ABSTRACT
Submitted: TP63 (p63), a member of the tumor suppressor TP53 (p53) gene family, is expressed in keratinocyte stem cells and well-differentiated squamous cell carcinomas to maintain cellular potential for growth and differentiation. Controversially, activation of the Wnt/β-catenin signaling by p63 (Patturajan M. et al., 2002, Cancer Cells) and inhibition of the target gene expression (Drewelus I. et al., 2010, Cell Cycle) have been reported. Upon p63 RNA-silencing in squamous cell carcinoma (SCC) lines, a few Wnt target gene expression substantially increased, while several target genes moderately decreased. Although ΔNp63α, the most abundant isoform of p63, appeared to interact with protein phosphatase PP2A, neither GSK-3β phosphorylation nor β-catenin nuclear localization was altered by the loss of p63. As reported earlier, ΔNp63α enhanced β-catenin-dependent luc gene expression from pGL3-OT having 3 artificial Wnt response elements (WREs). However, this activation was detectable only in HEK293 cells examined so far, and involved a p53 family-related sequence 5′ to the WREs. In Wnt3-expressing SAOS-2 cells, ΔNp63α rather strongly inhibited transcription of pGL3-OT. Importantly, ΔNp63α repressed WREs isolated from the regulatory regions of MMP7. ΔNp63α-TCF4 association occurred in their soluble forms in the nucleus. Furthermore, p63 and TCF4 coexisted at a WRE of MMP7 on the chromatin, where β-catenin recruitment was attenuated. The combined results indicate that ΔNp63α serves as a repressor that regulates β-catenin-mediated gene expression.
KEYWORDS: β-catenin, p63, TCF4, TP63, Wnt
Introduction
TP63 (p63), a TP53 (p53)-related gene, is expressed in keratinocyte stem cells to maintain the cellular potential of proliferation and keratinocyte differentiation.1-5 It is essential for morphogenesis of ectodermal tissues including skin, glands, head-and-neck, limb and urinary tracts.6,7 Mutations of this gene cause the EEC and related syndromes.8,9 Furthermore, the chromosome 3q amplification in head-and-neck squamous cell carcinomas (SCCs) underlies the high level expression of p63 (3q27-q29).10,11 This gene produces at least 6 proteins termed variant 1-6 (National Center for Biotechnology Information, USA). Due to the 2 different transcription initiation sites, the TA-isoforms (variant 1-3) with the trans-activation domain and the ΔN-isoforms (variant 4-6) without TA are produced. For each N-terminal variant, alternative RNA splicing causes 3 different C-terminal structures, α, β, and γ 1.
TAp63γ, originally identified as p51A, has a trans-activating ability similar to p53 to act on the same consensus sequences.1,3,12 Many of the direct target genes determined for TAp63γ are involved in cell-cell and cell-matrix interactions.12-17 In contrast to the lower level expression of TAp63γ, ΔNp63α (variant 4) is the most abundant isoform detected in the basal layer stem cells, SCCs, and cancers originating from basal cells of ectoderm.13,18-21 ΔNp63α comprises DNA binding domain, oligomerization domain and sterile α motif with which various proteins can interact.22 Although initial studies experimentally identified ΔNp63α as a dominant negative-type protein against TAp63γ and p53,1 the trans-activating ability of TAp63γ seems vital in keratinocytes and SCCs. The dominant negative-type action may not be the only function of ΔNp63α.23,24
Because invasion of SCCs coincides with a steep decline in p63 expression, maintenance of the well-differentiated status by p63 has been proposed.23-26 As well-documented for colorectal and hepatocellular carcinomas, carcinogenesis and malignant progression are often accompanied by somatic mutations resulting in Wnt signal activation.27-29 Head-and-neck SCCs, however, rarely have a mutation in the major factors such as APC and CTNNB1 (β-catenin).10 The possibilities of positive and negative regulation of Wnt/β-catenin signaling by p63 has been proposed in earlier studies. Patturajan M. et al. reported activation of the Wnt signaling to accumulate β-catenin through protein phosphatase 2A (PP2A) inhibition by ΔNp63α.30 On the other hand, Drewelus I. et al. proposed that p63 blocks β-catenin-induced transcription.31 The authors detected a specific interaction between ΔNp63α and the HMG box of TCF1, TCF3, TCF4, and LEF1 by a pulldown assay.
Confusingly, however, these reports concurred in one point that ΔNp63α enhances luc gene expression from the prototype reporter plasmids in HEK293 cells. TOPflash (referred to as Lef1:luciferase reporter plasmid by Patturajan et al.30) and pGL3-OT (referred to as TOPflash by Drewelus et al.31) have 3 copies of artificial Wnt response element (WRE),32 while superTOPflash has 8 repeats. Moreover, the impacts of ΔNp63α on the chromosomal WRE sequences and the assembly of TCFs/LEF and β-catenin at the transcriptionally functional WREs have not been investigated.
Our gene expression profiling of SCC lines showed substantial alterations in target genes of p53 and p63, and basal layer keratinocyte-specific genes by p63 knockdown. It was of interest that some Wnt target genes were activated by p63-silencing, while some others were down-regulated. These results, in conjunction with the above described conflicting reports, led us to deeply investigate the influence of p63 over the Wnt/β-catenin signaling pathway and the target gene expression. We reexamined the reporter gene expression assay, and the signaling proteins in the cytosol and nucleus. Furthermore, we tested endogenous WRE sequences upstream of the Wnt/β-catenin target genes for their sensitivity to β-catenin and p63. Eliminating the ambiguity caused by the reporter assay, our results strongly suggest that β-catenin-mediated gene expression is impaired by ΔNp63α in SCCs. This study provides new evidence for the prediction by Drewelus I. et al.,31 and offers deeper insights into the function of p63.
Results
Alteration of Wnt target gene expression by p63 RNA silencing
FaDu cells are derived from a hypopharyngeal carcinoma, and expresses ΔNp63α with other p63 isoforms.25,30 Based on the Catalogue of Somatic Mutations in Cancer (COSMIC) database (Sanger Institute, UK), this cell line has a missense mutation (c.743G>T, p.R248L) in TP53 and an intronic mutation (c.151-1G>T) in CDKN2A (cyclin-dependent kinase inhibitor 2A, also termed p14ARF/p16INK4a). No mutation related to the canonical Wnt signaling has been identified in these cells so far.
We performed gene expression profiling with FaDu cells transfected with p63-specific siRNA (p63si) and control siRNA (Csi). p63 RNA was decreased to 1/4 – 1/6.5 in p63si-transfected cells compared with Csi-transfected cells, indicating efficient RNA silencing (Table 1). NOTCH, JAG2, COL7A1, CLDN1, and DST among the reported p63-target genes,33 were obviously downregulated by p63 silencing in varied magnitudes. Concerning the TP53 target genes,34 suppression of GADD45A and BAX35 and activation of P53INP136 and P53INP237 were evident in p63 knockout cells, implying involvement of the trans-activating and dominant-negative functions of the p63 isoforms. Furthermore, expression of the basal cytokeratin genes, KER14 (K14), and KER5 (K5), decreased with p63-silencing, consistent with the notion that p63 is specifically expressed in the basal layer of keratinocytes and well-differentiated SCCs. Of interest was that some of the Wnt target genes, AXIN2/CONDUCTIN,38 MITF 39 and MMP7 (matrix metalloproteinase-7)40 were upregulated by p63-silencing, whereas some others including CCND2 (cyclin D2)41 and SNAI2/SLUG42 were down-regulated.
Table 1.
Alteration of gene expression by p63 silencing in squamous carcinoma FaDu cells.
| Experiment |
1 |
2 |
3 |
|---|---|---|---|
| Fold change (p63si versus Csi)* | |||
| p63 | |||
| TP63 | -6.519 | -3.904 | -4.355 |
| Reported p63 target genes | |||
| NOTCH1** | -3.187 | -1.609 | -1.967 |
| JAG2** | -3.339 | -1.583 | -1.654 |
| COL7A1** | -2.826 | -3.068 | -2.954 |
| JAG1** | -2.812 | -1.678 | -1.565 |
| CLDN1** | -2.082 | -1.048 | -1.207 |
| DST** | -1.339 | -3.071 | -3.391 |
| p53 target genes | |||
| GADD45A | -6.761 | -3.333 | -2.083 |
| BAX | -5.534 | -1.176 | -1.078 |
| TP53INP1 | ND | +3.518 | +3.978 |
| TP53INP2 | ND | +2.780 | +1.180 |
| Keratinocyte-specific genes | |||
| LCE5A | -6.338 | +1.502 | -1.819 |
| KRT5 (basal) | -3.250 | -2.849 | -2.849 |
| KER17(basal) | -3.382 | -2.108 | -2.108 |
| KER16 | -3.496 | -1.083 | -1.083 |
| KRT14** (basal) | -3.030 | -2.240 | -2.240 |
| Wnt target genes | |||
| AXIN2 | +14.411 | +7.302 | +4.311 |
| MMP7 | +5.469 | +7.221 | +5.989 |
| MITF | +4.432 | +2.681 | +2.914 |
| DKK1 | ND | +2.190 | +2.753 |
| SNAI2 (SLUG) | -5.877 | -3.323 | -3.045 |
| CCND2 | -3.489 | -2.353 | -2.391 |
| LBH | -3.415 | -1.822 | +1.039 |
| NEGFA | -3.087 | -2.897 | -2.426 |
| JAG1** | -2.812 | -1.678 | -1.565 |
| MYC | -2.628 | -1.760 | -2.228 |
| CLDN1 | -2.082 | -1.048 | -1.207 |
| TWIST1 | -1.343 | -1.327 | -4.047 |
| EDN3 | -1.746 | -2.188 | -1.400 |
*Upregulation (+) and downregulation (-) are indicated.
**reported p63 target gene.
ND, not determined.
We confirmed these changes in the Wnt target gene expression by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and Western blotting (Fig. 1). p63 proteins decreased to less than 15% in p63si-transfected cells, while total p63 RNA decreased to 40% when quantified with our primers. RNA of CONDUCTIN/AXIN2 and MMP7 was increased by 2.5-fold and 3-fold, respectively (Fig. 1A), while SNAI2/SLUG and CCND2 was reduced to 80% and 50%, respectively. Western blotting also showed 3.5-fold increase in the MMP-7 protein and 2.5-fold decrease in the CCND2 protein (Fig. 1B).
Figure 1.
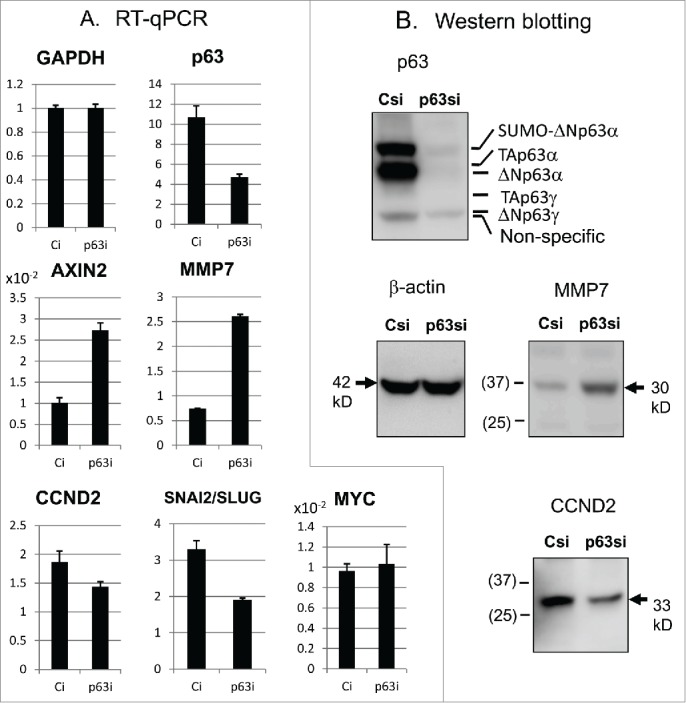
Alteration of the Wnt/β-catenin target gene expression by p63-silencing.(A) Expression of indicated genes was quantified by RT-qPCR. (B) Western blot analyses for the proteins. Positions of p63 isoforms and smolylated (SUMO) p63 are also shown.25 Molecular masses of the standard proteins are marked in parentheses (in kD).
Interaction of ΔNp63α with PP2A phosphatase
We next analyzed proteins in the Wnt/β-catenin signaling pathway focusing on GSK-3β and β-catenin phosphorylation by cell fractionation and immunoprecipitation (Fig. 2). The FaDu cell cytosol fractions were enriched in the PP2A catalytic subunit (PP2A-C), while the nuclear fractions showed localization of the PP2A B56α (PPP2R5A) regulatory subunit (Fig. 2A). An antibody against PP2A-C (clone ID6) precipitated not only PP2A-C but also PP2A-B56α. As reported earlier,43 ΔNp63α existed in the PP2A-C immune complex from the control (Csi) nucleus, but not from the p63 knockdown nucleus (p63si) (Fig. 2A). After PP2A-C gene (PPP2CA, PPP2CB) silencing with siRNA (PPsi), immunoprecipitation was carried out with the PP2A-C antibody (Fig. 2B). When the PP2A-C protein amount decreased to ˜25% in the immunoprecipitate (IP, PPsi) as well as in the nuclear fraction (input, PPsi), the amount of co-precipitatied ΔNp63α decreased concomitantly.
Figure 2.
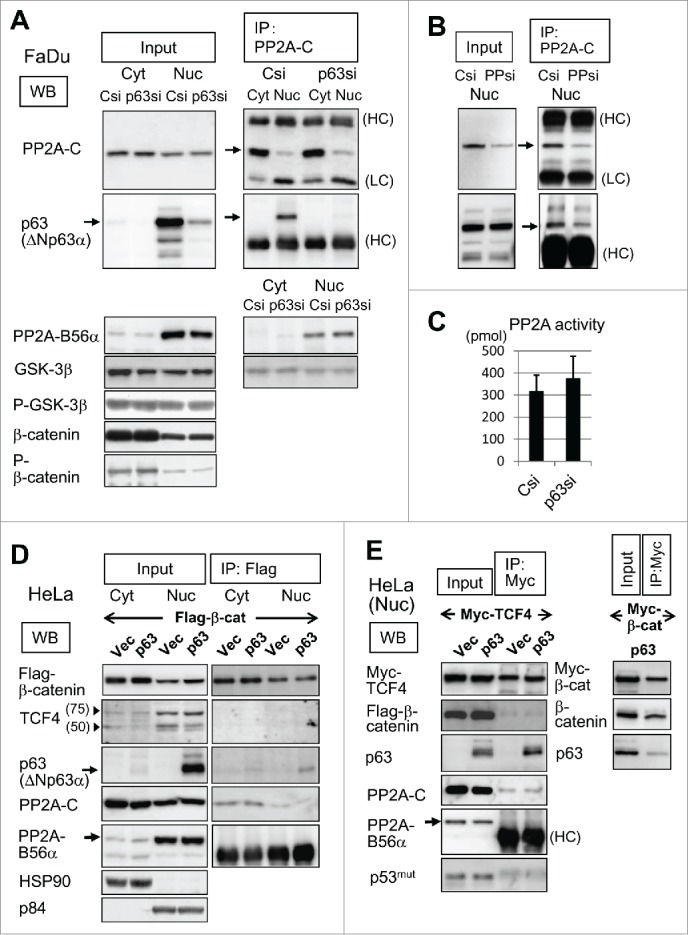
Analyses of the interactions of PP2A, p63 and Wnt/β-catenin signaling proteins. (A) Csi and p63si-transfected FaDu cells were fractionated to the cytosol (Cyt) and nuclear (Nuc) extracts. Cytosol and nuclear samples were loaded on the gel in a ratio of 1:2. Proteins detected in each fraction (Input) are shown. Immunoprecipitates (IP) by a PP2A-C antibody (mouse IgG) were subjected to Western blotting (WB) for proteins indicated. ΔNp63α detected by a p63 monoclonal antibody, 4A4, is marked by arrow. IgG heavy chain (HC) and light chain (LC) are shown. (B) PP2A-C gene silencing and immunoprecipitation. Nuclear extracts from FaDu cells transfected with siRNA targeting PP2A-C (PPsi) and Csi were analyzed as in (A). Western blotting for detection of PP2A-C and p63 are shown. (C) PP2A phosphatase activity in Csi and p63si-transfected FaDu cells. Released phosphate amounts are shown in picomoles (pmol). (D) HeLa cells were transfected with Flag-β-catenin in combination with ΔNp63α or the vector plasmid, fractionated, and immunoprecipitated with an anti-Flag antibody (rabbit IgG). Obtained fractions (Input) and the immunoprecipitates (IP) were analyzed for indicated proteins. Arrowheads indicate the positions of 75 kD and 50 kD standard proteins. (E) HeLa cells were cotransfected with ΔNp63α, Myc-TCF4 and Flag-β-catenin as in (D). The nuclear extracts (Input) and immunoprecipitates (IP) with an anti-Myc antibody (rabbit IgG) were analyzed. Myc-tagged TCF4 appeared as a single band of ˜80 kD. Control experiment was carried out by cotransfection of ΔNp63α and Myc-tagged β-catenin (Myc-β-cat) followed by nuclear fractionation and immunoprecipitation with the anti-Myc antibody (right panels)
We performed a PP2A phosphatase assay in which the release of phosphate from a phospho-threonine peptide substrate was quantified. The PP2A activity in the whole-cell extracts did not substantially change with p63-silencing (Fig. 2C). GSK-3β (a substrate of PP2A) and β-catenin (a substrate of GSK-3β) showed no significant alteration in the protein amount, phosphorylation or subcellular localization (Fig. 2A, lower panels). Immunofluorescence analysis showed dense stain of β-catenin at the cell periphery and its spread throughout the cytosol. The nuclei were stained only weakly. No significant difference appeared between the p63-knockdown and control cells regarding β-catenin fluorescence intensity and distribution (Supplementary Information-1).
Association of ΔNp63α with TCF4
To further analyze protein interactions between Wnt signaling proteins and p63, we introduced flag-tagged β-catenin in combination with ΔNp63α or the empty expression vector into HeLa cells. Flag-β-catenin was immunoprecipitated with an anti-flag antibody from the cytosol and nuclear extracts (Fig. 2D). Fractionation efficiency was assessed with HSP90 and p84, representing the cytosol and nuclear proteins, respectively. The flag-β-catenin immune complex did not contain endogenous TCF4 observed as a triplet including a major band of 75 kD corresponding to the longest form of the splicing variants, nor was ΔNp63α associated with flag-β-catenin. Furthermore, ΔNp63α did not influence β-catenin nuclear translocation. In addition, PP2A-C and PP2A-B56α interacted with Flag-β-catenin only poorly, if at all.
When Myc-tagged TCF4 expression plasmid was co-transfected with ΔNp63α or the vector plasmid, immunoprecipitation with an anti-Myc antibody revealed a strong association of Myc-TCF4 with ΔNp63α in the nucleus (Fig. 2E). PP2A-B56α and PP2A-C were poorly detectable in the precipitates regardless of ΔNp63α. Furthermore, we did not found p53 or Flag-β-catenin by the anti-myc immunoprecipitation. As a control experiment, Myc-tagged β-catenin expression plasmid was cotransfected with ΔNp63α (Fig. 2E, right panels). The anti-Myc antibody efficiently precipitated Myc-β-catenin, but not ΔNp63α. We thus confirmed the specific interaction between ΔNp63α and TCF4.31
luc reporter assay with pGL3-OT
To assess the influence of p63 on the β-catenin-mediated gene expression, a luc gene reporter assay was performed with pGL3-OT, pGL3-OF and the truncated forms (Fig. 3A-C). Consistent with the previous reports, ΔNp63α dose-dependently enhanced luc expression from pGL3-OT only when activated by β-catenin in HEK293 cells (Fig. 3D). Cotransfection of TCF4 (25 ng of the plasmid in each well) neither positively nor negatively influenced the response to β-catenin (25 ng) with p63 (0-50 ng) (Supplementary Information 2A). Among the p63 isoforms transfected, ΔNp63α exhibited the strongest β-catenin-enhancing ability with pGL3-OT (Supplementary Information-2B). When the maximum transcriptional activation (˜50-fold) was achieved with a S33Y β-catenin mutant (20-50 ng/105 cells), ΔNp63α could not further increase the luc expression (Supplementary Information-2C). pGL3-OF which has 2 nucleotide mutations in each WRE did not respond to β-catenin or ΔNp63α (Fig. 3E).
Figure 3.
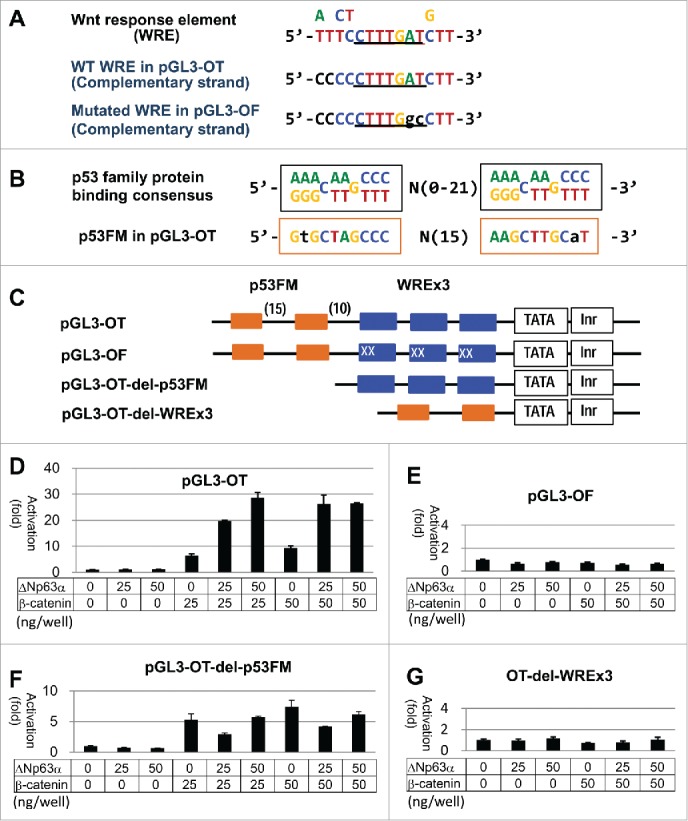
luc expression assay with pGL3-OT in HEK293. (A) Proposed WRE consensus sequences32 and WREs in pGL3-OT and pGL3-OF are aligned. (B) p53 binding consensus sequence and p53FM are aligned. (C) Enhancer structures of pGL3-OT and the mutants are shown. Blue boxes represent WREs and related stretches, and orange boxes p53 consensus half-sites. Letter X indicates the mutated nucleotide in pGL3-OF. Nucleotide numbers between the elements are in parentheses. (D)-(G) Results of the luc assays with indicated plasmids at 48 hr of transfection. Amounts (ng) of regulator plasmids (p63, β-catenin) are indicated for each reaction. Luciferase activities are shown in relation to the control reaction with the empty vector (1.0).
Intriguingly, we found a repeat of 10 nucleotide sequences corresponding to the originally proposed half site of p53 binding motif 44 immediately 5′ to the WREs. We tentatively termed it p53FM (p53 family protein binding motif), because it was recognizable by p63 proteins12,33 (Fig. 3B). Deletion of p53FM from pGL3-OT preserved its sensitivity to β-catenin, but canceled the transcriptional activation by ΔNp63α (Fig. 3F). The p53FM sequence per se showed neither positive nor negative response to ΔNp63α (Fig. 3G).
SAOS-2 is a p53-null osteosarcoma cell line,45 by which functions of p53 family proteins are sensitively monitored (Supplementary Information-3A).46 In addition, these cells constantly express a higher level of Wnt3a, β-catenin and LEF1, which maintain the cellular signaling and growth potential.47 luc expression from pGL3-OT was boosted to 10-fold and 50-fold by transfection of 50 and 67 ng of β-catenin-encoded plasmid, respectively, in each well (Fig. 4A). TCF4-expression plasmid (25 ng in each reaction) was contained in every reaction with SAOS-2 cells, by which the level of transcriptional activation by β-catenin (50 ng) was 3-fold elevated. (Compare panel A with Supplementary Information-3B.) pGL3-OT-del-p53FM responded to β-catenin at nearly the same sensitivity (Fig. 4B). Unexpectedly, ΔNp63α strongly inhibited luc expression from pGL3-OT and pGL3-OT-del-p53FM when induced by β-catenin transfection in SAOS-2 cells. ΔNp63α also decreased the basal level transcription from these luc plasmids without β-catenin transfection.
Figure 4.
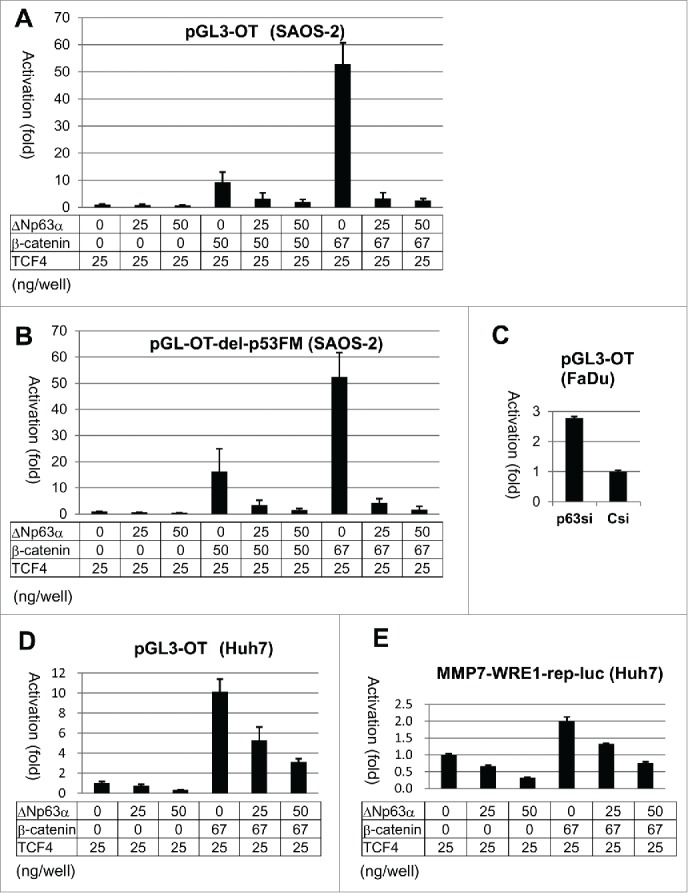
luc expression assay with pGL3-OT in SAOS-2, Huh7 and FaDu cells. Results of the luc assays with pGL3-OT (A) and del-p53FM (B) in SAOS-2 cells. After transfection of FaDu cells with Csi and p63si (for 24 hr), pGL3-OT was introduced. Luciferase activity was measured at 48 hr of the luc plasmid transfection (C). Huh7 cells were transfected with pGL3-OT (D) and MMP7-WRE1-rep-luc (E) in combination with ΔNp63α, β-catenin and TCF4. Luc activities are indicated as in Figure 3.
In hepatocellular carcinoma Huh7 cells, luc expression from pGL3-OT was also decreased by ΔNp63α in the persistent and β-catenin-induced conditions (Fig. 4D). Furthermore, p63-silencing in FaDu cells caused a 2.8-fold increase in the luciferase activity (Fig. 4C). Thus, these results obtained with pGL3-OT in SAOS-2, Huh7 and FaDu cells were consistent with the result that endogenous MMP7 and AXIN2 were activated by p63-silencing (Table 1, Fig. 1).
Repression of chromosomal WREs by p63
To explore the transcriptional control of Wnt target gene by ΔNp63α, we analyzed endogenous WREs for their activities and regulation. Using the proposed consensus sequences,32 we detected several WREs in the 10 kb regulatory regions of MMP7 and CCND2, and cloned 3 segments containing repetitive WREs into a reporter plasmid (pGL3-basic promoter vector). The MMP7-WRE1 segment located 5.6 kb upstream from the initiation site had a pair of WRE core sequences, 5′-YCTTTGAT-3′, with one nucleotide mismatch in the second WRE (Fig. 5A). The region containing the known WREs at −109 and −194 (reference 40) was referred to as MMP7-WRE2 in this study (Fig. 5B). At 2.2 kb upstream of CCND2, we identified an interesting segment termed CCND2-WREb, which comprised a palindromic WRE repeat preceded by 3 half-sites (10 nucleotides) of p53 binding consensus (Fig. 5C). We also constructed MMP7-WRE1-rep-luc and CCND2-WREb-rep-luc by inserting an additional copy of the corresponding WRE pair.
Figure 5.
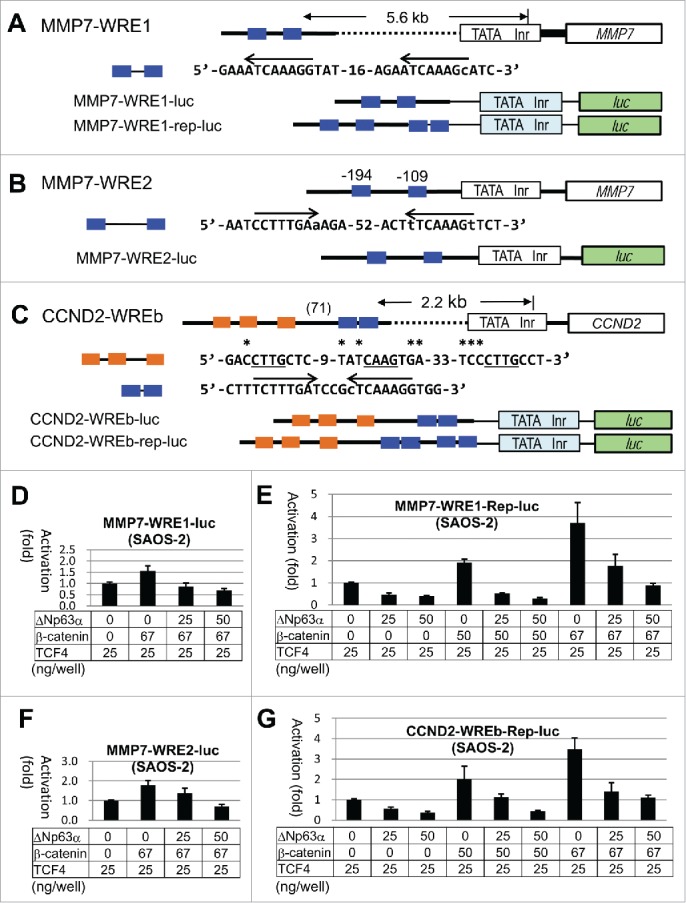
Structural and functional analyses of WREs of MMP7 and CCND2.(A)-(C), Line drawings and nucleotide sequences for the WREs analyzed in this study. Endogenous promoter region containing TATA box (TATA) and initiator site (Inr), and the body of the gene are shown by white boxes. Bold lines mark the endogenous sequences, while the thin lines mark sequences in the plasmids. The light blue and green boxes signify the promoter and the luc gene in pGL3-promoter, respectively. Nucleotides matching the WRE consensus 32 in the positive and negative strands are indicated by arrows. Nucleotides deviated from the consensus are in lower case. (D)-(G), Results of the luc assays with SAOS-2 cells using the reporter plasmids shown in (A)-(C). Transfected plasmids and the DNA amounts are shown for each experiment.
In SAOS-2 cells, both MMP7-WRE1-luc and MMP7-WRE2-luc caused only a 1.5-1.8-fold increase when co-transfected with the β-catenin expression plasmid (67 ng in each well) (Fig. 5D, F). This moderate activation was canceled by ΔNp63α. The basal level transcription was also affected. The MMP7-WRE1-rep-luc and CCND2-WREb-rep-luc plasmids showed 2-fold and 3-4-fold activation by cotransfection with 50 and 67 ng of the β-catenin plasmid, respectively (Fig. 5E, G). ΔNp63α suppressed the luc expression from these plasmids in a dose-related manner.
Huh7 with an endogenous Wnt signaling activity48 caused only 2-fold activation of MMP7-WRE1-rep-luc when transfected with β-catenin (67 ng). ΔNp63α decreased the luciferase activity in the persistent and β-catenin-transfected conditions (Fig. 4E). In HEK293, luc expression from MMP7-WRE1-luc was slightly (20%) increased by β-catenin, which was inhibited by ΔNp63α. Only a 50% increase was found with CCND2-WREb-rep-luc upon induction by active form S33Y β-catenin, which was also blocked by ΔNp63α (data not shown). Thus, ΔNp63α repressed the endogenous WREs in SAOS-2, Huh7 and HEK293 cells. The sequences related to p53 half-sites in CCND-WREb did not act like p53FM that allowed positive regulation of WREs by ΔNp63α in HEK293.
Association of p63 with MMP7-WRE1 on the chromatin
Chromatin immunoprecipitation was carried out with Csi and p63si-transfected FaDu cells. Sheared chromatin containing 200-800 bp DNA fragments were immunoprecipitated with anti-p63, anti-TCF4, anti-β-catenin and control IgG. Recovered DNA fragments were quantified by qPCR with primers targeting MMP7-WRE1 and CCND2-WREb (Fig. 6). Both p63 and TCF4 were detected at the MMP7-WRE1 site in Csi-transfected cells (Fig. 6A). By p63 RNA silencing, p63 protein at MMP7-WRE1 was significantly decreased, which was accompanied by an approximately 2.5-fold increase of β-catenin protein bound to the site. Thus, p63 coexisted with TCF4 at MMP7-WRE1, by which recruitment of β-catenin was impaired. However, none of the immunoprecipitates from Csi or p63si-transfected cells contained the CCND2-WREb segment, suggesting that this site on the chromatin does not function as a Wnt/β-catenin target site in FaDu cells (Fig. 6B).
Figure 6.
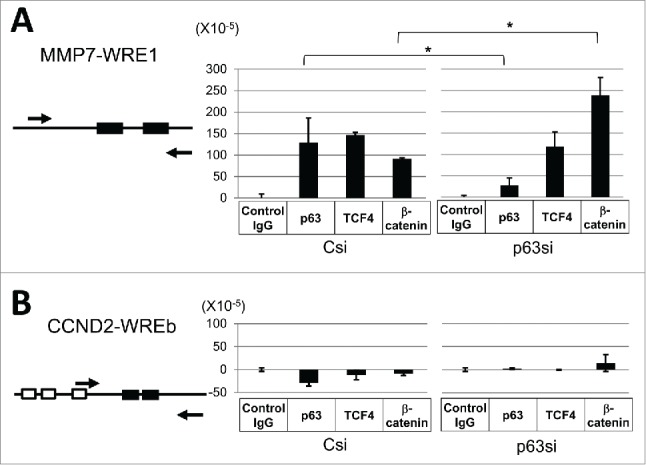
ChIP analysis for the MMP7-WRE1 and CCND2-WREb sites in FaDu.Relative positions of the primers used in the PCR are shown for MMP7-WRE1 (A, left) and CCND2-WREb (B, left). Filled and open boxes indicate WREs and p53 consensus half-sites, respectively. The amount of DNA precipitated by each antibody was quantified by PCR. After subtraction of the background value obtained with control IgG, DNA copy numbers relative to the input DNA (1.0) were determined (A, B, right). Statistic significance: *, 0.01 < P < 0.05
Discussion
This study strongly suggests that p63 serves as a repressor of WREs to attenuate Wnt/β-catenin target gene expression. Gene expression profiling of p63 knockdown cells implied negative regulation of Wnt/β-catenin target genes by p63. ΔNp63α suppressed luc expression driven by cloned endogenous WREs with β-catenin. Furthermore, p63-TCF4 association in the nuclear extract, coexistence of p63 with TCF4 at a chromosomal WRE site, and interference of β-catenin binding to the target site by p63 may explain the mechanism of transcriptional downregulation.
As used in many studies, pGL3-OT in combination of HEK293 cells provides a standard reporter assay for analyses of Wnt/β-catenin signaling pathway activation. With this system, however, we observed the opposite outcome regarding the p63 function. At least p53FM in the plasmid seemed to contribute to the reaction. TOPflash and SuperTOPflash, the earlier versions of pGL3-OT, also have sequences related to p53FM at a position close to the WRE repeats, which may have caused the contradiction in previous studies.30,31 Drewelus et al. hypothesized a switching mechanism between the positive and negative regulation depending on the p63 concentration.31 However, we failed to detect it in the luc assays with different amounts of the ΔNp63α expression plasmid.
HEK293 cells were transformed by adenovirus type-5 E1A and E1B genes and constitutively express them. The small E1A oncoprotein interacts with Rb, p300/CBP, etc. to cause genome-wide transcriptional and epigenetic changes,49 while E1B 55 kD protein binds p53 to block the function. These viral proteins potentially influence p63 and nuclear factors of the Wnt signaling pathway. Hypothetically, the (p53FM)-(WREs) structure of pGL3-OT might allow ΔNp63α, TCF and β-catenin to form an activating complex under the influence of E1A and/or E1B. However, there has been so far no evidence for a chromosomal gene controlled by the (p53FM)-(WREs)-type enhancer. Intriguingly, the twin gene of Xenopus laevis has a tandem arrangement of (Smad binding sites)-(Lef1/Tcf binding sites), which is activated synergistically by Smad, Lef1 and β-catenin.50
Wnt3a-expressing p53-null SAOS-2 cells provided a sensitive assay system for β-catenin-driven luc expression. ΔNp63α evidently repressed luc expression from pGL3-OT. Importantly, the WRE repeats cloned from the MMP7 and CCND2 regulatory regions also positively responded to β-catenin albeit weakly (4-fold at most), and were repressed by ΔNp63α. Experiments with Huh7 and FaDu cells supported this result. In addition to the previously identified WREs at −109 and −194 (collectively termed WRE2 in this study) of MMP7, WRE1 at −5.4 kb was also able to mediate β-catenin-induced transcriptional activation.
In the previously proposed model, formation of a quadruple protein complex of p63, TCF/LEF, β-catenin and unidentified repressor molecule was hypothesized.31 Our ChIP experiment suggests that interaction of p63 with TCF4 bound to MMP7-WRE1 reduces the accessibility of β-catenin to TCF4. Although the antibody against p63 does not discriminate p63 isoforms, ΔNp63α most likely represents the precipitated p63 isoforms based on its abundance and ability to form a complex with TCF4 as detected by immunoprecipitation (Fig. 2D). It remains to be investigated whether or not ΔNp63α requires corepressor Groucho/TLE1 for the control of β-catenin.
Among the TCF/LEF family proteins, we focused on TCF4. Tcf3 and Tcf4 are expressed in keratinocyte stem cells, and play essential roles for skin homeostasis in mice.51 Furthermore, TCF4 binding sequences were extensively studied in colon cancer cells52,53 and found to match the proposed WREs.32,54
The Wnt/β-catenin target genes moderately downregulated in p63 knockdown cells, including CCND2 and JAG1, may not be governed by the Wnt signaling pathway in FaDu. Whereas CCND2-WREb responded to β-catenin in the reporter assay, TCF4, p63 and β-catenin were missing at the site on the chromosome of FaDu. JAG1 is not only a Wnt target gene but also a p63 target gene.16 The interaction between ΔNp63α and PP2A B56α found in the previous 30 study might be functional in some way, apart from the Wnt signaling. Ruptier et al. reported that the ΔNp63 promoter is activated by β-catenin in human hepatocellular carcinomas.55 In fact, HepG2 and Huh7 cells express ΔNp63 isoforms poorly in comparison with SCC lines. In some cellular contexts, ΔNp63α might play a negative feedback function to limit the ΔNp63 transcription.
MMP7 was identified as a target gene of TCF-4/β-catenin, being overexpressed in colorectal cancers with Wnt signaling pathway activation.40 Matrix metalloproteinase-7 catalyzes breakdown of extracellular matrix proteins, and is frequently expressed in the phase of invasion and metastasis of gastric and renal carcinomas.56 57 In SCCs, its transcriptional suppression may be abolished by the loss of p63 with the malignant conversion. Although Axin2 was originally found as a negative regulator of Wnt/β-catenin signaling,58 a recent study observed a tumor promoting activity of the protein in colorectal cancers.59 Thus, the WRE-repressing function of p63 provides an explanation for the generally accepted notion that SCCs gain a malignant phenotype when p63 is diminished.
Materials and methods
Cell lines
FaDu (HTB-43) and SAOS-2 (HTB-85) were from American Type Culture Collection. HEK293 (JCRB9068), HeLa (JCRB9004) and Huh7 (JCRB0403) were from Japanese Collection of Research Bioresources.
RNA interference
siRNA transfection was described.25 Anti-p63 siRNA (p63si) consisted of IMXRU (sense RNA, 5′-ggacguauuccacugaacutt-3′; antisense RNA, 5′-aguucaguggaauacgucctt-3′) and CUBCP (sense RNA, 5′-gcacugaauucacgacagutt-3′; antisense RNA, 5′-acugucgugaauucagugctt-3′). Control siRNA Csi (AM4636, Ambion) was from (Thermo Fisher Scientific). Stealth siRNA (HSS108360, HSS108361) targeting PP2A-C genes (PPP2CA, PPP2CB) were from Invitrogen (Thermo Fisher Scientific).
Gene expression profiling
RNA was obtained from cells transfected with Csi and p63si, and clarified with RNeasy MinElute Cleanup Kit (Qiagen). DNA microarray analysis was performed by Oncomics with the system from Agilent technologies including Quick Amp labeling kit, 2-color (5190-0444), Agilent RNA Spike-In kit, 2-color (5188-5279), Whole Human Genome Microarray kit (version 2.0), Gene Expression Hybridization kit (5188-5242) and GeneSpring GX software (version12.5.0).
qRT-PCR
RNA was purified with High Pure RNA Isolation kit (Roche). Random primed reverse transcription was with RevertAid Reverse Transcriptase (Thermo Fisher Scientific). PCR was performed with DyNAmo ColorFlash SYBR Green qPCR kit in the PikoReal Real-Time PCR System (Thermo Fisher Scientific). Primers were: hGAPDH-F2 (5′-acaactttggtatcgtggaagg-3′), hGAPDH-R2 (5′-gccatcacgccacagtttc-3′), p63ALL-F (5′-ccctccaacaccgactaccc-3′), p63ALL-R (5′-caccgcttcaccacctccgt-3′), MMP7-F2 (5′-gagtgagctacagtgggaaca-3′), MMP7-R2 (5′-ctatgacgcgggagtttaacat-3′), AXIN2-F1 (5′-caacaccaggcggaacgaa-3′), AXIN2-R1 (5′-gcccaataaggagtgtaaggact-3′), CCND2-F (5′-accttccgcagtgctccta-3′), CCND2-R(5′- cccagccaagaaacggtcc-3′), SNAI2-F2 (5′-cgaactggacacacatacagtg-3′), SNAI2-R2 (5′-ctgaggatctctggttgtggt-3′) c-Myc-F (5′-aaaggcccccaaggtagtta-3′) and c-Myc-F (5′-aaaggc ccccaaggtagtta-3′).
Antibodies used for immunoprecipitation and Western blotting
A PP2A-C subunit antibody (clone ID6, 05-421) and a TCF4 antibody (05-511) were from Merck Millipore. A PPR2R5A (B56α) antibody (Ab72028), a nuclear protein p83 antibody (Ab487) and a Myc tag antibody (ab9106) were from Abcam. Cell Signaling Technology supplied antibodies against GSK-3β (#9315), phospho-GSK-3β (Ser9) (#9323), C-terminal β-catenin (#9587), phospho-β-Catenin (Ser33/37) (#2009) and DYKDDDDK (#8146). Also supplied were alkaline phophatase-conjugated secondary antibodies against mouse IgG (#7056) and rabbit IgG (#7054). Santa Cruz Biotechnology (Dallas, TX) supplied anti-MMP7 (sc-80205), anti-Cyclin D2 (sc-181) and anti-p53 (Pab1801, sc-98) antibodies. An anti-p63 monoclonal antibody (4A4) was from Santa Cruz Biotechnology (sc-8431) and also from Abcam (Ab735). An anti-DDDK tag antibody from Origene (TA50011-5) was also used. Images obtained with the Immun-Star Chemiluminescent Protein Detection system (Bio-Rad) were captured by ECL minicamera and ImageQuant LAS 4000mini (GE Healthcare).
PP2A enzyme assay
PP2A Immunoprecipitation Phosphatase Assay Kit (Millipore) was used for immunoprecipitation and enzyme activity quantification. One reaction contained PP2A from sonicated cell lysate (aliquot corresponding to 2.5×105 cells). Released phosphate amounts were measured by a malachite green colorimetric assay. Experiments were performed 4 times with 2 technical replicates.
Plasmids
pGL3-OT, pGL3-OF, wild-type β-catenin cloned in pCIneo, S33Y β-catenin cloned in pCIneo, Flag-tagged β-catenin cloned in pSG5 (MF66), and Myc-TCF4 cloned in pcDNA were described.60 CMV promoter-driven p63 expression plasmids were described.13 Deletion mutants, del-p53FM and del-WREx3 were constructed by MluI-PstI and PstI-BglII digestion, respectively, followed by ligation with synthetic nucleotides minimizing the sequences to be deleted. The MMP7-WRE1 sequences were obtained from genomic DNA by PCR with primers MluI-MMP7-WRE1-F1 (5′-cttacgcgtaaccggggctgaataactct-3′) and BglII-MMP7-WRE1-R1 (5′-gaaagatctactgccaaatccaaggtcac-3′), and inserted at the MluI-BglII sites of the pGL3-promoter vector (Promega, Madison, WI). The CCND2-WREb sequences were amplified with primers MluI-CCND2-WREb-F1 (5′-cttacgcgtgggtggaagagaccttgctc-3′) and BglII-CCND2-WREb-R1 (5′-gaaagatcttttgagtcaccccggataag-3′) to be inserted at the same sites. The region covering MMP7-WRE2, TATA box and initiation site was amplified with MMP7-Amp-F3 (5′-cttacgcgtaatttatgcagcagacagaaaaa-3′) and MMP7-Amp-R3 (5′-cgcagatcttgttcttggacctatggttga-3′), and inserted at the MluI-BglII site of pGL3-OT after excising the p53FM-WREx3-promoter region.
Luciferase reporter assay
Plasmids were transfected with Effectene transfection reagent (Qiagen). Cells were plated in 24-well plates at 5×104−105 cells/well 24 hr before transfection. At 48 hr of transfection cells were harvested. Luciferase assay was performed with the luciferase assay systems and Steady-Glo luciferase assay system in combination with Glo lysis buffer (Promega). The enzyme activity was quantified with Lumat LB9507 (Perkin Elmer,). Experiments were performed in 2 biological replicates with 3 technical replicates.
Chromatin Immunoprecipitation (ChIP)
We used the ChIP-IT High Sensitivity kit (Active Motif, Carlsbad, CA). ChIP-validated antibodies for p63 (39739, Active Motif), TCF4 (17-10109, Millipore) and β-catenin (#9587, Cell Signaling Technology) were applied. Pretreated ChIP IgG (4 μg) and sheared chromatin (10 μg) were incubated overnight at 4ºC, combined with washed protein G-agarose beads, and incubated for additional 3 hr. Purified DNA was quantified by qPCR with primers: MMP7-WRE1-F1 (5′-aaccggggctgaataactct-3′), MMP7-WRE1-R1 (5′-actgccaaatccaaggtcac-3′), CCND2-WREb-F2 (5′-ttgcctgtcgggttagattt-3′) and CCND2-WREb-R2 (5′-tttgagtcaccccggataag-3′).
Supplementary Material
Abbreviations
- SCC
squamous cell carcinoma
- WRE
Wnt response element
- PP2A
protein phosphatase 2A
- ChIP
chromatin immunoprecipitation
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by Japan Society for Promotion of Sciences (JSPS) in Grant Numbers 24592827, 15K11090 (to SK) and 25460475 (to IK).
References
- [1].Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2:305-16; PMID:9774969; http://dx.doi.org/ 10.1016/S1097-2765(00)80275-0 [DOI] [PubMed] [Google Scholar]
- [2].Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, Ponzin D, McKeon F, De Luca M. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci U S A 2001; 98:3156-61; PMID:11248048; http://dx.doi.org/ 10.1073/pnas.061032098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, et al.. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med 1998; 4:839-43; PMID:9662378; http://dx.doi.org/ 10.1038/nm0798-839 [DOI] [PubMed] [Google Scholar]
- [4].Senoo M, Pinto F, Crum CP, McKeon F. p63 Is Essential for the Proliferative Potential of Stem Cells in Stratified Epithelia. Cell 2007; 129:523-36; PMID:17482546; http://dx.doi.org/ 10.1016/j.cell.2007.02.045 [DOI] [PubMed] [Google Scholar]
- [5].Keyes WM, Pecoraro M, Aranda V, Vernersson-Lindahl E, Li W, Vogel H, Guo X, Garcia EL, Michurina TV, Enikolopov G, et al.. ΔNp63α is an oncogene that targets chromatin remodeler lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 2011; 8:164-76; PMID:21295273; http://dx.doi.org/ 10.1016/j.stem.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al.. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398:714-8; PMID:10227294; http://dx.doi.org/ 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- [7].Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999; 398:708-13; PMID:10227293; http://dx.doi.org/ 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- [8].Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, et al.. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 1999; 99:143-53; PMID:10535733; http://dx.doi.org/ 10.1016/S0092-8674(00)81646-3 [DOI] [PubMed] [Google Scholar]
- [9].Brunner HG, Hamel BC, Van Bokhoven H. The p63 gene in EEC and other syndromes. J Med Gen 2002; 39:377-81; PMID:12070241; http://dx.doi.org/ 10.1136/jmg.39.6.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cancer Genome Atlas N . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015; 517:576-82; PMID:25631445; http://dx.doi.org/ 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci U S A 2000; 97:5462-7; PMID:10805802; http://dx.doi.org/ 10.1073/pnas.97.10.5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Osada M, Park HL, Nagakawa Y, Yamashita K, Fomenkov A, Kim MS, Wu G, Nomoto S, Trink B, Sidransky D. Differential Recognition of Response Elements Determines Target Gene Specificity for p53 and p63. Mol Cell Biol 2005; 25:6077-89; PMID:15988020; http://dx.doi.org/ 10.1128/MCB.25.14.6077-6089.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kurata S, Okuyama T, Osada M, Watanabe T, Tomimori Y, Sato S, Iwai A, Tsuji T, Ikawa Y, Katoh I. p51/p63 Controls subunit alpha3 of the major epidermis integrin anchoring the stem cells to the niche. J Biol Chem 2004; 279:50069-77; PMID:15361520; http://dx.doi.org/ 10.1074/jbc.M406322200 [DOI] [PubMed] [Google Scholar]
- [14].Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci 2007; 104:3255-60; http://dx.doi.org/ 10.1073/pnas.0611376104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 2006; 8:551-61; PMID:16715076; http://dx.doi.org/ 10.1038/ncb1420 [DOI] [PubMed] [Google Scholar]
- [16].Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T. The p53 Family Member Genes Are Involved in the Notch Signal Pathway. J Biol Chem 2002; 277:719-24; PMID:11641404; http://dx.doi.org/ 10.1074/jbc.M108080200 [DOI] [PubMed] [Google Scholar]
- [17].Osada M, Nagakawa Y, Park HL, Yamashita K, Wu G, Sook Kim M, Fomenkov A, Trink B, Sidransky D. p63-Specific Activation of the BPAG-1e Promoter. J Investig Dermatol 2005; 125:52-60; PMID:15982302; http://dx.doi.org/ 10.1111/j.0022-202X.2005.23801.x [DOI] [PubMed] [Google Scholar]
- [18].Quade BJ, Yang A, Wang Y, Sun D, Park J, Sheets EE, Cviko A, Federschneider JM, Peters R, McKeon FD, et al.. Expression of the p53 homologue p63 in early cervical neoplasia. Gynecol Oncol 2001; 80:24-9; PMID:11136565; http://dx.doi.org/ 10.1006/gyno.2000.5953 [DOI] [PubMed] [Google Scholar]
- [19].Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR, Pietenpol JA, Carbone DP, et al.. Significance of p63 Amplification and Overexpression in Lung Cancer Development and Prognosis. Cancer Res 2003; 63:7113-21; PMID:14612504 [PubMed] [Google Scholar]
- [20].Parsa R, Yang A, McKeon F, Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J Invest Dermatol 1999; 113:1099-105; PMID:10594758; http://dx.doi.org/ 10.1046/j.1523-1747.1999.00780.x [DOI] [PubMed] [Google Scholar]
- [21].Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. PNAS 2000; 97:5462-7; PMID:10805802; http://dx.doi.org/ 10.1073/pnas.97.10.5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sathyamurthy A, Freund SMV, Johnson CM, Allen MD, Bycroft M. Structural basis of p63α SAM domain mutants involved in AEC syndrome. FEBS Journal 2011; 278:2680-8; PMID:21615690; http://dx.doi.org/ 10.1111/j.1742-4658.2011.08194.x [DOI] [PubMed] [Google Scholar]
- [23].Giacobbe A, Compagnone M, Bongiorno-Borbone L, Antonov A, Markert EK, Zhou JH, Annicchiarico-Petruzzelli M, Melino G, Peschiaroli A. p63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene 2015; PMID:26119942; http://dx.doi.org/ 10.1038/onc.2015.230 [DOI] [PubMed] [Google Scholar]
- [24].Higashikawa K, Yoneda S, Tobiume K, Taki M, Shigeishi H, Kamata N. Snail-Induced Down-Regulation of {Delta}Np63{alpha} Acquires Invasive Phenotype of Human Squamous Cell Carcinoma. Cancer Res 2007; 67:9207-13; PMID:17909026; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-0932 [DOI] [PubMed] [Google Scholar]
- [25].Fukunishi N, Katoh I, Tomimori Y, Tsukinoki K, Hata R, Nakao A, Ikawa Y, Kurata S. Induction of DeltaNp63 by the newly identified keratinocyte-specific transforming growth factor beta Signaling Pathway with Smad2 and IkappaB Kinase alpha in squamous cell carcinoma. Neoplasia 2010; 12:969-79; PMID:21170261; http://dx.doi.org/ 10.1593/neo.101054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 Leads to Increased Cell Migration and Up-regulation of Genes Involved in Invasion and Metastasis. Cancer Res 2006; 66:7589-97; PMID:16885358; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-2020 [DOI] [PubMed] [Google Scholar]
- [27].Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006; 25:7531-7; PMID:17143297; http://dx.doi.org/ 10.1038/sj.onc.1210059 [DOI] [PubMed] [Google Scholar]
- [28].Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J, et al.. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013; 23:1422-33; PMID:23788652; http://dx.doi.org/ 10.1101/gr.154492.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yeang C-H, McCormick F, Levine A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J 2008; 22:2605-22; PMID:18434431; http://dx.doi.org/ 10.1096/fj.08-108985 [DOI] [PubMed] [Google Scholar]
- [30].Patturajan M, Nomoto S, Sommer M, Fomenkov A, Hibi K, Zangen R, Poliak N, Califano J, Trink B, Ratovitski E, et al.. DeltaNp63 induces beta-catenin nuclear accumulation and signaling. Cancer Cell 2002; 1:369-79; PMID:12086851; http://dx.doi.org/ 10.1016/S1535-6108(02)00057-0 [DOI] [PubMed] [Google Scholar]
- [31].Drewelus I, Gopfert C, Hippel C, Dickmanns A, Damianitsch K, Pieler T, Dobbelstein M. p63 antagonizes Wnt-induced transcription. Cell Cycle (Georgetown, Tex) 2010; 9:580-87; PMID:20107313; http://dx.doi.org/ 10.4161/cc.9.3.10593 [DOI] [PubMed] [Google Scholar]
- [32].Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol 2007; 27:8352-63; PMID:17893322; http://dx.doi.org/ 10.1128/MCB.02132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Perez CA, Pietenpol JA. Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle (Georgetown, Tex) 2007; 6:246-54; PMID:17297308; http://dx.doi.org/ 10.4161/cc.6.3.3801 [DOI] [PubMed] [Google Scholar]
- [34].Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9:402-12; PMID:18431400; http://dx.doi.org/ 10.1038/nrm2395 [DOI] [PubMed] [Google Scholar]
- [35].Bian J, Sun Y. p53CP, a putative p53 competing protein that specifically binds to the consensus p53 DNA binding sites: A third member of the p53 family? Proc Nat Acad Sci 1997; 94:14753-8; http://dx.doi.org/ 10.1073/pnas.94.26.14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou Y, Zhang E, Berggreen C, Jing X, Osmark P, Lang S, Cilio CM, Göransson O, Groop L, Renström E, et al.. Survival of pancreatic beta cells is partly controlled by a TCF7L2-p53-p53INP1-dependent pathway. Hum Mole Gen 2012; 21:196-207; PMID:21965303; http://dx.doi.org/ 10.1093/hmg/ddr454 [DOI] [PubMed] [Google Scholar]
- [37].Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pébusque M-J, Vaccaro MI, Velasco G, Dagorn J-C, Iovanna JL. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell 2009; 20:870-81; PMID:19056683; http://dx.doi.org/ 10.1091/mbc.E08-07-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lim X, Tan SH, Koh WLC, Chau RMW, Yan KS, Kuo CJ, van Amerongen R, Klein AM, Nusse R. Interfollicular Epidermal Stem Cells Self-Renew via Autocrine Wnt Signaling. Science 2013; 342:1226-30; PMID:24311688; http://dx.doi.org/ 10.1126/science.1239730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Takeda K, Yasumoto K, Takada R, Takada S, Watanabe K, Udono T, Saito H, Takahashi K, Shibahara S. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J Biol Chem 2000; 275:14013-6; PMID:10747853; http://dx.doi.org/ 10.1074/jbc.C000113200 [DOI] [PubMed] [Google Scholar]
- [40].Brabletz T, Jung A, Dag S, Hlubek F, Kirchner T. β-Catenin Regulates the Expression of the Matrix Metalloproteinase-7 in Human Colorectal Cancer. Am J Pathol 1999; 155:1033-8; PMID:10514384; http://dx.doi.org/ 10.1016/S0002-9440(10)65204-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Huang W, Chang HY, Fei T, Wu H, Chen YG. GSK3 beta mediates suppression of cyclin D2 expression by tumor suppressor PTEN. Oncogene 2007; 26:2471-82; PMID:17043650; http://dx.doi.org/ 10.1038/sj.onc.1210033 [DOI] [PubMed] [Google Scholar]
- [42].Lambertini E, Franceschetti T, Torreggiani E, Penolazzi L, Pastore A, Pelucchi S, Gambari R, Piva R. SLUG: a new target of lymphoid enhancer factor-1 in human osteoblasts. BMC Mol Biol 2010; 11:13; PMID:20128911; http://dx.doi.org/ 10.1186/1471-2199-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Patturajan M, Nomoto S, Sommer M, Fomenkov A, Hibi K, Zangen R, Poliak N, Califano J, Trink B, Ratovitski E, et al.. [Delta]Np63 induces [beta]-catenin nuclear accumulation and signaling. Cancer Cell 2002; 1:369-79; PMID:12086851; http://dx.doi.org/ 10.1016/S1535-6108(02)00057-0 [DOI] [PubMed] [Google Scholar]
- [44].el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet 1992; 1:45-9; PMID:1301998; http://dx.doi.org/ 10.1038/ng0492-45 [DOI] [PubMed] [Google Scholar]
- [45].Berglind H, Pawitan Y, Kato S, Ishioka C, Soussi T. Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol Ther 2008; 7:699-708; PMID:18277095; http://dx.doi.org/ 10.4161/cbt.7.5.5712 [DOI] [PubMed] [Google Scholar]
- [46].Tomimori Y, Katoh I, Kurata S, Okuyama T, Kamiyama R, Ikawa Y. Evolutionarily conserved expression pattern and trans-regulating activity of Xenopus p51/p63. Biochem Biophys Res Commun 2004; 313:230-6; PMID:14684151; http://dx.doi.org/ 10.1016/j.bbrc.2003.11.113 [DOI] [PubMed] [Google Scholar]
- [47].Ma Y, Ren Y, Han EQ, Li H, Chen D, Jacobs JJ, Gitelis S, O'Keefe RJ, Konttinen YT, Yin G, et al.. Inhibition of the Wnt-beta-catenin and Notch signaling pathways sensitizes osteosarcoma cells to chemotherapy. Biochem Biophys Res Commun 2013; 431:274-9; PMID:23291185; http://dx.doi.org/ 10.1016/j.bbrc.2012.12.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Austinat M, Dunsch R, Wittekind C, Tannapfel A, Gebhardt R, Gaunitz F. Correlation between beta-catenin mutations and expression of Wnt-signaling target genes in hepatocellular carcinoma. Mol Cancer 2008; 7:21; PMID:18282277; http://dx.doi.org/ 10.1186/1476-4598-7-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. Epigenetic reprogramming by adenovirus e1a. Science 2008; 321:1086-8; PMID:18719284; http://dx.doi.org/ 10.1126/science.1155546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nishita M, Hashimoto MK, Ogata S, Laurent MN, Ueno N, Shibuya H, Cho KWY. Interaction between Wnt and TGF-[beta] signalling pathways during formation of Spemann's organizer. Nature 2000; 403:781-5. [DOI] [PubMed] [Google Scholar]
- [51].Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet 2009; 41:1068-75; PMID:19718027; http://dx.doi.org/ 10.1038/ng.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, et al.. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol 2008; 28:2732-44; PMID:18268006; http://dx.doi.org/ 10.1128/MCB.02175-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of β-catenin binding regions in colon cancer cells using ChIP-Seq. Nucl Acids Res 2010; 38:5735-45; PMID:20460455; http://dx.doi.org/ 10.1093/nar/gkq363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cadigan KM, Waterman ML. TCF/LEFs and Wnt Signaling in the Nucleus. Cold Spring Harbor Perspect Biol 2012; 4:a007906; PMID:23024173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ruptier C, De Gasperis A, Ansieau S, Granjon A, Taniere P, Lafosse I, Shi H, Petitjean A, Taranchon-Clermont E, Tribollet V, et al.. TP63 P2 promoter functional analysis identifies [beta]-catenin as a key regulator of [Delta]Np63 expression. Oncogene 2011; 30:4656-65; PMID:21643019; http://dx.doi.org/ 10.1038/onc.2011.171 [DOI] [PubMed] [Google Scholar]
- [56].Yamashita K, Azumano I, Mai M, Okada Y. Expression and tissue localization of matrix metalloproteinase 7 (matrilysin) in human gastric carcinomas. Implications for vessel invasion and metastasis. Int J Cancer 1998; 79:187-94; http://dx.doi.org/ 10.1002/(SICI)1097-0215(19980417)79:2%3c187::AID-IJC15%3e3.0.CO;2-7 [DOI] [PubMed] [Google Scholar]
- [57].Miyata Y, Iwata T, Ohba K, Kanda S, Nishikido M, Kanetake H. Expression of Matrix Metalloproteinase-7 on Cancer Cells and Tissue Endothelial Cells in Renal Cell Carcinoma: Prognostic Implications and Clinical Significance for Invasion and Metastasis. Clin Cancer Res 2006; 12:6998-7003; PMID:17145820; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-1626 [DOI] [PubMed] [Google Scholar]
- [58].Jho E-H, Zhang T, Domon C, Joo C-K, Freund J-N, Costantini F. Wnt/β-Catenin/Tcf Signaling Induces the Transcription of Axin2, a Negative Regulator of the Signaling Pathway. Mol Cell Biol 2002; 22:1172-83; PMID:11809808; http://dx.doi.org/ 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wu Z-Q, Brabletz T, Fearon E, Willis AL, Hu CY, Li X-Y, Weiss SJ. Canonical Wnt suppressor, Axin2, promotes colon carcinoma oncogenic activity. Proceed Natl Acad Sci 2012; 109:11312-7; http://dx.doi.org/ 10.1073/pnas.1203015109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fujimuro M, Hayward SD. The Latency-associated nuclear antigen of kaposi's sarcoma-associated herpesvirus manipulates the activity of glycogen synthase kinase-3β. J Virol 2003; 77:8019-30; PMID:12829841; http://dx.doi.org/ 10.1128/JVI.77.14.8019-8030.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


