0.0042) | 2 (0.0005) | 9.0 (1.9 – 42.5) | 0.002 | 14 (0.0004) | 10.9 (4.6 – 26.0) | <0.001 |
| BARD1 | 4 (0.0021) | 4 (0.0021) | 0 | 20.3 (1.1 – 377) | 0.009 | 18 (0.0005) | 4.2 (1.4 – 12.5) | 0.02 |
| Other known or suspected cancer associated genes | ||||||||
| CHEK2 | 11 (0.0057) | 7 (0.0036) | 25 (0.0058) | 0.6 (0.3 – 1.5) | 0.37 | 297 (0.0082) | 0.4 (0.2 – 0.9) | 0.04 |
| ATM | 11 (0.0057) | 10 (0.0052) | 9 (0.0021) | 2.5 (1.0 – 6.2) | 0.07 | 79 (0.0022) | 2.4 (1.2 – 4.7) | 0.01 |
| NBN | 9 (0.0047) | 6 (0.0031) | 6 (0.0014) | 2.2 (0.7 – 7.0) | 0.26 | 49 (0.0014) | 2.3 (0.99 – 5.4) | 0.09 |
| TP53 | 6 (0.0031) | 6 (0.0031) | 4 (0.0009) | 3.4 (0.95 – 12.0) | 0.08 | 39 (0.0011) | 2.9 (1.2 – 6.9) | 0.03 |
| RAD50 | 3 (0.0016) | 3 (0.0016) | 11 (0.0026) | 0.6 (0.2 – 2.2) | 0.57 | 87 (0.0024) | 0.7 (0.2 – 2.1) | 0.63 |
| FAM175A | 3 (0.0016) | 3 (0.0016) | 5 (0.0012) | 1.3 (0.3 – 5.6) | 0.71 | 30 (0.0008) | 1.9 (0.6 – 6.2) | 0.23 |
| MRE11A | 2 (0.0010) | 1 (0.0005) | 1 (0.0002) | 2.2 (0.1 – 36.0) | 0.52 | 25 (0.0007) | 0.8 (0.1 – 5.6) | 1.00 |
Mutation numbers in EXAC are adjusted for racial groups using weighted frequencies
Abbreviations: Mut (Mutations), Freq (Frequency), CNV (copy number variant), OR (odds ratio)
In total, 347 women with OC (18.1%) had 352 mutations in OC-associated genes: 280 (14.6%) in BRCA1 or BRCA2, 64 (3.3%) in another BRCA-Fanconi anemia OC-associated gene (BRIP1, PALB2, RAD51C, RAD51D, or BARD1), and 8 (0.4%) in an MMR gene. 5/347 (1.4%) had more than one mutation. Individual mutations and associated clinical data for OC-associated genes other than BRCA1 and BRCA2 are provided in eTable 1.
Clinical characteristics and mutation status
OC-associated mutations were categorized as: BRCA1, BRCA2, other BRCA-Fanconi anemia OC-associated (BRIP1, PALB2, RAD51C, RAD51D, BARD1), mismatch repair (MSH6, PMS2, MSH2, MLH1), and no mutation (either a mutation in a gene not clearly associated with OC or no mutation). Women with BRCA1 mutations had an earlier age of onset of OC (median 52 [27 – 77]) than those with no mutation (median 62 [23 – 91]), p<0.001. BRCA2 mutation carriers had a median age of 59 (41 – 83). Those with mutations in other BRCA-Fanconi anemia OC-associated genes had a median age of 60 (34 – 79). The 8 patients with mutations in MMR genes had a median age of 50 (47 – 69). Table 3 summarizes clinical information by each OC-associated gene. In the 54/570 (9.5%) UW patients with a previous history of breast cancer, 28/54 (51.8%) had mutations in OC associated genes, 17 (31.5%) in BRCA1, 9 (16.7%) in BRCA2, and 2 (3.7%) in other OC genes (1 BRIP1, 1 RAD51D).
Table 3.
Clinical Characteristics by OC-associated Gene
| Gene | N | Median Age (Range) |
Stagea | Histologyb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | HGS | LGS | HGE | LGE | Carc | CC | CS | M | T | |||
| BRCA1 | 182 | 52 (27 – 77) | 1 | 4 | 130 | 47 | 155 | 3 | 4 | 14 | 4 | 1 | 1 | ||
| BRCA2 | 98 | 59 (41 – 83) | 1 | 75 | 22 | 85 | 1 | 3 | 9 | ||||||
| BRIP1 | 26 | 65.5 (43 – 79) | 1 | 12 | 13 | 22 | 1 | 3 | |||||||
| PALB2 | 12 | 56 (49 – 65) | 10 | 2 | 9 | 2 | 1 | ||||||||
| RAD51C | 11 | 64 (47 – 70) | 1 | 7 | 3 | 7 | 1 | 2 | 1 | ||||||
| RAD51D | 11 | 54 (34 – 75) | 1 | 5 | 5 | 7 | 1 | 3 | |||||||
| BARD1 | 4 | 55.5 (53 – 60) | 1 | 2 | 1 | 3 | 1 | ||||||||
| PMS2 | 4 | 52.5 (48 – 69) | 2 | 2 | 4 | ||||||||||
| MSH6 | 3 | 49 (47 – 62) | 2 | 1 | 1 | 2 | |||||||||
| MLH1 | 1 | 47 | 1 | 1 | |||||||||||
| None | 1568 | 62 (23 – 91) | 37 | 35 | 1117 | 376 | 1208 | 66 | 55 | 11 | 131 | 53 | 20 | 16 | 8 |
If more than one mutation, (N=5), patients are listed in each gene mutated.
Stage was not available for 3 GOG patients.
Abbreviations: HGS (high-grade, grade 2–3, serous), LGS (low-grade, grade 1, serous), HGE (high-grade, grade 2–3, endometrioid), LGE (low-grade, grade 1, endometrioid), Carc (carcinoma, unspecified), CS (carcinosarcoma), CC (clear cell), M (mucinous), T (transitional cell)
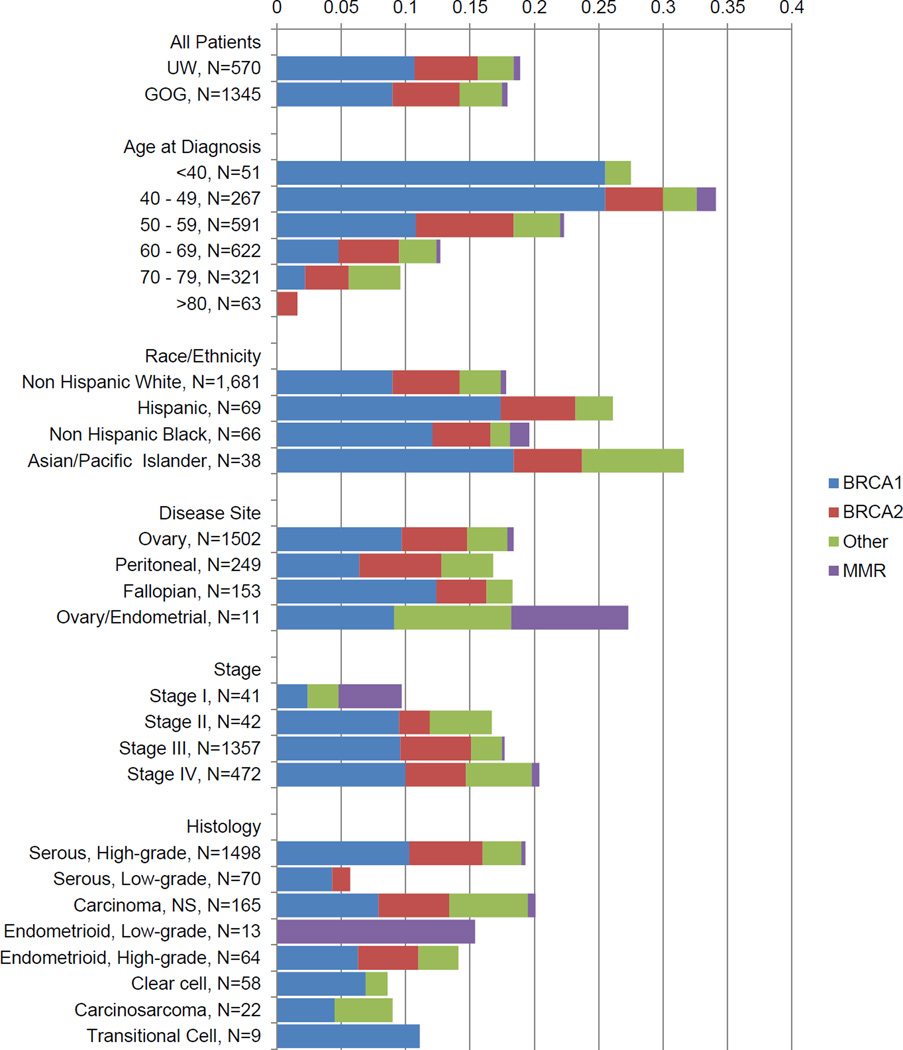
Figure 1 summarizes mutation frequency by clinical characteristics. Disease site, race and ethnicity were not associated with mutation frequency. Patients with high-grade (grade 2–3) serous carcinomas had an overall mutation frequency of 19.4% and a combined BRCA1 and BRCA2 frequency of 16.0%. Patients with unspecified carcinomas, endometrioid, carcinosarcoma, and transitional cell histology had similar mutation rates to high-grade serous histology, both for all OC genes and for BRCA1 and BRCA2. Patients with clear cell histology had a lower overall mutation frequency (8.6% versus 19.4%, p=0.04, Fisher’s exact) relative to high-grade serous cases, but differences in the BRCA1 and BRCA2 mutation frequency were of borderline significance (6.9% versus 16.0%, p=0.07). There were fewer mutations in patients with low-grade (grade 1) serous histology compared to high-grade serous (5.7% versus 19.4%, p=0.003 for any mutation; 5.7% versus 16.0% for BRCA1 and BRCA2, p=0.02). In 16 women with mucinous histology, no OC-associated mutations were found.
Figure 1.
Mutation status by clinical characteristics in OC patients.
“Other” indicates the genes BRIP1, PALB2, RAD51C, RAD51D, and BARD1, and “MMR” indicates mismatch repair genes, PMS2, MSH6, and MLH1. 61 had unknown or other race/ethnicity and are not shown. Three GOG patients had no listed stage. In the histology section 16 mucinous (all no mutation) are not shown.
Survival
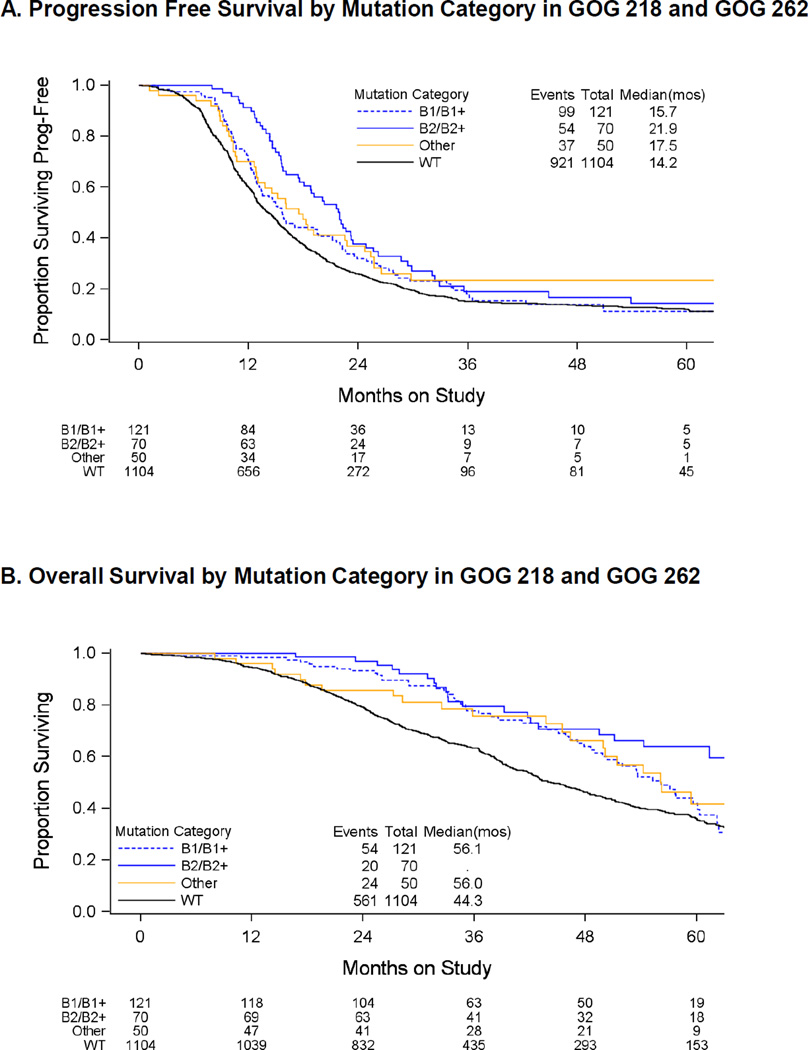
In the GOG patients, hazard ratios (HR) for progression and death were adjusted for protocol, study treatment, stage, residual disease, and initial performance status. The GOG patients were followed for 5 years. Both PFS and OS were longer for BRCA2 mutation carriers (median 21.9 months and not yet reached) than for those with no mutations (median 14.2 and 44 months), with HR 0.60 (95% CI 0.45 – 0.79, p<0.001) for PFS, and HR 0.39 (95% CI 0.25 – 0.60, p<0.001) for OS (Figure 2A and 2B). OS in BRCA1 mutation carriers (median 56 months) was intermediate between BRCA2 mutation carriers and women without mutations. Relative to those without mutations, the HR for BRCA1 mutations carriers was 0.75 (95% CI: 0.56–1.00, p=0.05, [Figure 2B]). In the UW group, OS for women with stage III and IV disease was longer in both BRCA2 mutation carriers (median 70 months, p=0.004, Log-rank) and BRCA1 mutation carriers (median 46 months, p=0.04, Log-rank), when compared to women with no mutations (median 36 months). Exploratory analyses of the individuals from the GOG studies suggest that compared to those without mutations the PFS event rates for those individuals with BRCA1 mutations (p=0.009) or BRCA2 (p<0.001) mutations are time-dependent (Figure 2A). Specifically, the PFS event rate is initially lower for those with BRCA1 or BRCA2 mutations but this advantage declines overtime. There is a similar time-dependent decline in the relative death rate apparent for those with BRCA1 mutations compared to those without mutations (p<0.001, Figure 2B). In both GOG and UW patients (Figure 1 and data not shown), survival was similar for women with mutations in BRCA1 and other OC genes in the BRCA-Fanconi anemia pathway, but the analysis was limited due to low non-BRCA mutation frequency.
Figure 2.
Survival by mutation category in GOG 218 and GOG 262.
A. PFS by mutation category in GOG 218 and GOG 262. B. OS by mutation category in GOG 218 and GOG 262.
DISCUSSION
BRCA1 and BRCA2 are part of the BRCA-Fanconi anemia DNA repair pathway, which controls DNA repair via homologous recombination.23–25 It is plausible that damaging mutations in other genes in this pathway would also confer a risk of OC. Indeed, our data demonstrate that mutations in the BRCA-Fanconi anemia OC genes BRIP1, RAD51C, and RAD51D together account for an additional 2.5% of unselected OC. Previous studies of high risk families or specific founder mutations have suggested that damaging mutations in BRIP1, RAD51C, and RAD51D confer a relative risk for OC of 6–8 fold and an absolute lifetime risk of 10–15%.8–11,26,27 Our data from unselected OC patients confirm BRIP1, RAD51C, and RAD51D as important OC genes. The Fanconi anemia gene BRIP1 (FANCJ), mutated in 1.4% of women with OC, is the next most commonly mutated gene in OC after BRCA1 and BRCA2.
In addition to identifying mutations in genes already implicated in OC, we identified mutations in the BRCA-Fanconi anemia genes PALB2 and BARD1 more frequently in women with OC compared to population frequencies in the ESP and ExAC (Table 2). PALB2 (FANCN) is a Fanconi anemia gene whose protein binds BRCA1 and BRCA2 at sites of DNA damage.28 Mutations in PALB2 are associated with an elevated risk of breast cancer and have been identified in families with both breast and OC29–31, but have not been clearly associated with OC risk.28,29 An analysis of germline mutations in high grade serous OC from the Cancer Genome Atlas project (TCGA) identified PALB2 as the only gene other than BRCA1 and BRCA2 to be significantly more frequently mutated compared to a subset of ESP controls from the Women’s Health Initiative. The PALB2 mutation frequency in our larger series was highly significant compared to population rates and was associated with similar odds ratios for OC as were RAD51C, RAD51D, and BRIP1 (Table 2). The corrected frequency of PALB2 mutations in the ESP is consistent with PALB2 mutation rates from other previously published control sets in which full sequencing of PALB2 was performed30,32, supporting the use of the ESP as a comparison population.
Three recent publications by the Ovarian Cancer Association Consortium (OCAC) have examined rates of mutations in BRCA1, BRCA2, mismatch repair genes, RAD51C, RAD51D, BRIP1, PALB2, and BARD1 compared to controls.12–14 The sequencing methodology reported by OCAC is different from our study. As the authors acknowledged, their sequencing methods resulted in relatively low coverage, were unable to detect genomic rearrangements, and likely lead to underestimation of mutation frequencies12. The impact of less sensitive sequencing methods is demonstrated by OCAC’s low BRCA1 mutation rate of 3.8%14, which is significantly lower than our BRCA1 mutation rate of 9.5% (84/2222 versus 182/1915, p<0.001, Fisher’s exact), and lower than that reported in other population-based series2,3. Even if we exclude genomic rearrangements, which OCAC could not detect, our BRCA1 mutation rate remains significantly higher (166/1915, 8.7%, versus 84/2222, p<0.001, Fisher’s exact). Despite a mutation rate for RAD51C and RAD51D that was approximately 30% lower than ours, OCAC identified a similar odds ratio to our calculation for these genes. OCAC did not find a difference in mutation frequency for PALB2 or BARD1 in OC cases versus controls13, but their lower mutation rates in cases and an unexpectedly high PALB2 mutation rate in controls impacted their power to detect such a difference.
BARD1 forms a heterodimer with BRCA1 mediated by their homologous ring finger motifs.33 This heterodimerization is critical to several tumor suppressor functions of BRCA1, and mutations in BRCA1 that affect binding to BARD1 are associated with increased cancer risk.34BARD1 mutations were rare in OC patients, but significantly more common than in the general population, leading to wide confidence intervals for the generated odds ratio for OC (Table 2). However, the significant p-value and the shared homology to BRCA1 support BARD1 as a rare OC susceptibility gene. These results are interpreted with some caution as two of the BARD1 mutation carriers also had mutations in BRCA1 (eTable1). Mutations in BARD1 have also been identified in women with triple negative breast cancer, another phenotype associated with BRCA1 mutations.35 Additional studies are needed to define the absolute risk of BARD1 for both breast and OC. The addition of BARD1 and PALB2 to other known OC genes (BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2, MSH2, MLH1, MSH6, and PMS2) brings the total suspected hereditary OC genes to 11.
In contrast to PALB2 and BARD1, mutations in other genes in the BRCA-Fanconi anemia pathway previously implicated in breast but not OC risk (CHEK2, NBN, RAD50, FAM175A, and MRE11A) were not more commonly mutated in OC patients. Mutations in TP53 (which cause early onset breast cancer and Li Fraumeni syndrome) and ATM were slightly more frequent in OC than in population controls, which appeared significant when compared to the larger ExAC database. Given that mutations in all of these genes are rare, confidence intervals are wide, and we cannot fully exclude that some of these genes, and in particular ATM and TP53, are associated with some risk for OC. However, a relatively high risk of OC is unlikely.
Mutations in mismatch repair (MMR) genes (MSH2, MLH1, MSH6, and PMS2), which cause Lynch syndrome are often cited as the other major cause of hereditary OC in addition to BRCA1 and BRCA2.36 However, MMR mutations were infrequent (0.4%) in this series. Interestingly, 88% of MMR mutations in women with OC occurred in PMS2 or MSH6, in contrast to patients with colon cancer, in which MSH2 and MLH1 mutations predominate.37 Notably, two of three OCs with MSH6 mutations were endometrioid and low stage, but all four PMS2-mutated cases were advanced stage, high-grade serous carcinomas. Our data are consistent with a population-based study of 1,638 women with invasive OC who were sequenced for mutations in MLH1, MSH2, and MSH6, where 0.5% had mutations, 5/9 in MSH6 (PMS2 not assessed).38
The National Comprehensive Cancer Network (NCCN) recommends genetic testing for all women affected by OC, but some authors have proposed limiting BRCA1 and BRCA2 testing to high-grade serous OC and suggested that BRCA1 and BRCA2 mutations found in non-serous cases represent pathological misclassification.39 In our study, all cases were centrally reviewed by a panel of gynecologic pathologists, minimizing pathological misclassification. The overall mutation rate for high-grade serous histology was not significantly different from the mutation frequency in undifferentiated carcinoma, endometrioid, or carcinosarcoma histologies. While clear cell and low-grade serous OC did have fewer mutations compared to high-grade serous OC, the mutation rates (8.5% and 5.7%, respectively) are probably still high enough to warrant genetic testing. Therefore, our data do not support the restriction of genetic testing to women with high-grade serous OC. Race and ethnicity also did not predict mutation status and should not be used to guide testing. Consistent with previous studies, OC survival was correlated with mutation status, with BRCA2 mutation carriers having the longest progression-free and overall survival.2,40 This study is unique in that cancer treatments were standardized within the confines of clinical trials, reducing potential bias in how mutation status affected survival. Mutation status should be considered when analyzing outcomes of OC clinical trials.
There are currently no published guidelines for managing unaffected women found to have mutations in BRIP1, PALB2, RAD51C, RAD51D, and BARD1. We found similar odds ratios for OC conferred by damaging mutations in these genes to the previously identified 6–8 fold relative risk in BRIP1, RAD51C, and RAD51D8–11,26,27 and a similar age distribution of OC diagnosis for women with mutations in these genes compared to BRCA2 (Table 2 and Table 3). If a lifetime risk of 10–15% is confirmed for these genes, it would be reasonable to consider risk-reducing salpingo-oophorectomy by age 45.
There are several limitations to this study. This was not a population-based study and the GOG cases were limited to stage III and IV cancers. However, all patients were enrolled at the time of diagnosis, which reduces survival bias, and they were unselected for age or family history. The ESP is thought to reflect US population mutation rates as opposed to cancer-free controls, as cancer status is not fully characterized. ExAC has the advantage of sequencing data on a large number of people, characterized by racial and ethnic group, but also contains people with known malignancies such as those in the Cancer Genome Atlas (TCGA) studies. The inclusion of known cancer patients in ExAC could falsely lower our calculated OR for OC. Ideally we would use a larger, well-characterized control group with age and race matching, with known cancer status. However, it is reassuring that mutation frequencies in the ESP for these genes were similar to previously published controls.8,9,30,32,41 Further research is needed to clarify the roles of inherited mutations in PALB2 and BARD1 in ovarian cancer risk.
In summary, we present data from a large, comprehensively sequenced group of unselected OC patients and provide data to implicate two new suspected hereditary OC genes, BARD1 and PALB2. Overall, 347/1915 (18.1%) OC patients had 352 germline mutations in 11 OC genes (BRCA1, BRCA2, BRIP1, RAD51C, RAD51D, PALB2, BARD1, PMS2, MSH6, MLH1, and MSH2). Mutations were identified in all histologies of ovarian carcinoma, with the exception of mucinous carcinoma, and 72/352 (20.5%) OC-associated mutations occurred in a gene other than BRCA1 or BRCA2.
Supplementary Material
Acknowledgments
Funding/Support:
This study was supported by National Cancer Institute grants to the Gynecologic Oncology Group Administrative Office (CA 27469), the Gynecologic Oncology Group Statistical and Data Center (CA 37517), the Gynecologic Oncology Group Tissue Bank (U10 CA27469, U24 CA114793, and U10 CA180868), NRG Oncology Grant (1 U10 CA180822), R01CA131965 (ES), R01CA157744 (MCK), RO1CA175716 (TW and MCK), and P50CA083636 (ES), and by the Ovarian Cancer Research Foundation, Women’s Reproductive Health Research Career Development Award (5K12HD001264-13 (BN)), The Liz Tilberis Early Career Award from the Ovarian Cancer Research Foundation (BN), The Breast Cancer Research Foundation, the Department of Defense Ovarian Cancer Research Program (OC093285 (TW) and OC120312 (ES)), and the Wendy Feuer Research Fund for Prevention and Treatment of Ovarian Cancer.
Participating Institutions:
The following institutions participated in GOG 218 and 262: Roswell Park Cancer Institute, University of Alabama at Birmingham, Duke University Medical Center, Abington Memorial Hospital, Walter Reed Army Medical Center, Wayne State University, University of Minnesota Medical School, Mount Sinai School of Medicine, Northwestern Memorial Hospital, University of Mississippi Medical Center, Colorado Gynecologic Oncology Group P.C., University of California at Los Angeles, University of Washington, University of Pennsylvania Cancer Center, Milton S. Hershey Medical Center, University of Cincinnati, University of North Carolina School of Medicine, University of Iowa Hospitals and Clinics, University of Texas Southwestern Medical Center at Dallas, Indiana University School of Medicine, Wake Forest University School of Medicine, University of California Medical Center at Irvine, Rush-Presbyterian-St. Luke’s Medical Center, Magee Women’s Hospital, SUNY Downstate Medical Center, University of Kentucky, University of New Mexico, The Cleveland Clinic Foundation, State University of New York at Stony Brook, Washington University School of Medicine, Memorial Sloan-Kettering Cancer Center, Cooper Hospital/University Medical Center, Columbus Cancer Council, MD Anderson Cancer Center, University of Massachusetts Medical School, Fox Chase Cancer Center, Women’s Cancer Center, University of Oklahoma, University of Virginia Health Sciences Center, University of Chicago, Mayo Clinic, Case Western Reserve University, Tampa Bay Cancer Consortium, Yale University, GOG Japan-Saitama Medical University International Medical Center, University of Wisconsin Hospital, Cancer Trials Support Unit, University of Texas - Galveston, Women and Infants Hospital, Korean Gynecologic Oncology Group, The Hospital of Central Connecticut, Georgia Core, GYN Oncology of West Michigan, PLLC, Aurora Women’s Pavilion of West Allis Memorial Hospital, Fred Hutchinson Cancer Research Center, UCSF – Mount Zion, St. Joseph’s Hospital and Medical Center, Carolinas Medical Center/Levine Cancer Institute and Community Clinical Oncology Program.
Other Acknowledgements:
The authors would like to thank Barry E. Storer, Ph.D., Department of Biostatistics, University of Washington School of Public Health, Seattle, WA for reviewing the manuscript, and Kim M. Blaser with NRG Oncology for providing assistance with manuscript preparation.
The authors would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010)
The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Role of The Sponsor:
Genentech had no role in the design or conduct of this study, including data collection, management, analysis, interpretation of the data, or preparation of the manuscript. Genentech reviewed and commented on the manuscript, and approved it prior to submission for publication.
Footnotes
Author Contributions:
Dr. Norquist had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Dr. Birrer and Dr. Swisher served as co-senior authors.
Study concept and design: Norquist, Harrell, Walsh, King, Swisher, Birrer
Acquisition, analysis, or interpretation of the data: Norquist, Harrell, Brady, Walsh, Lee, Gulsuner, Bernards, Casadei, Yi, Burger, Chan, Davidson, Mannel, DiSilvestro, Lankes, Ramirez, King, Swisher, Birrer
Drafting of the manuscript: Norquist, Swisher
Critical revision of the manuscript for important intellectual content: Harrell, Brady, Walsh, Lee, Gulsuner, Bernards, Casadei, Yi, Burger, Chan, Davidson, Mannel, DiSilvestro, Lankes, Ramirez, King, Birrer
Statistical analysis: Brady, Norquist
Obtaining funding: Swisher, Birrer, King, Walsh
Administrative, technical, or material support: Harrell, Walsh, Lee, Gulsuner, Bernards, Casadei, Yi, Burger, Chan, Davidson, Mannel, DiSilvestro, Lankes, Ramirez, King
Study supervision: Norquist, Swisher, Birrer
Conflict of Interest Disclosures: Dr. Mannel reports participation in advisory boards for Endocyte, Astra Zeneca, MedImmune, Oxigene, Advaxis, and Amgen; and his institution was reimbursed for his time. Dr. Burger reports that he previously participated as a consultant to Genentech/Roche during advisory board meetings. No other disclosures are reported.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics 2015. CA Cancer J Clin. 2015 Jan;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Alsop K, Fereday S, Meldrum C, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. 2012 Jul 20;30(21):2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang S, Royer R, Li S, et al. Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected patients with invasive ovarian cancer. Gynecol Oncol. 2011 May 1;121(2):353–357. doi: 10.1016/j.ygyno.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011 Nov 1;108(44):18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003 Oct 24;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 6.Domchek SM, Friebel TM, Neuhausen SL, et al. Mortality after bilateral salpingo-oophorectomy in BRCA1 and BRCA2 mutation carriers: a prospective cohort study. Lancet Oncol. 2006 Mar;7(3):223–229. doi: 10.1016/S1470-2045(06)70585-X. [DOI] [PubMed] [Google Scholar]
- 7.Finch AP, Lubinski J, Moller P, et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014 May 20;32(15):1547–1553. doi: 10.1200/JCO.2013.53.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loveday C, Turnbull C, Ramsay E, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011 Sep;43(9):879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loveday C, Turnbull C, Ruark E, et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet. 2012 May;44(5):475–476. doi: 10.1038/ng.2224. author reply 476. [DOI] [PubMed] [Google Scholar]
- 10.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010 May;42(5):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 11.Rafnar T, Gudbjartsson DF, Sulem P, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat. Genet. 2011;43(11):1104–1107. doi: 10.1038/ng.955. [DOI] [PubMed] [Google Scholar]
- 12.Song H, Dicks E, Ramus SJ, et al. Contribution of Germline Mutations in the RAD51B, RAD51C, and RAD51D Genes to Ovarian Cancer in the Population. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015 Sep 10;33(26):2901–2907. doi: 10.1200/JCO.2015.61.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramus SJ, Song H, Dicks E, et al. Germline Mutations in the BRIP1, BARD1, PALB2, and NBN Genes in Women With Ovarian Cancer. Journal of the National Cancer Institute. 2015 Nov;107(11) doi: 10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song H, Cicek MS, Dicks E, et al. The contribution of deleterious germline mutations in BRCA1, BRCA2 and the mismatch repair genes to ovarian cancer in the population. Hum Mol Genet. 2014 Sep 1;23(17):4703–4709. doi: 10.1093/hmg/ddu172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh T, Lee MK, Casadei S, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010 Jul 13;107(28):12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abel HJ, Duncavage EJ, Becker N, Armstrong JR, Magrini VJ, Pfeifer JD. SLOPE: a quick and accurate method for locating non-SNP structural variation from targeted next-generation sequence data. Bioinformatics. 2010 Nov 1;26(21):2684–2688. doi: 10.1093/bioinformatics/btq528. [DOI] [PubMed] [Google Scholar]
- 17.Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12:184. doi: 10.1186/1471-2164-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouwman P, van der Gulden H, van der Heijden I, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013 Oct;3(10):1142–1155. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- 19.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Human mutation. 2007 Jun;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 20.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) [Accessed October, 2014]; http://evs.gs.washington.edu/EVS/
- 21.Exome Aggregation Consortium (ExAC) [Accessed May, 2015]; http://exac.broadinstitute.org.
- 22.Burger RA, Brady MF, Bookman MA, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011 Dec 29;365(26):2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 23.Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002 Jul 26;297(5581):606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 24.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999 Oct;4(4):511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 25.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001 Feb;7(2):263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 26.Pelttari LM, Heikkinen T, Thompson D, et al. RAD51C is a susceptibility gene for ovarian cancer. Hum Mol Genet. 2011 Aug 15;20(16):3278–3288. doi: 10.1093/hmg/ddr229. [DOI] [PubMed] [Google Scholar]
- 27.Pelttari LM, Kiiski J, Nurminen R, et al. A Finnish founder mutation in RAD51D: analysis in breast, ovarian, prostate, and colorectal cancer. J Med Genet. 2012 Jul;49(7):429–432. doi: 10.1136/jmedgenet-2012-100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JY, Zhang F, Andreassen PR. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochimica et biophysica acta. 2014 Aug;1846(1):263–275. doi: 10.1016/j.bbcan.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. The New England journal of medicine. 2014 Aug 7;371(6):497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007 Feb;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casadei S, Norquist BM, Walsh T, et al. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011 Mar 15;71(6):2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janatova M, Kleibl Z, Stribrna J, et al. The PALB2 gene is a strong candidate for clinical testing in BRCA1- and BRCA2-negative hereditary breast cancer. Cancer Epidemiol Biomarkers Prev. 2013 Dec;22(12):2323–2332. doi: 10.1158/1055-9965.EPI-13-0745-T. [DOI] [PubMed] [Google Scholar]
- 33.Brzovic PS, Keeffe JR, Nishikawa H, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci U S A. 2003 May 13;100(10):5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001 Oct;8(10):833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 35.Couch FJ, Hart SN, Sharma P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015 Feb 1;33(4):304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miesfeldt S, Lamb A, Duarte C. Management of genetic syndromes predisposing to gynecologic cancers. Curr Treat Options Oncol. 2013 Mar;14(1):34–50. doi: 10.1007/s11864-012-0215-3. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Carbonell L, Ruiz-Ponte C, Guarinos C, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012 Jun;61(6):865–872. doi: 10.1136/gutjnl-2011-300041. [DOI] [PubMed] [Google Scholar]
- 38.Pal T, Akbari MR, Sun P, et al. Frequency of mutations in mismatch repair genes in a population-based study of women with ovarian cancer. Br J Cancer. 2012 Nov 6;107(10):1783–1790. doi: 10.1038/bjc.2012.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrader KA, Hurlburt J, Kalloger SE, et al. Germline BRCA1 and BRCA2 mutations in ovarian cancer: utility of a histology-based referral strategy. Obstetrics and gynecology. 2012 Aug;120(2 Pt 1):235–240. doi: 10.1097/AOG.0b013e31825f3576. [DOI] [PubMed] [Google Scholar]
- 40.Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014 Feb 1;20(3):764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006 Nov;38(11):1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.