ABSTRACT
It has been reported that Wnt/β-catenin is critical for dedifferentiation of differentiated epidermal cells. Cyclin D1 (CCND1) is a β-catenin target gene. In this study, we provide evidence that overexpression of CCND1 induces reprogramming of epidermal cells into stem cell-like cells. After introducing CCND1 gene into differentiated epidermal cells, we found that the large flat-shaped cells with a small nuclear-cytoplasmic ratio changed into small round-shaped cells with a large nuclear-cytoplasmic ratio. The expressions of CK10, β1-integrin, Oct4 and Nanog in CCND1 induced cells were remarkably higher than those in the control group (P < 0.01). In addition, the induced cells exhibited a high colony-forming ability and a long-term proliferative potential. When the induced cells were implanted into a wound of laboratory animal model, the wound healing was accelerated. These results suggested that overexpression of CCND1 induced the reprogramming of differentiated epidermal cells into stem cell-like cells. This study may also offer a new approach to yield epidermal stem cells for wound repair and regeneration.
KEYWORDS: Cyclin D1, dedifferentiation, epidermal stem cells, reprogramming, wound repair
Introduction
Epidermal stem cells have an important role in wound repair and tissue engineering of replacement skin.1,2 But the number of these stem cells, which only consist of 1–10% of the basal layer in normal epidermis,3 is limited, which has hampered the widely clinical applications. So, the new sources of epidermal stem cells need to be found. There are a large number of differentiated epidermal cells which have lost their regenerative ability. How to make these cells reverse to a less differentiated form and regain the ability to regenerate the injured skin may resolve the problem of a shortage of epidermal stem cells.
Cellular reprogramming which includes horizontal and vertical reprogramming has been used to generate patient specific cells for therapeutic purposes.4 Horizontal reprogramming is called transdifferentiation and vertical reprogramming is called dedifferentiation. Cellular dedifferentiation is the progression of cells from a more differentiated to a less differentiated state and the major process underlying totipotency, regeneration and formation of new stem cell lineages. In 2006, Yamanaka et al.5 reported that mouse embryonic fibroblasts could be reprogrammed into embryonic stem-like cells by transduction of 4 transcription factors, Oct4, Sox2, c-Myc, and Klf4.1, which proved that differentiated, perhaps nondividing cells, could be reprogrammed into highly proliferative embryonic cells. Induction of these induced pluripotent stem (iPS) cells is a typical example of dedifferentiation. The dedifferentiation phenomenon of epidermal cells has already been reported in the wound treated with recombinant human epidermal growth factor (rhEGF).6 In succedent research, we provided evidence that Wnt/β-catenin pathway was necessary for dedifferentiation of epidermal cells in vivo and in vitro. In addition, CCND1 as β-catenin target gene was also upregulated during the process of dedifferentiation.7,8
Recently, it has been shown that the loss of p53 function can enhance the efficiency of reprogramming, suggesting that the cell cycle is an important step in the reprogramming process.9-11 CCND1 is a critical gene involved in the regulation of progression through the G1 phase of the cell cycle, thereby contributing to cell proliferation. Repression of CCND1 gene expression is a hallmark of cell differentiation.12 Edel et al.13 found that CCND1 protein levels were highly expressed in iPSCs compared to ESCs. Overexpression of CCND1 with 3 factors (Oct4/sox2/Klf4) increased the efficiency of reprogramming more than threefold, by increasing the number of cells in S phase of the cell cycle, demonstrating that CCND1 up-regulation was sufficient for enhancing reprogramming. In our previous study, we had found increased expression of CCND1 in dedifferentiation-derived epidermal stem cells. Thus, we speculate that CCND1 might be involved in dedifferentiation of epidermal cells. With overexpression of CCND1 in differentiated epidermal cells, we herein offered more evidences to support the hypothesis that the cell cycle is a rate-limiting step in epidermal cell dedifferentiation.
Results
CCND1 expression in cells
pEGFP-N1-empty and pEGFP-N1-CCND1 plasmids were transfected into differentiated epidermal cells. After two days, transfected cells were collected for flow sorting. All of the obtained cells expressed green fluorescent protein (GFP) (Fig. 1A). The results of RT-PCR showed that the expression of CCND1 in G-CCND1 was much higher than that in G-empty (P < 0.01) (Fig. 1B).
Figure 1.
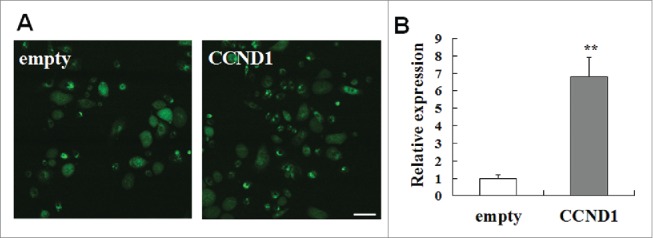
Cell transfection and the expression of pEGFP-N1-CCND1. A: Cell transfection of pEGFP-N1-CCND1. Scale bar = 50 μm. B: The expression of CCND1 detected by quantitative real-time PCR. The CT data of empty group (control) were seen as “1” and the relative expression of the other group was calculated according to the empty group by the CT data. The data are the means ± SD (n = 10). **P < 0.01, as compared with empty vector control. CCND1, cyclin D1; EGFP, enhanced green fluorescent protein; SD, standard deviation.
Morphologic characteristics
Transfected cells were plated again into the culture dish after flow sorting. Three days later, morphologic characteristics of transfected cells including G-empty and G-CCND1 were photographed along with non transfected cells including G-non and G-positive. The morphology of cells in G-empty and G-CCND1 groups had striking differences. The former were large flat-shaped cells with a small nuclear-cytoplasmic ratio and the latter were small round shaped cells with a large nuclear-cytoplasmic ratio. This demonstrated that the large flat-shaped cells had changed into small round-shaped cells along with the increase in the nuclear-cytoplasmic ratio after a 5-day induction by CCND1. There were no differences in morphology between G-non and G-empty and also between G-CCND1 and G-positive (Fig. 2). This result demonstrated that the CCND1-induced cells had morphologic characteristics of epidermal stem cells.
Figure 2.
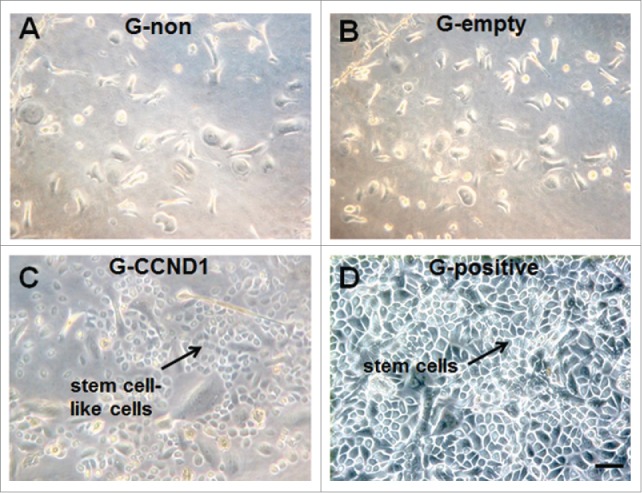
Morphological characteristics of epidermal cells in the 4 groups. A: Non transfection (G-non) group; B: Empty vector transfection (G-empty) group; C: CCND1 transfection (G-CCND1) group; D: Positive control (G-positive) group. Scale bar = 50 μm. CCND1, cyclin D1; G, group.
CK10 and β1 integrin expression
The expressions of CK10 and β1 integrin in cultured epidermal cells from the 4 groups were observed by using immunofluorescence. We found that overexpression of CCND1 in differentiated epidermal cells significantly decreased the number and proportion of CK10 positive cells (Fig. 3A and B). Just as G-positive (Fig. 3C), there was no CK10 positive cells in G-CCND1. In contrast, the expression of β1 integrin was enhanced by the transfection of recombinant plasmid pEGFP-N1-CCND1 into differentiated epidermal cells (Fig. 3D and E). Moreover, red staining indicated very intense β1 integrin expression in the membrane and cytoplasm of epidermal stem cells (Fig. 3F) and CCND1-induced cells. G-non had CK10 positive cells, but no β1 integrin positive cells were shown in G-non (data not shown). This result demonstrated that the CCND1-induced cells had phenotypic characteristics of epidermal stem cells.
Figure 3.
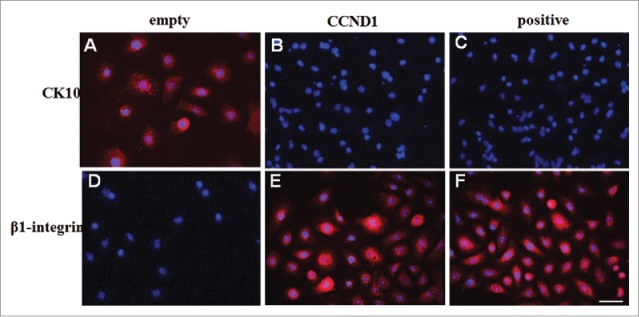
CK10 and β1-integrin expressions in epidermal cells from G-empty, G-CCND1 and G-positive groups. A-C: Representative photographs of CK10 expression; D-F: Representative photographs of β1-integrin expression. PE signals (red) were examined under fluorescence microscopy. The nuclei were counterstained with DAPI (blue). Scale bar = 50 μm. CCND1, cyclin D1; CK10, cytokeratin 10; PE, phycoerthrin; DAPI, 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride.
Oct4 and Nanog expression
Recently, transcription factors Oct4 and Nanog have been found to be expressed in stem cells from different adult human tissues. Thus, their expressions have been considered general markers of self-renewal and pluripotency in stem cells. To further confirm the stem cell-like nature of CCND1-induced cells, we investigated the expressions of Oct4 and Nanog. Real-time PCR analysis revealed that CCND1-induced cells, as well as epidermal stem cells, were > 4-5 fold enriched for both Oct4 and Nanog compared with G-empty and G-non groups (P < 0.01; Fig. 4). This finding is consistent with observations reporting Oct4 and Nanog expression in epidermal stem cells cultured in vitro7,8,14 and Oct4 expression in rare interfollicular basal cells of human epidermis in situ.15
Figure 4.
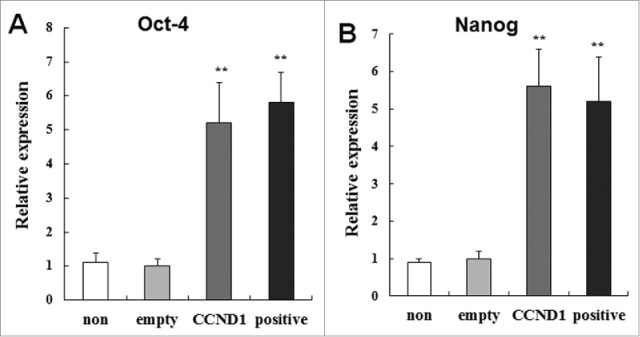
Relative expression of self-renewal and pluripotency genes Oct4 and Nanog in the 4 groups. A: Relative expression of Oct4; B: Relative expression of Nanog. The data are the means ± SD (n = 10). **P < 0.01, as compared with empty vector control. CCND1, cyclin D1; G, group; SD, standard deviation.
Cell cycle
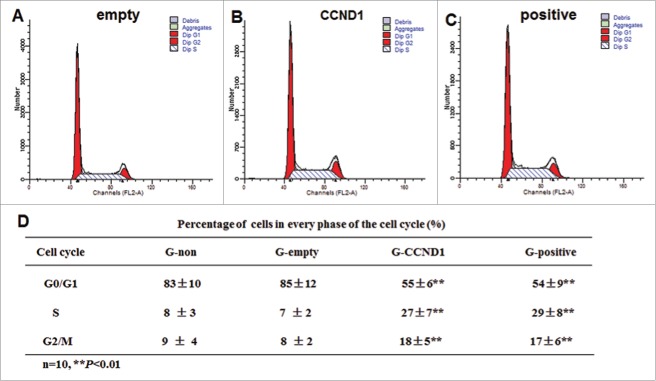
To study the changes in cell cycle of the induced cells, 3 cell subpopulations (G0/G1, S and G2/M) were estimated by performing a flow cytometric measurement of the DNA distributions of the cells. In CCND1-induced and positive control groups, more cells were found in S phase and G2 /M phase of the cell cycle compared with the G-empty and G-non groups (P < 0.01; Fig. 5). There were no differences in 3 cell subpopulations between G-non and G-empty. The data suggested that there were more CCND1-induced cells in the proliferative phase.
Figure 5.
Cell cycle detection of epidermal cells in the 4 groups. A: Representative flow cytometric analysis of G-empty; B: Representative flow cytometric analysis of G-CCND1; C: Representative flow cytometric analysis of G-positive; D: Statistics data of cell cycle. The data are the means ± SD (n = 10). **P < 0.01, as compared with empty vector control. CCND1, cyclin D1; G, group; SD, standard deviation.
Long-term growth potential
Experiments were performed to examine whether CCND1-induced cells and epidermal stem cells had long-term growth potential. Days in culture in G-empty group ranged from 7 to 11, with an average of 9 ± 2 days, and there was no significant difference compared with G-non. However, for the CCND1-induced group, days in culture ranged from 56 to 80, with an average of 68 ± 12 days, and there was no significant difference compared with the positive control group (Fig. 6A). The number of passages in G-empty group ranged from 1 to 3, with an average of 2 ± 1, and there was no significant difference compared with G-non. For the CCND1-induced group, the number of passages ranged from 9 to 19, with an average of 14 ± 5, and there was no significant difference compared with the positive control group (Fig. 6B). Total cell outputs of G-empty and G-CCND1 groups were 1.0 × 105 and 2.0 × 1010, respectively (Fig. 6C). These results demonstrated that CCND1-induced cells and epidermal stem cells had similar long-term growth potentials.
Figure 6.
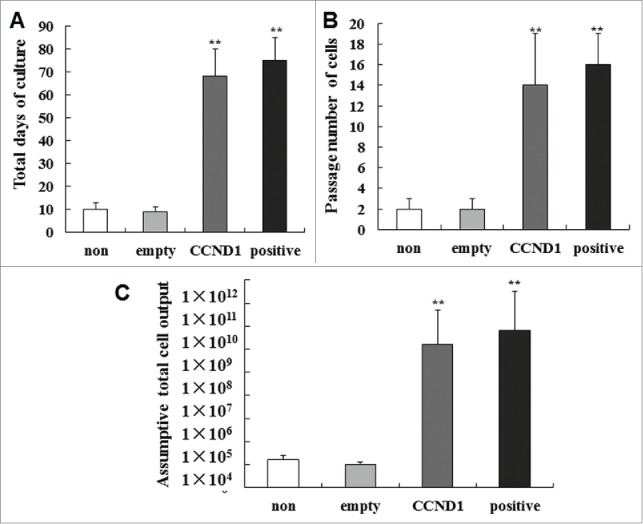
Long-term growth potential of epidermal cells in the 4 groups. (A) Total days in culture of each group. (B) Number of passages in each group. (C) Assumptive total cell output in each group. The data are the means ± SD (n = 10). **P < 0.01, as compared with empty vector control. SD, standard deviation.
Colony-forming ability
Epidermal stem cells are clonogenic and have high colony-forming ability. To determine whether CCND1-induced cells also have this ability, the number of colonies consisting of 10 or more cells was counted blindly. We found that the CCND1-induced cells possessed higher clonogenic capacity than that of G-empty group (Fig. 7A and B). The number of colonies consisting of 10 or more cells in G-CCND1 group increased 6-fold compared with that in G-empty (P < 0.01), and there was no significant difference compared with the positive control (Fig. 7C and D). These data showed a highly significant capacity of CCND1-induced cells to give rise to colonies.
Figure 7.
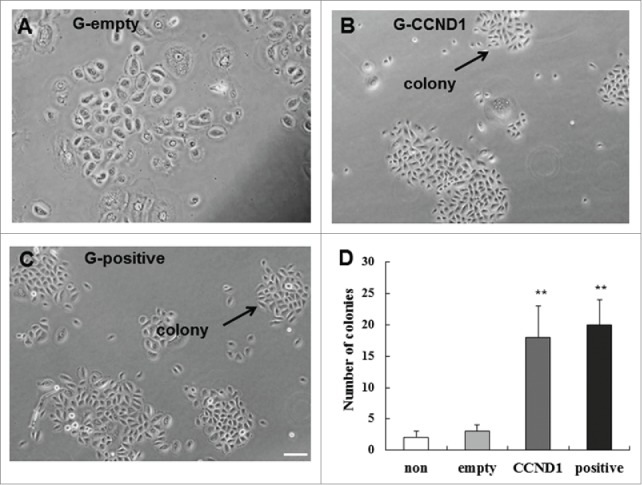
Colony-forming efficiency of of epidermal cells in the 4 groups. A: Colonies in G-empty group; B: Colonies in G-CCND1 group; C: Colonies in G-positive group; D: Cartogram of Colony-forming efficiency. The data are the means ± SD (n = 10). **P < 0.01, as compared with empty vector control. Scale bar = 50 μm. CCND1, cyclin D1; G, group; SD, standard deviation.
Evaluation of healing
After anaesthetization, the full-thickness cutaneous wounds were created on the back of BALB/c nude mice. The wound area was measured at 2 and 4 weeks after implantation (Fig. 8). At 2 weeks, wound closure in G-CCND1 and G-positive groups were enhanced obviously compared with that in G-non and G-empty groups. At 4 weeks, all wounds in G-CCND1 and G-positive groups healed up and no completely closed wounds were observed in G-non, G-empty and control (no graft) groups. Wound closure in G-non, as well as in G-empty, appeared similar to that in control. To trace the implanted cells, immunofluorescence was performed on tissue sections with primary mouse monoclonal anti-human β1-integrin antibody and Hoechst 33342. We found that the implanted cells in G-CCND1 and G-positive groups formed a stratified epidermal layer on the wound surface, but not in G-non and G-empty groups (Fig. 9).
Figure 8.
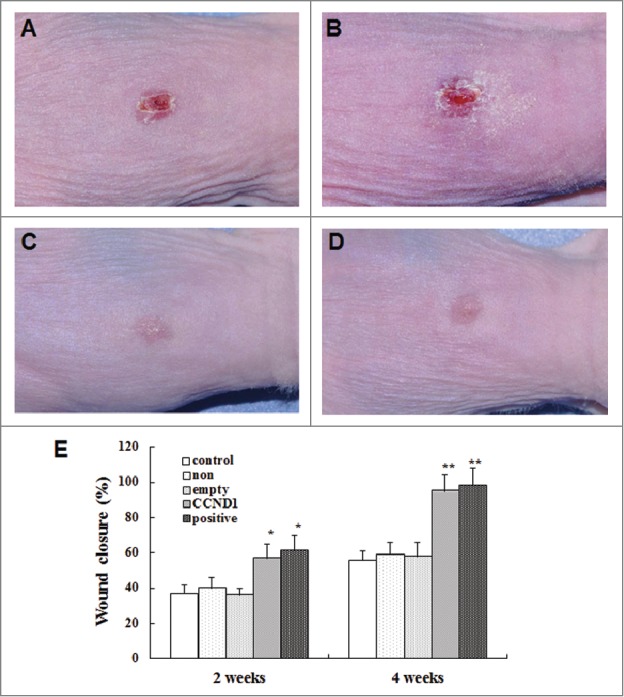
Acceleration of wound closure by epidermal cells in the 4 groups. A: Representative photograph of G-non group; B: Representative photograph of G-empty group; C: Representative photograph of G-CCND1 group; D: Representative photograph of G-positive group; E: Cartogram of wound closure at days 14 and 28 after transplantation. The data are the means ± SD (n = 8). *P < 0.05, **P < 0.01, as compared with empty vector control. Scale bar = 50 μm. CCND1, cyclin D1; G, group; SD, standard deviation.
Figure 9.
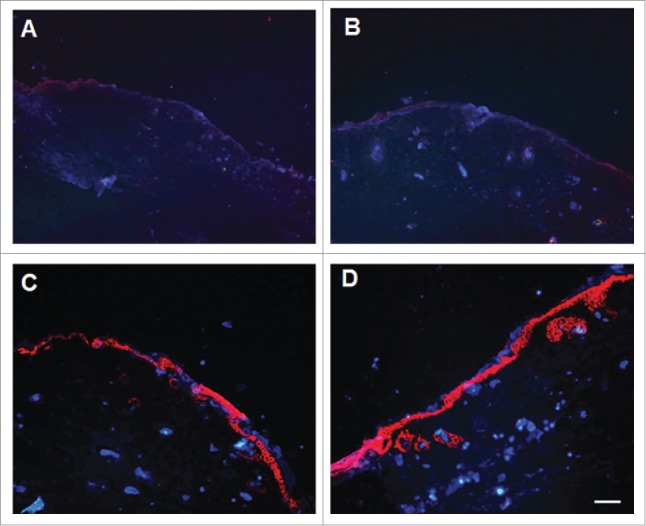
Stratified epidermal layer on the wound surface formed by the implanted cells. A: Representative photograph of G-non group; B: Representative photograph of G-empty group; C: Representative photograph of G-CCND1 group; D: Representative photograph of G-positive group. Scale bar = 50 μm. CCND1, cyclin D1; G, group.
Discussion
In the present study, we demonstrated that high expression of CCND1 induced dedifferentiation (vertical reprogramming) of differentiated epidermal cells into stem cell-like cells. PEGFP-CCND1 plasmids were transfected into differentiated epidermal cells. After 5 days, CCND1 overexpression induced differentiated epidermal cells to exhibit positive expression of CK19, β1 integrin, Oct4, and Nanog. CCND1-induced cells and epidermal stem cells exhibited a high colony-forming efficiency, a long-term proliferative potential, and the ability to repair wounds. These results suggest that abundant epidermal stem cells can be generated by overexpression of CCND1, maybe through manipulating the cell cycle.
It has long been thought that once mammalian cells are committed to a specific lineage, they can no longer change their fate, thus they become the so-called terminally differentiated cells. But recent studies suggest that differentiated mammalian cells can be induced to assume new fates under appropriate conditions. It has been shown that a combination of 3 or 4 factors of Oct4, Sox2, and KLf4, with or without Myc, can reprogram somatic cells to generate iPSCs.5,16 Analysis of partially reprogrammed iPSCs reveals temporal and separable contributions of the 4 factors and indicates that ectopic c-Myc acts earlier than the pluripotency regulators.17 Indeed, overexpression of Myc is known to regulate cyclin D1, promoting cell cycle progression, although it remains unknown if the cell cycle function of c-Myc plays a separate role to the pluripotency genes (Oct4/sox2/Klf4) in the reprogramming process.18 Rapid cell cycling is a feature of PSCs and is central in promoting pluripotency and self-renewal.19,20 G1/S arrest resulted in smaller hESC colonies, the down-regulation of the pluripotency marker Oct4, and rapid differentiation of the hESCs.21,22,23 CCND1 is a critical gene regulating cells through the G1 phase of the cell cycle and was found to play an important role in pluripotency maintenance.24 Edel et al.13 also reported that overexpression of wild-type cyclin D1 was responsible for reprogramming human somatic cells toward iPSCs.
We have obtained evidence of the dedifferentiation of epidermal cells to stem cells or stem cell-like cells in vivo6,25 and in vitro.26 We demonstrated further evidence to demonstrate that this dedifferentiation phenomenon happened when the Wnt/β-catenin pathway was activated. Cyclin D1 and c-Myc are β-catenin target genes whose expressions were upregulated during the process of dedifferentiation.27,28 In this experiment, the effects of CCND1 overexpression on morphologic, phenotypic, growth and functional characteristics of the differentiated epidermal cells were observed. The shape changed as discribed in results and the phenotypic assays revealed that the CCND1-induced cells exhibited positive expressions of β1 integrin, Oct4 and Nanog and negative expression of CK10. It has been reported that CK10 and β1 integrin are markers of differentiated epidermal cells and epidermal stem cells in adult human skin, respectively.29 The self-renewal genes Oct4 and Nanog, known to be highly expressed in self-renewing embryonic stem cells,30,31 are also expressed in several adult stem cells including epidermal stem cells, but not in normal differentiated cells.15 So, Oct4 and Nanog expressions have been considered general markers of self-renewal and pluripotency in stem cells. In addition, the growth and functional assays showed that the CCND1-induced cells, as well as epidermal stem cells,32,33 were able to develop significantly larger, active colonies than were non-induced cells (G-non and G-empty). In differentiated epidermal cells, the proportion of cells in S and G2M phases remained lower, since they had reached their postmitotic stage. In contrast, the CCND1-induced cells consisted of the rapidly proliferating cell population (cells in S and G2M phases). Furthermore, the total period of culture, the passage number of cells, and assumptive total cell output all suggested that the CCND1-induced cells had long-term growth potential. These results demonstrated that the morphologic, phenotypic and growth characteristics of the CCND1-induced cells were very similar to that of epidermal stem cells, that was to say the overexpression of CCND1 induced the dedifferention of differentiated epidermal cells. This is in agreement with the earlier study that primary cultured keratinocytes in which Cyclin D1 expression was induced resistant to calcium-induced terminal differentiation and continued cell growth in vitro.34
In our laboratory, we have done a significant amount of work in wound repair and regeneration with stem cells.35-37 This work includes a relatively complete system for the isolation, culture, identification, and transplant of certain stem cells. We have also induced these stem cells to differentiate into many cell types, both in vitro and in vivo, and explored the possibilities of enhancing the healing and regeneration of skin injury by cell transplantation.7,38,39 The most significant characteristic of the epidermal stem cells is their ability to enhance the wound healing. In this experiment, CCND1-induced cells promoted wound closure by forming a stratified epidermal layer on the wound surface. In addition, tumor formation was not been observed in the wound. These results demonstrated that dedifferention-derived epidermal stem cells induced by CCND1 were the safe seed cells for wound treatment.
Collectively, we have reproduced the phenomenon of epidermal cell dedifferentiation by transgenic method. The differentiated epidermal cells induced by CCND1, have the morphological, phenotypical, and functional characteristics of epidermal stem cells. Furthermore, the dedifferention-derived epidermal stem cells can accelerate the wound healing safely. This provides us with a promising method of generating epidermal stem cells for wound repair and skin tissue engineering.
Materials and methods
Ethics statement: Tissues were obtained from human subjects after they gave their informed consent. The protocol was approved by the national ethics committee in China.
Cell culture
Differentiated human epidermal cells were isolated from epidermal sheets by differential adhesion. Human foreskin specimens were digested at 4°C with 2 mg/ml protease (Sigma, St. Louis, MO) for 10-12 h and then the epidermis was isolated from dermis. The isolated epidermis sheets were cut into pieces, digested with 0.25% trypsinase for 20 minutes at 37°C and made into single cell suspension. After centrifugation, the cells were gently resuspended in Epilife Medium supplemented with 1% human keratinocyte growth supplement (HKGS) (Cascade Biologics, Portland, OR) and seeded on collagen IV-coated 25 cm2 flasks (Costar, Corning, NY) at a density of 5 × 105 cells/cm2. After 3 h, non-adherent cells were gently removed to new collagen IV-coated culture flasks and cultured for another 6 h in a 37°C/5% CO2 incubator. A series of experiments were performed on subsequent adherent cells. Epidermal stem cells dispersed from full-thickness epidermal sheets by adhering to collagen IV for 10 min.40,41
Vector construction, cell transfection and flow sorting
We retrieved the CCND1 gene sequence from GenBank and designed the following primers: CCND1-F (Bgl2): GAagatctATGGAACACCAGCTCCTGTG, CCND1-R (Kpn1): GGggtaccGTGATGTCCACGTCCCGCAC. The eukaryotic expression plasmid pEGFP-N1 was digested with the restriction endonucleases Bgl2 and Kpn1. The CCND1 insert and linearized plasmid were joined by T4-DNA ligase to construct the plasmid pEGFP-N1-CCND1. The recombinant plasmids were identified by Bgl2/Kpn1 and gel electrophoresis. One fragment of approximately 1,000 bp was observed after digestion, which is the CCND1 gene.
Differentiated epidermal cells were divided into non transfection (G-non), empty vector transfection (G-empty) and CCND1 transfection (G-CCND1) groups. Epidermal stem cells were used as the positive control (G-positive). Untransfected cells were used as the negative control. Transfection was carried out using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After two days, transfected cells were collected for flow sorting and used to detect the expression of CCND1 by quantitative real-time PCR (RT-PCR).
Microscope
Cells from the 4 groups were plated into a 6-well plate (Costar, Corning, NY) whith 4 × 105 cells/ml. Inverted phase contrast microscope was used to image these cells.
Immunofluorescence
Cells from the 4 groups were fixed with 4% paraformaldehyde and then incubated with primary mouse monoclonal anti-human CK10 and β1-integrin antibodies (1:500; Santa Cruz) for 1 hour at room temperature. The primary antibodies were removed, and the cells were incubated for 30 min with phycoerthrin (PE)-conjugated goat anti-mouse IgG secondary antibody (1:1,000; Santa Cruz). The nuclei were counterstained with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) (Roche Diagnostics, Basel, Switzerland). Fluorescence images were visualized with a fluorescence microscope (Eclipse 80I; Nikon, Tokyo, Japan).
Quantitative real-time PCR
mRNA was isolated from the cultured cells in each group using the TRIzol kit (Invitrogen) and reverse-transcribed into cDNA with SuperScript III (Invitrogen) according to the manufacturer's instructions. Gene-specific primers for Oct4 (forward, 5′-GAAGTTGGAGAAGGTGGAACCA-3′; reverse, 5′-GCTTCAGCAGCTTGGCAAA-3′) and Nanog (forward, 5′-TCTTCCTGGTCCCCACAGTTT-3′; reverse, 5′-GCAAGAATAGTTCTCGGGATGAA-3′) CCND1 (forward, 5′-CTGTCCCACTCCTACGATACG-3′; reverse, 5′-CAGCATCTCATAAACAGGTCACTAC-3′),42 were optimized for expression analyses by real-time PCR on an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA). The reactions comprised ABsolute QPCR SYBR Green ROX mix (ABgene, Epsom, UK) and were incubated for 2 min at 50°C, 15 min at 95°C, and then 40 cycles of 15 s at 95°C, and 1 min at 60°C, followed by dissociation-curve analysis to confirm specificity. Expression of elongation factor (EF)-1α (forward, 5′-ATTCGAGACCAGCAAATACTATGTGA-3′; reverse, 5′-AGCCTGGGATGTGCCTGTAA-3′) was used for normalization, and the relative expression was calculated by using the comparative CT method.43,44 The CT data of control group are seen as “1” and the relative expressions of the target genes were calculated according to the control group by the CT data.
Cell cycle
We plated 6 × 104 cells/ml, from the above 4 groups, onto a 60mm culture dish (Costar), and cultured them for another 3 and 6 d. After fixation with 75% ethanol, single cell suspensions were digested with DNase-free RNase in PBS containing 5 μg/ml propidium iodide (Sigma) for DNA staining (45 min at 37°C). The propidium iodide fluorescence and forward light scattering were detected with a flow cytometer (FACS scan; Beckton Dickinson) equipped with cellquest (Largo, FL, USA) software. The percentage of cells in every phase of the cell cycle was calculated.
Long-term growth potential
Harvested cells from the 4 groups were seeded onto culture vessels at a density of 6.0 × 103 cells/cm2. When cell density reached 70-90% confluence, they were serially passaged (up to 1.5 × 106 cells seeded) until cells lost their proliferative capacity. The total days of culture, the passage number of cells, and assumptive total cell output were determined. As up to 1.5 × 106 cells were re-plated at each passage, the cell outputs were calculated based on the assumption that all the cells from the previous passage had been re-plated.
Colony-forming efficiency
We plated 4 × 103 cells, from the above 4 groups, into a 6-well plate (Costar), cultured them for another 7 d in Epilife Medium (changed 4 d post-plating). Under the microscope, colonies consisting of 10 or more cells were counted blindly.
Transplantation of human cells into BALB/c nude mice
BALB/c nude mice (No. 01030101) were supplied by the Institute of Experimental Animals, Chinese Academy of Science (CAS). Mice were housed in standard animal cages under controlled temperature and humidity with 12-hour light/dark cycles. Animal handling and experimentation was approved by the Animal Care and Use Review Committee of IOZ, CAS, and the Institute of Biological Products of Beijing. Mice were anesthetized with xylazine (5 mg/kg) and prepared for the wound model. Full-thickness circular wounds (1.5 cm) were generated down to the muscular layer on the back using a 1 cm punch biopsy. A total of 40 animals were randomized to 5 groups for treatment: control (no graft) and G-non, G-empty, G-CCND1 and G-positive grafting. Cells (6 × 106 cells) were suspended in 0.1 ml Epilife Medium, mixed with 0.1 ml cold Matrigel (BD Biosciences, USA), and incubated at 37°C for 30 minutes to allow gel formation. After gel formation, the cell mixtures were grafted to the wounds and overlaid with a piece of nonadherent dressing placed orthotopically. After 2 weeks, the dressings were removed and the mice were left undisturbed until 4 weeks postoperatively, when they were killed.
Evaluation of healing
The wound area was determined by computer planimetry. After 4 weeks, biopsies of tissue were performed and the specimens were harvested. To monitor the transplanted cells in the wound, tissue sections were incubated with primary mouse monoclonal anti-human β1-integrin antibody and stained with Hoechst 33342 (SigmaeAldrich, St. Louis, MO) according to the manufacturer's protocol.
Statistical analysis
The data were analyzed with SPSS software, Version 12.0 (SPSS Inc., Chicago, IL). All values are expressed as the mean ± standard deviation. The data were analyzed using one-way analysis of variance (ANOVA) and Newman–Keuls–Student's t test. A tied-P value of < 0.05 was considered statistically significant.
Abbreviations
- CCND1
cyclin D1
- CK10
cytokeratin 10
- DAPI
2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride
- EF-1α
elongation factor −1α
- EGFP
enhanced green fluorescent protein
- HKGS
human keratinocyte growth supplement
- iPSCs
induced pluripotent stem cells
- PE
phycoerthrin
- rhEGF
recombinant human epidermal growth factor
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported in part by the National Nature Science Foundation of China 81171798, 81571905, 81272104, 81471882, 81421064, 81230041), Beijing Municipal Natural Science Foundation (Grant No. 7142124) and the National Basic Science and Development Programme (973 Programme, 2012CB518105).
References
- [1].Kellouche S, Martin C, Korb G, Rezzonico R, Bouard D, Benbunan M, Dubertret L, Soler C, Legrand C, Dosquet C. Tissue engineering for full-thickness burns: a dermal substitute from bench to bedside. Biochem Biophys Res Commun 2007; 363:472-8; PMID:17888881; http://dx.doi.org/ 10.1016/j.bbrc.2007.08.155 [DOI] [PubMed] [Google Scholar]
- [2].Xie JL, Li TZ, Qi SH, Huang B, Chen XG, Chen JD. A study of using tissue-engineered skin reconstructed by candidate epidermal stem cells to cover the nude mice with full-thickness skin defect. J Plast Reconstr Aesthet Surg 2007; 60:983-90; PMID:17662463; http://dx.doi.org/ 10.1016/j.bjps.2005.12.062 [DOI] [PubMed] [Google Scholar]
- [3].Cotsarelis G, Kaur P, Dhouailly D, Hengge U, Bickenbach J. Epithelial stem cells in the skin: definition, markers, localization and functions. Exp Dermatol 1999; 8:80-8; PMID:10206725; http://dx.doi.org/ 10.1111/j.1600-0625.1999.tb00351.x [DOI] [PubMed] [Google Scholar]
- [4].Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008; 451:141-6; PMID:18157115; http://dx.doi.org/ 10.1038/nature06534 [DOI] [PubMed] [Google Scholar]
- [5].Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663-76; PMID:16904174; http://dx.doi.org/ 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- [6].Fu X, Sun X, Li X, Sheng Z. Dedifferentiation of epidermal cells to stem cells in vivo. Lancet 2001; 358:1067-8; PMID:11589942; http://dx.doi.org/ 10.1016/S0140-6736(01)06202-X [DOI] [PubMed] [Google Scholar]
- [7].Zhang CP, Chen P, Fei Y, Fu XB, Zhao ZL, Sun TZ, Sheng ZY. Wnt/b-catenin signaling is critical for dedifferentiation of aged epidermal cells in vivo and in vitro. Aging Cell 2012; 11:14-23; PMID:26009897; http://dx.doi.org/ 10.1111/j.1474-9726.2011.00753.x [DOI] [PubMed] [Google Scholar]
- [8].Zhao ZL, Zhang CP, Fu XB, Xang RY, Chen P, Gu TM, Sui ZF, Wang CM, Liu C. Differentiated epidermal cells regain the ability to regenerate a skin equivalent by increasing β-catenin expression. Cells Tissues Organs 2012; 196:353-61; PMID:22538966; http://dx.doi.org/ 10.1159/000335474 [DOI] [PubMed] [Google Scholar]
- [9].Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisúa Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009; 460:1140-4; PMID:19668186; http://dx.doi.org/ 10.1038/nature08311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009; 460:1136-9; PMID:19668188; http://dx.doi.org/ 10.1038/nature08290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009; 460:1145-8; PMID:19668190; http://dx.doi.org/ 10.1038/nature08285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Alteri A, De Vito F, Messina G, Pompili M, Calconi A, Visca P, Mottolese M, Presutti C, Grossi M. Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle 2013; 12:3781-90; PMID:24107628; http://dx.doi.org/ 10.4161/cc.26674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Edel MJ, Menchon C, Menendez S, Consiglio A, Raya A, Izpisua Belmonte JC. Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev 2010; 24:561-73; PMID:20231315; http://dx.doi.org/ 10.1101/gad.1876710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Redvers RP, Li A, Kaur P. Side population in adult murine epidermis exhibits phenotypic and functional characteristics of keratinocyte stem cells. Proc Natl Acad Sci U S A 2006; 103:13168-73; PMID:16920793; http://dx.doi.org/ 10.1073/pnas.0602579103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis 2005; 26:495-502; PMID:15513931; http://dx.doi.org/ 10.1093/carcin/bgh321 [DOI] [PubMed] [Google Scholar]
- [16].Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861-72; PMID:18035408; http://dx.doi.org/ 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- [17].Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell 2009; 136:364-77; PMID:19167336; http://dx.doi.org/ 10.1016/j.cell.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Daksis JI, Lu RY, Facchini LM, Marhin WW, Penn LJ. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene 1994; 9:3635-45; PMID:7526316 [PubMed] [Google Scholar]
- [19].Edel MJ, Izpisua Belmonte JC. The cell cycle and pluripotency: is there a direct link?. Cell Cycle 2010; 9:2694-5; PMID:20581443; http://dx.doi.org/ 10.4161/cc.9.14.12456 [DOI] [PubMed] [Google Scholar]
- [20].Hindley C, Philpott A. The cell cycle and pluripotency. Biochem J 2013; 451:135-43; PMID:23535166; http://dx.doi.org/ 10.1042/BJ20121627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 2002; 21:8320-33; PMID:12447695; http://dx.doi.org/ 10.1038/sj.onc.1206015 [DOI] [PubMed] [Google Scholar]
- [22].Ballabeni A, Park IH, Zhao R, Wang W, Lerou PH, Daley GQ, Kirschner MW. Cell cycle adaptations of embryonic stem cells. Proc Natl Acad Sci U S A 2011; 108:19252-7; PMID:22084091; http://dx.doi.org/ 10.1073/pnas.1116794108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang VS, Carter SA, Hyland SJ, Tachibana-Konwalski K, Laskey RA, Gonzalez MA. Geminin escapes degradation in G1 of mouse pluripotent cells and mediates the expression of Oct4, Sox2, and Nanog. Curr Biol 2011; 21:692-9; PMID:21497086; http://dx.doi.org/ 10.1016/j.cub.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen CL, Wang LJ, Yan YT, Hsu HW, Su HL, Chang FP, Hsieh PC, Hwang SM, Shen CN. Cyclin D1 acts as a barrier to pluripotent reprogramming by promoting neural progenitor fate commitment. FEBS Lett 2014; 588:4008-17; PMID:25261251; http://dx.doi.org/ 10.1016/j.febslet.2014.08.039 [DOI] [PubMed] [Google Scholar]
- [25].Li H, Fu X, Zhang L, Sun T, Wang J. In vivo dedifferentiation of human epidermal cells. Cell Biol Int 2007; 31:1436-41; PMID:17689109; http://dx.doi.org/ 10.1016/j.cellbi.2007.05.016 [DOI] [PubMed] [Google Scholar]
- [26].Sun X, Fu X, Han W, Zhao Y, Liu H, Sheng Z. Dedifferentiation of human terminally differentiating keratinocytes into their precursor cells induced by basic fibroblast growth factor. Biol Pharm Bull 2011; 34:1037-45; PMID:21720010; http://dx.doi.org/ 10.1248/bpb.34.1037 [DOI] [PubMed] [Google Scholar]
- [27].Huelsken J, Behrens J. The Wnt signalling pathway. J Cell Sci 2002; 115:3977-8; PMID:12356903; http://dx.doi.org/ 10.1242/jcs.00089 [DOI] [PubMed] [Google Scholar]
- [28].Takayama S, Rogatsky I, Schwarcz LE, Darimont BD. The glucocorticoid receptor represses cyclin D1 by targeting the Tcf-beta-catenin complex. J Biol Chem 2006; 281:17856-63; PMID:16644723; http://dx.doi.org/ 10.1074/jbc.M602290200 [DOI] [PubMed] [Google Scholar]
- [29].Larouche D, Hayward C, Cuffley K, Germain L. Keratin 19 as a stem cell marker in vivo and in vitro. Methods Mol Biol 2005; 289:103-10; PMID:15502175 [DOI] [PubMed] [Google Scholar]
- [30].Pesce M, Schöler HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 2001; 19:271-8; PMID:11463946; http://dx.doi.org/ 10.1634/stemcells.19-4-271 [DOI] [PubMed] [Google Scholar]
- [31].Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003; 113:631-42; PMID:12787504; http://dx.doi.org/ 10.1016/S0092-8674(03)00393-3 [DOI] [PubMed] [Google Scholar]
- [32].Papini S, Cecchetti D, Campani D, Fitzgerald W, Grivel JC, Chen S, Margolis L, Revoltella RP. Isolation and clonal analysis of human epidermal keratinocyte stem cells in long-term culture. Stem Cells 2003; 21:481-94; PMID:12832701; http://dx.doi.org/ 10.1634/stemcells.21-4-481 [DOI] [PubMed] [Google Scholar]
- [33].Islam MS, Zhou H. Isolation and characterization of putative epidermal stem cells derived from Cashmere goat fetus. Eur J Dermatol 2007; 17:302-8; PMID:17540636 [DOI] [PubMed] [Google Scholar]
- [34].Yamamoto H, Ochiya T, Takeshita F, Toriyama-Baba H, Hirai K, Sasaki H, Sasaki H, Sakamoto H, Yoshida T, Saito I, et al.. Enhanced skin carcinogenesis in cyclin D1-conditional transgenic mice: cyclin D1 alters keratinocyte response to calcium-induced terminal differentiation. Cancer Res 2002; 62:1641-7; PMID:11912134 [PubMed] [Google Scholar]
- [35].Wang Y, Liu ZY, Zhao Q, Sun TZ, Ma K, Fu XB. Future application of hair follicle stem cells: capable in differentiation into sweat gland cells. Chin Med J (Engl) 2013; 126:3545-52; PMID:24034106 [PubMed] [Google Scholar]
- [36].Dong L, Hao H, Xia L, Liu J, Ti D, Tong C, Hou Q, Han Q, Zhao Y, Liu H, et al.. Treatment of MSCs with Wnt1a-conditioned medium activates DP cells and promotes hair follicle regrowth. Sci Rep 2014; 4:5432; PMID:24961246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Fu X. Wound care study and translation application: a team's work in China. Int J Low Extrem Wounds 2014; 13:84-87; PMID:24872470; http://dx.doi.org/ 10.1177/1534734614535406 [DOI] [PubMed] [Google Scholar]
- [38].Zhang C, Chen Y, Fu X. Sweat gland regeneration after burn injury: is stem cell therapy a new hope?. Cytotherapy 2015; 17:526-35; PMID:25533933; http://dx.doi.org/ 10.1016/j.jcyt.2014.10.016 [DOI] [PubMed] [Google Scholar]
- [39].Zhang Y, Hao H, Liu J, Fu X, Han W. Repair and regeneration of skin injury by transplanting microparticles mixed with Wharton's jelly and MSCs from the human umbilical cord. Int J Low Extrem Wounds 2012; 11:264-70; PMID:23089966; http://dx.doi.org/ 10.1177/1534734612463577 [DOI] [PubMed] [Google Scholar]
- [40].Zhang C, Fu X, Chen P, Bao X, Li F, Sun X, Lei Y, Cai S, Sun T, Sheng Z. Dedifferentiation derived cells exhibit phenotypic and functional characteristics of epidermal stem cells. J Cell Mol Med 2010; 14:1135-45; PMID:19426155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu Y, Zhou H, Gao F. Isolation and identification of stem cells from adult cashmere goat skin. Int J Dermatol 2008; 47(6):551-6; PMID:18477142; http://dx.doi.org/ 10.1111/j.1365-4632.2008.03647.x [DOI] [PubMed] [Google Scholar]
- [42].Han K, Chen X, Bian N, Ma B, Yang T, Cai C, Fan Q, Zhou Y, Zhao TB. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget 2015; 6:8875-89; PMID:25823925; http://dx.doi.org/ 10.18632/oncotarget.3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- [44].Zhao Z, Xu M, Wu M, Ma K, Sun M, Tian X, Zhang C, Fu X. Direct reprogramming of human fibroblasts into sweat gland-like cells. Cell Cycle 2015; 14:3498-505; PMID:26566868; http://dx.doi.org/ 10.1080/15384101.2015.1093707 [DOI] [PMC free article] [PubMed] [Google Scholar]