ABSTRACT
Intestinal divisions in Caenorhabditis elegans take place in 3 stages: (1) cell divisions during embryogenesis, (2) binucleations at the L1 stage, and (3) endoreduplications at the end of each larval stage. Here, we report that CDC-25.2, a C. elegans ortholog of Cdc25, is required for these specialized division cycles between the 16E cell stage and the onset of endoreduplication. Results of our genetic analyses suggest that CDC-25.2 regulates intestinal cell divisions and binucleations by counteracting WEE-1.3 and by activating the CDK-1/CYB-1 complex. CDC-25.2 activity is then repressed by LIN-23 E3 ubiquitin ligase before the onset of intestinal endoreduplication, and this repression is maintained by LIN-35, the C. elegans ortholog of Retinoblastoma (Rb). These findings indicate that timely regulation of CDC-25.2 activity is essential for the progression of specialized division cycles and development of the C. elegans intestine.
KEYWORDS: Binucleation, C. elegans, cdc-25.1, cdc-25.2, cell-cycle regulation, endoreduplication, intestinal division, lin-23, lin-35, nuclear division
Introduction
Cell cycle regulation is essential during development. Therefore, defects in cell cycle regulation are often teratogenic and may cause various diseases such as cancer. Understanding of the molecular mechanisms regulating the cell cycle may potentially lead to development of treatments for such diseases. Caenorhabditis elegans is an ideal model organism for studying cell cycle regulation during organ development because it has an invariant cell lineage and a well-defined anatomy. The intestine is involved in multiple fundamental biological processes, such as food digestion, nutrition distribution, immunity, stress response, and aging.1-4 All intestinal cells of C. elegans originate from a single embryonic blastomere, E, which undergoes 4 rounds of nearly symmetrical cell divisions to produce 16 intestinal cells, the 16E cells. 16E cells exist as pairs of bilateral cells, and the structure of each cell pair is called an “int ring.” Thus, there are 8 int rings, int1 to int8, at the 16E cell stage. The structural framework of the intestine is established at this 16E cell stage.5-7 Thereafter, additional cell divisions occur in the anterior-most int1 cells and the posterior-most int8 cells. As a result, the int1 ring consists of 4 cells through dorsal/ventral division, and the int8 ring divides into 2 new int rings, int8 and int9, through anterior/posterior division; in total, the intestine comprises 20 cells in 9 int rings before hatching.6-8 After hatching, intestinal cells further undergo specialized division cycles. At the first larval stage (L1), some intestinal nuclei undergo nuclear divisions without cytokinesis, resulting in binucleated cells, during a process called binucleation. Int1 and int2 cells never binucleate, while int3-7 cells always binucleate, and int8 and int9 cells occasionally binucleate.9 Therefore, worms in the late L1 stage have 30 to 34 intestinal nuclei in 20 cells. Subsequently, all intestinal nuclei undergo endoreduplication before each larval molting, which double DNA content without nuclear division and cytokinesis.10 Since intestinal nuclei endoreduplicate 4 times in the 4 larval stages, they finally contain 32C DNA content (1C is the amount of DNA content equivalent to a haploid genome set) in the adult stage. Taken together, the intestinal divisions of C. elegans involve transition of the cell cycle mode from cell division, binucleation, to endoreduplication. Therefore, studies of intestinal divisions in C. elegans may reveal not only the regulatory mechanisms by which respective division modes are controlled, but also the mechanisms by which their timely transitions are regulated.
Cdc25 phosphatase promotes cell cycle progression through dephosphorylation of cyclin-dependent kinases (Cdks) that are phosphorylated, and thus inactivated, by Wee1/Myt1 kinases.11,12 cdc-25.1 gain-of-function (gf) mutants displayed intestinal hyperplasia, while cdc-25.1 loss-of-function (lf) mutants exhibited defects in embryonic intestinal divisions and in germline proliferation.13-18 Prolonged CDC-25.1 activity in cdc-25.1(gf) mutants caused extra cell divisions between the 8E and the 16E cell stages of embryonic intestinal development.16,19 In contrast, division patterns were not affected before the 8E and after the 16E cell stages. This indicates that timely inactivation of CDC-25.1 is required for normal intestinal development. The cdc-25.1(gf) mutant phenotype also suggests that, after the 16E cell stage, another member of the CDC-25 family plays a pivotal role in intestinal division. There are 4 cdc25 homologs in C. elegans: cdc-25.1, cdc-25.2, cdc-25.3, and cdc-25.4.20 We previously reported that cdc-25.2 regulates oocyte maturation.21 Also it has been reported that RNAi of cdc-25.2 resulted in embryonic defects in neuroblasts, ABarp lineage, and C and E lineages.22 cdc-25.3 and cdc-25.4 mRNAs are expressed but their functions are yet largely unknown. Cdc25 phosphatases are evolutionarily conserved from yeast to mammals.20,23-26 Although yeast has one cdc25, fruit flies, mice, and humans have multiple cdc25 family members that function in distinct developmental stages. Drosophila has 2 cdc25 homologs, string and twine.23,26 string regulates mitotic cell cycles during embryogenesis and germline development, while twine regulates meiotic cell cycles in the germline.27,28 In mammals, there are 3 cdc25 family members, CDC25A, CDC25B, and CDC25C.24,25 CDC25A predominantly functions during the G1 to S phase transition,29 while CDC25B and CDC25C regulate the G2 to M phase transition.30,31 Nevertheless, mice lacking both Cdc25B and Cdc25C develop almost normally without any obvious abnormalities except female sterility caused by lack of Cdc25B. This indicates that, although the functions of Cdc25B and Cdc25C are distinct from that of Cdc25A during development, Cdc25A can largely compensate for their functions when they are absent.32
In this study, we report that C. elegans CDC-25.2 solely regulates intestinal cell divisions and binucleations after the 16E cell stage by counteracting WEE-1.3 and by activating the CDK-1/CYB-1 complex. After that, CDC-25.2 activity is repressed by LIN-23 before the onset of intestinal endoreduplications, and this negative regulation is maintained by LIN-35. These findings indicate that CDC-25.2 has a unique function during C. elegans intestinal development and its timely regulation is essential for the transition between specialized division cycles during the development.
Results
Deletion of cdc-25.2 causes an arrest of intestinal division at the 16E cell stage
To explore the roles of cdc-25 family genes during C. elegans intestinal development, we scored the number of intestinal nuclei in each deletion mutant (Table 1). To facilitate the analysis, an intestine-specific reporter transgene, rrIs01[elt-2::GFP],15,16,33 was introduced into each mutant strain used in this study. When each cdc-25 gene was knocked out, only the deletion mutant of cdc-25.2, ok597, showed a decreased number of intestinal nuclei compared to wild type (Table 1).
Table 1.
The number of intestinal nuclei per worm in respective genotypes.
| Number of intestinal nuclei |
||
|---|---|---|
| Genotype | L1 | Adult |
| wild type | 20.0 ± 0.4 (n = 22) | 31.5 ± 1.8 (n = 22) |
| cdc-25.1(nr2036) | n.d. | 33.2 ± 1.0 (n = 12) |
| cdc-25.2(ok597) | 16.0 ± 0 (n = 22) | 16.0 ± 0 (n = 23) |
| cdc-25.3(ok358) | n.d. | 31.3 ± 1.8 (n = 15) |
| cdc-25.4(tm4088) | n.d. | 32.0 ± 1.3 (n = 16) |
| M+ Z+cdc-25.2(ok597)/nT1[qIs51] | n.d. | 29.2 ± 3.3 (n = 32) |
| M+ Z−cdc-25.2(ok597)/cdc-25.2(ok597) | n.d. | 16.0 ± 0 (n = 48) |
| M− Z−cdc-25.2(ok597)/cdc-25.2(ok597) | n.d. | 16.0 ± 0.2 (n = 27) |
M, maternally loaded product; Z, zygotically expressed product. Most of the worm strains examined contained a transgene, rrIs01[elt-2::GFP], to specifically mark intestinal nuclei with GFP. GFP-positive nuclei were counted as intestinal nuclei in each worm. n.d., not determined.
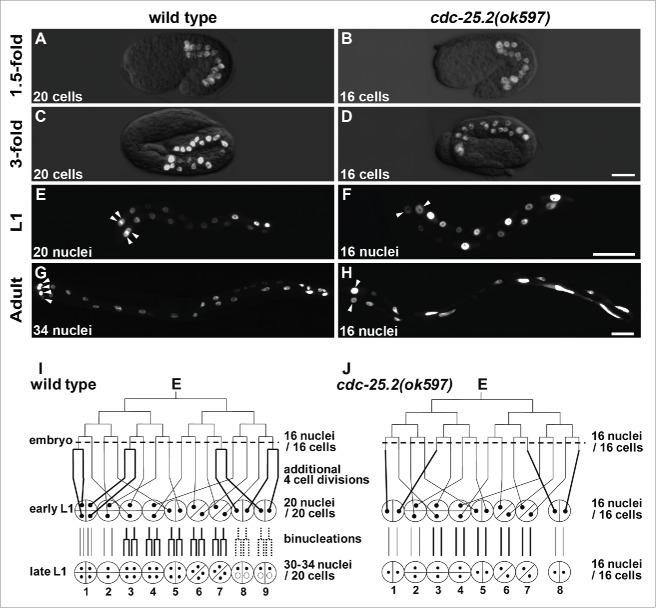
The intestinal development originating from the single E blastomere increased the number of cells to 20 by the “1.5-fold” embryonic stage in wild type (Fig. 1A). This number remained the same until the early L1 stage (Fig. 1C, E, and I).5,6 Post-embryonic nuclear divisions in the L1 stage increased the number of intestinal nuclei to 30–34, although the int2 nuclei remained undivided (Fig. 1G and I).9 After the L1 nuclear divisions, all intestinal nuclei doubled their DNA content before each larval molting via endoreduplication. In contrast, there were 16 intestinal cells at the “1.5-fold” embryonic stage in cdc-25.2(ok597) mutants (Fig. 1B), and this number remained the same during the later developmental stages until the adult stage due to lack of additional cell divisions and binucleations (Fig. 1D, F, H and J). As a result, the int1 ring contained only 2 nuclei (Fig. 1F and H, arrowheads), and the int9 ring was not generated (Fig. 1J). Because binucleations also failed to occur in the int3-7 and the int8, the cdc-25.2 mutant adults contained only 16 intestinal nuclei that extended along the body axis (Fig. 1H). On the other hand, int2 division patterns were identical between wild type and the cdc-25.2 mutants because additional cell divisions and binucleations do not occur after the 16E cell stage in int2 nuclei (Fig. 1I and J). To evaluate the contribution of cdc-25.2 to endoreduplication, we took advantage of the fact that the int2 division pattern was identical between wild type and cdc-25.2 mutants. “Relative DNA content of int2 nuclei” was determined as the average DNA staining intensity of int2 nuclei in each genotype at the adult stage, which was normalized with that of int2 nuclei in controls. We found that the relative DNA content of int2 nuclei was not different between wild type and cdc-25.2 mutants (Table 2). This result suggests that the cdc-25.2 mutants underwent the same endoreduplications as did the wild type. Taken together, these findings indicate that the cdc-25.2 mutants had defective intestinal divisions beginning in the 16E cell stage, but the mutants' endoreduplications before each larval molting were normal. Specifically, the 4 cell divisions after the 16E cell stage during embryogenesis and all the binucleations at the L1 stage were omitted in the cdc-25.2 mutants (Fig. 1J, thick lines). This intestinal defect was also observed in cdc-25.2(ok597) mutant males (data not shown), indicating that the function of cdc-25.2 is not sex-dependent.
Figure 1.
Intestinal development was arrested at the 16E cell stage in cdc-25.2 mutants. (A-H) GFP-marked intestinal nuclei in wild type (A, C, E and G) and cdc-25.2(ok597) mutants (B, D, F and H) at different developmental stages: (A, B) 1.5-fold embryos, (C, D) 3-fold embryos, (E, F) L1-stage larvae, (G, H) adult worms. Left, the anterior side. Arrowheads in (E-H) indicate locations of int1 nuclei. Scale bars, 10 μm in (D), 25 μm in (F) and 50 μm in (H). (I, J) Schematic diagrams of intestinal divisions in wild type (I) and cdc-25.2(ok597) mutants (J). Vertical and horizontal lines indicate division patterns of the E lineage. Broken horizontal lines indicate the 16E cell stage. Bold lines indicate different division patterns between wild type and cdc-25.2(ok597) mutants. Small black dots indicate intestinal nuclei. Half circles and quarter circles indicate int ring cells. Bottom numbers indicate the numbering of each int ring. In wild type, int8-9 nuclei do not always divide at the L1 stage. Therefore, they are indicated by dotted lines and small dotted circles.
Table 2.
Relative DNA content of int2 nuclei in respective genotypes.
| Genotype | Relative DNA content of int2 nuclei | p value |
|---|---|---|
| wild type | 1.0 ± 0.1 (n = 14) | |
| cdc-25.2(ok597) | 1.0 ± 0.1 (n = 14) | = 0.5 |
| mock RNAi | 1.0 ± 0.3 (n = 21) | |
| cdk-1 RNAi | 1.1 ± 0.3 (n = 19) | = 0.23 |
| cdk-2 RNAi | 0.6 ± 0.3 (n = 19) | < 0.05 |
| cdk-4 RNAi | 0.7 ± 0.4 (n = 13) | < 0.05 |
| cyb-1 RNAi | 1.1 ± 0.3 (n = 22) | = 0.48 |
| cyd-1 RNAi | 1.0 ± 0.2 (n = 16) | = 0.85 |
| cye-1 RNAi | 0.4 ± 0.2 (n = 24) | < 0.05 |
Intestine-specific RNAi was performed using the OLB11 strain. The relative DNA content of int2 nuclei in each genotype was determined as the average DNA staining intensity of int2 nuclei in each genotype normalized with that of int2 nuclei in the control (wild type or mock RNAi) measured at the adult stage. n, number of examined nuclei. P values are calculated against the appropriate controls.
cdc-25.2 is transiently expressed and functions in the intestine
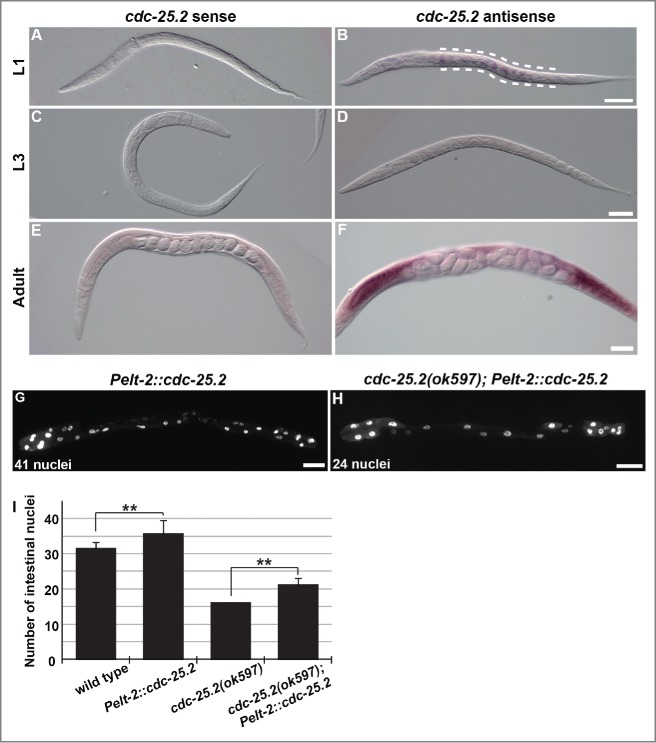
Because cdc-25.2 activity is required for normal intestinal development, the spatiotemporal expression of cdc-25.2 during larval development was examined by whole-mount in situ hybridization.34 As expected from the mutant phenotype, we found that cdc-25.2 mRNA was expressed in the intestine at the L1 stage (Fig. 2B, between the broken lines). In contrast, a cdc-25.2 mRNA signal was not detected in the L2- to L3-stage larvae (Fig. 2D), but reappeared after the L4 stage in the gonad (Fig. 2F). These results indicate that cdc-25.2 is transiently expressed in the intestine at the L1 stage.
Figure 2.
In situ expression pattern of cdc-25.2 mRNA and partial rescue of the cdc-25.2 mutant phenotype by intestinal cdc-25.2 transgene expression. (A-F) In situ hybridization of cdc-25.2 during larval development with a cdc-25.2 sense probe (A, C and E) and a cdc-25.2 antisense probe (B, D and F): (A, B) L1 stage; broken lines in (B) indicate the region of cdc-25.2 mRNA expression, (C, D) L3 stage, (E, F) adult stage; cdc-25.2 mRNA expression was observed in the gonads. Scale bars, 25 μm in (B), 50 μm in (D), and 100 μm in (F). (G) GFP-marked intestinal nuclei in a Pelt-2::cdc-25.2 transgenic adult worm, in which a cdc-25.2 transgene under the control of the intestine-specific elt-2 promoter was expressed in a wild-type background. (H) GFP-marked intestinal nuclei in a cdc-25.2(ok597); Pelt-2::cdc-25.2 transgenic adult worm, in which the Pelt-2::cdc-25.2 transgene was expressed in the cdc-25.2 mutant background. Left, the anterior side. Scale bars, 50 μm. (I) Average numbers of intestinal nuclei in wild-type adults (n = 22), Pelt-2::cdc-25.2 adults (n = 19), cdc-25.2(ok597) adults (n = 23), and cdc-25.2(ok597); Pelt-2::cdc-25.2 adults (n = 30). ** p < 0.001.
Furthermore, when a cdc-25.2 transgene was expressed under the control of the intestine-specific elt-2 promoter, the transgene induced intestinal hyperplasia in the wild-type background (Fig. 2G and I). In addition, when expressed in the cdc-25.2 mutant background, the transgene partially rescued the intestinal hypoplasia phenotype of the mutants (Fig. 2H and I). These results support the idea that cdc-25.2 functions in the intestine to regulate intestinal divisions.
cdc-25.2 is not required for intestinal divisions up to the 16E cell stage
Although embryonic cell divisions after the 16E cell stage and postembryonic binucleations were omitted, the E blastomere divided normally up to the 16E cell stage in the cdc-25.2 mutants (Fig. 1). This suggests that E cell divisions up to the 16E cell stage are regulated either by maternally loaded cdc-25.2 or by the activity of another cdc-25 family member. Because many aspects of early embryonic development are controlled by maternally loaded gene products (M) instead of zygotic gene products (Z) in C. elegans,35 maternal cdc-25.2 may be required for early intestinal divisions. Although the cdc-25.2 mutants are sterile, the penetrance of the sterility is approximately 85% at 20°C.21 Thus, a few homozygous cdc-25.2 mutant hermaphrodites produced from heterozygous mothers (M+ Z−) can produce homozygous cdc-25.2 mutant progeny in which neither the maternal load nor the zygotic product of cdc-25.2 is present (M− Z−). When the intestinal phenotype of M− Z− cdc-25.2 homozygous mutant progeny was examined, we found that they produced 16 intestinal nuclei, as did their M+ Z− cdc-25.2 homozygous mutant mothers (Table 1). This result indicates that maternally loaded cdc-25.2 is not required for intestinal divisions up to the 16E cell stage. Rather, it suggests that another cdc-25 family member is involved in the intestinal divisions before the 16E cell stage.
cdc-25.1(gf) mutation suppresses the embryonic but not the postembryonic intestinal defect of cdc-25.2(ok597) mutants
The phenotype of cdc-25.1(rr31gf) mutants suggests that cdc-25.1 functions around the 8E cell stage or earlier during embryonic intestinal development.16 The observation that RNAi against cdc-25.1 in adult hermaphrodites caused embryonic lethality of the progeny also implies that maternally loaded cdc-25.1 is essential for early embryonic cell cycles.13,15 To examine whether the prolonged presence of cdc-25.1 activity can compensate for the lack of cdc-25.2 activity, we introduced cdc-25.1(rr31gf) mutation into cdc-25.2 mutants. The intestinal divisions of this double mutant were then examined (Fig. 3). The cdc-25.1(gf) single mutants contained an increased number of intestinal nuclei at the L1 stage (Fig. 3A and E, on average 39.6 ± 1.1); this number further increased to almost 60 nuclei at the adult stage (Fig. 3C and E, on average 59.0 ± 3.4), as previously described.16,19 On the other hand, although the cdc-25.1(gf); cdc-25.2 double mutants contained more than the wild-type number (20) of intestinal nuclei at the L1 stage (Fig. 3B and E, on average 24.1 ± 1.9), the double mutants ended up containing fewer than the wild-type number (30–34) of intestinal nuclei at the adult stage (Fig. 3D and E, on average 27.6 ± 5.3). These results indicate that, although extra cell divisions occurred during embryogenesis, postembryonic nuclear divisions did not occur properly in the double mutant intestine (Fig. 3E). In other words, although the cdc-25.1(gf) mutation suppressed the defect of embryonic intestinal cell divisions, it failed to suppress the defect of postembryonic intestinal nuclear divisions of the cdc-25.2 mutants. This finding supports the view that cdc-25.1 regulates intestinal divisions before the 16E cell stage, and then cdc-25.2 takes over this role until binucleations are accomplished at the L1 stage.
Figure 3.
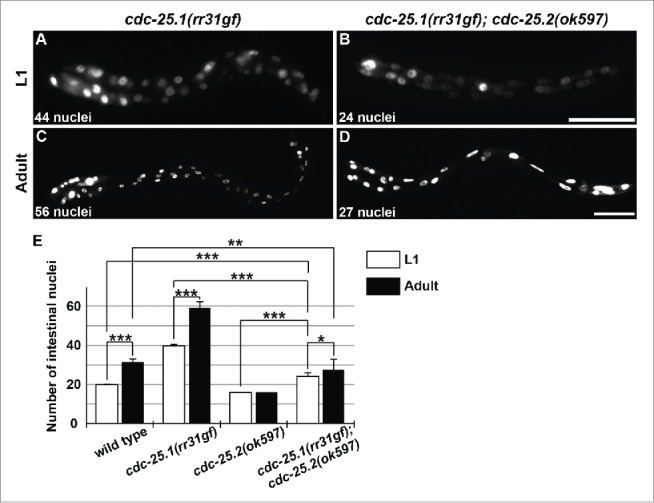
cdc-25.1(rr31gf) mutation partially suppressed the cdc-25.2 mutant intestinal phenotype. GFP-marked intestinal nuclei in (A) a cdc-25.1(rr31gf) mutant L1 larva, (B) a cdc-25.1(rr31gf); cdc-25.2(ok597) double-mutant L1 larva, (C) a cdc-25.1(rr31gf) mutant adult hermaphrodite, and (D) a cdc-25.1(rr31gf); cdc-25.2(ok597) double-mutant adult hermaphrodite. Left, the anterior side. Scale bars, 25 μm in (B), and 50 μm in (D). (E) Average numbers of intestinal nuclei in wild-type L1 and adult (n = 22 and 22, respectively), in cdc-25.1(rr31gf) mutant L1 and adult (n = 22 and 19, respectively), in cdc-25.2(ok597) mutant L1 and adult (n = 22 and 23, respectively), and in cdc-25.1(rr31gf); cdc-25.2(ok597) double-mutant L1 and adult (n = 19 and 59, respectively). * p = 0.0068, ** p = 0.0015, *** p < 0.001.
CDC-25.2 counteracts WEE-1.3 and positively regulates CDK-1/CYB-1 to promote intestinal divisions
In C. elegans, there are 3 Wee1/Myt1 homolog genes: wee-1.1, wee-1.2, and wee-1.3. Among them, wee-1.2 is most likely a pseudogene.36 It was previously shown that RNAi depletion of wee-1.3 partially suppressed the oocyte maturation defect of the cdc-25.2 mutants.21 We found that RNAi against wee-1.3, but not wee-1.1, partially suppressed the defect in intestinal division in the cdc-25.2 mutants (Table 3).
Table 3.
RNAi depletion of wee-1 family genes in wild type and cdc-25.2 mutants.
| Genotype | L4440 | wee-1.1(RNAi) | wee-1.3(RNAi) | p value |
|---|---|---|---|---|
| wild type | 30.6 ± 1.4 (n = 14) | 30.7 ± 1.4 (n = 14) | 29.5 ± 1.9 (n = 13) | = 0.1 |
| cdc-25.2(ok597) | 15.9 ± 0.6 (n = 15) | 15.9 ± 0.5 (n = 17) | 22.4 ± 3.0 (n = 9) | < 0.001 |
After RNAi treatment by the feeding method,53 adult progeny were fixed, stained with Hoechst 33342, and the numbers of their intestinal nuclei were counted. n, number of examined worms. P values were calculated by comparing L4440 controls and wee-1.3(RNAi) worms.
To identify a possible target CDK regulated by CDC-25.2 and WEE-1.3, as well as its cyclin partner in intestinal development, we tested RNAi depletion of C. elegans cdk and cyclin homolog genes. CDKs and cyclins are essential in many tissues. Thus, RNAi depletion of most of them in wild-type N2 induced pleiotropic defects leading to developmental arrest or retardation, such as embryonic lethality (Emb), larval lethality (Lvl), growth retardation (Sck, Dpy, Slo), and germline sterility (Ste) (Table 4), as previously reported.37-40 This hindered our ability to identify which CDKs and cyclins function during intestinal development. To overcome this obstacle, we used the C. elegans strain OLB11 and its derivative. Because OLB11 is an rde-1 loss-of-function mutant but expresses RDE-1 under the control of the intestine-specific elt-2 promoter, only the intestine in this strain is susceptible to RNAi.41 An elt-2::gfp transgene, rrIs01, was also introduced into OLB11 to facilitate observation of the intestinal nuclei (Table S1). Among the cdk homolog genes, RNAi depletion of cdk-1, cdk-2, and cdk-4 reduced the number of intestinal nuclei in OLB11 (Table 4). cdk-1 RNAi reduced the number of nuclei to approximately 20. However, the relative DNA content of int2 nuclei measured by fluorescence intensity did not differ from that of mock RNAi controls (Table 2). Although RNAi depletion of cdk-2 and cdk-4 also reduced the number of intestinal nuclei, the level of reduction was not nearly as significant (Table 4). Furthermore, the relative DNA content of int2 nuclei was also significantly reduced after RNAi depletion of cdk-2 and cdk-4 compared to mock RNAi controls (Table 2). Among the cyclin homolog genes, RNAi depletion of cyb-1, cyd-1, cye-1, and cyl-1 decreased the number of intestinal nuclei in OLB11 (Table 4). Like cdk-1 RNAi, although cyb-1 RNAi and cyd-1 RNAi reduced the number of intestinal nuclei, the relative DNA content of int2 nuclei was not different from that of mock RNAi controls (Table 2). In contrast, RNAi against cye-1 not only reduced the number of intestinal nuclei but also significantly decreased the relative DNA content of int2 nuclei, similar to RNAi against cdk-2 and cdk-4. On the other hand, cyb-3 RNAi induced early-stage embryonic lethality in both the N2 and OLB11 strains for unknown reasons (Table 4). It should be noted that the RNAi phenotypes of the cdk homolog and the cyclin homolog genes observed in the OLB11 background were much weaker than the corresponding mutant phenotypes or the RNAi phenotypes reported in previous studies.38,42-44 The incomplete RNAi phenotypes observed in OLB11 were possibly caused by insufficient recovery of the Rde-1(+) phenotype in OLB11 intestines during early embryogenesis (see Discussion). Although the RNAi phenotypes were incomplete, it can still be assumed that zygotic gene activities expressed during postembryonic intestinal development were effectively abrogated by RNAi in OLB11. Therefore, the results are still sufficient to conclude that CDK-1 and CYB-1 are the most likely candidates for CDK and cyclin that function in intestinal divisions after the 16E cell stage under the control of CDC-25.2 and WEE-1.3.
Table 4.
RNAi depletion of genes encoding cell-cycle regulators, cdks, and cyclins in N2 and OLB11.
| Phenotype |
|||
|---|---|---|---|
| Gene | N2 | OLB11 | Number of intestinal nuclei in OLB11 |
| mock | wild type | wild type | 29.7 ± 1.2 (n = 20) |
| cdc-25.1 | Emb | wild type | 31.6 ± 1.7 (n = 28) |
| cdc-25.2 | Emb | number reduced | 19.2 ± 5.3 (n = 29)** |
| cdc-25.3 | wild type | wild type | 32.1 ± 1.5 (n = 31) |
| cdc-25.4 | wild type | wild type | 29.8 ± 0.9 (n = 21) |
| wee-1.3 | P0 sterile | wild type | 33.3 ± 1.9 (n = 28) |
| cdk-1 | 1-cell Emb | number reduced | 19.5 ± 1.6 (n = 24)** |
| cdk-2 | Ste | number/DNA reduced | 27.9 ± 1.9 (n = 34)** |
| cdk-4 | Sck/ Dpy/ Lvl/ Emb | number/DNA reduced | 26.8 ± 3.3 (n = 29)** |
| cdk-5 | wild type | wild type | 31.2 ± 1.6 (n = 29) |
| cdk-7 | Slo/ Ste/ Lvl/ Emb | wild type | 31.5 ± 1.2 (n = 32) |
| cdk-8 | Ste | wild type | 31.8 ± 1.6 (n = 16) |
| cdk-9 | Emb | wild type | 30.5 ± 1.4 (n = 25) |
| cyb-1 | Emb | number reduced | 25.1 ± 1.7 (n = 25)** |
| cyb-2.1 | wild type | wild type | 31.5 ± 1.7 (n = 25) |
| cyb-2.2 | Lvl/ Emb | wild type | 31.3 ± 1.5 (n = 27) |
| cyb-3 | Emb | Emb | n.d. |
| cyd-1 | Lvl | number reduced | 25.1 ± 2.3 (n = 18)** |
| cye-1 | Emb | number/DNA reduced | 27.7 ± 2.4 (n = 20)* |
| cyh-1 | Ste/ Emb | wild type | 31.6 ± 1.5 (n = 25) |
| cyl-1 | Emb | L3 arrest | 27.0 ± 3.8 (n = 22)* |
The indicated dsRNA solutions were injected into N2 or OLB11: rde-1(ne219) V; dusIs[Pelt-2::rde-1 + rol-6(+)] worms, and the resulting phenotype and the number of intestinal nuclei in OLB11 were examined in their progeny. Only the intestine is susceptible to RNAi in OLB11. Buffer injection was used as the mock control. Abbreviations used are as follows: Sck, sick; Dpy, dumpy; Ste, sterile; Lvl, larval lethal; Slo, slow growth; Emb, embryonic lethal; number reduced, the number of intestinal nuclei reduced; DNA reduced, the DNA content of intestinal nuclei reduced; number/DNA reduced, both the number and the DNA content of intestinal nuclei reduced; n.d., not determined.
p < 0.05,
p < 0.001.
Extra nuclear divisions in lin-23 and lin-35 mutants are suppressed by cdc-25.2 RNAi
Because extra intestinal nuclear divisions were observed in place of endoreduplication in lin-23(e1883) L1 larvae,45 it is thought that LIN-23 is required during the L1 stage and has a role in restricting intestinal nuclear divisions. In other words, LIN-23 activity may be required for the shift from binucleation to endoreduplication, probably through ubiquitin-mediated degradation of a cell-cycle regulator essential for intestinal divisions. We hypothesized that CDC-25.2 is negatively regulated by LIN-23 during this shift. To test this possibility, we examined whether the intestinal hyper-nucleation of lin-23(e1883) mutants is suppressed by cdc-25.2 RNAi. If the hyper-nucleation is caused by a failure to repress CDC-25.2 activity, cdc-25.2 RNAi should suppress the lin-23 mutant phenotype. Indeed, we observed that the intestinal hyper-nucleation of lin-23(e1883) mutants was strongly suppressed by cdc-25.2 RNAi (Fig. 4A, B and E). In contrast, cdc-25.1 RNAi had virtually no effect on the lin-23 mutant phenotype (Fig. 4E).
Figure 4.
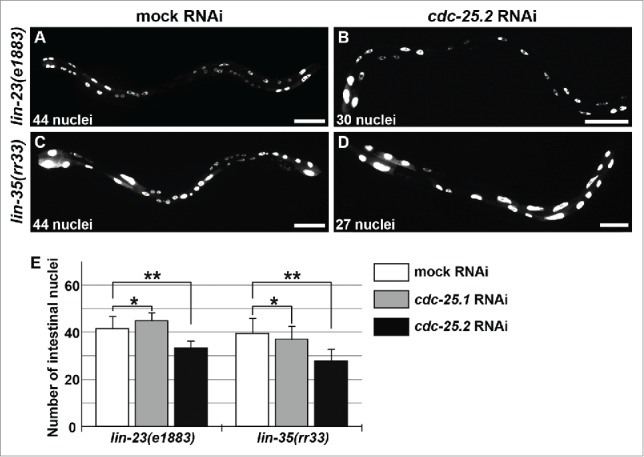
cdc-25.2 RNAi suppressed the intestinal hyper-nucleation phenotype of the lin-23 and lin-35 mutants. (A) Mock RNAi-treated lin-23(e1883) mutant adult. (B) cdc-25.2 RNAi-treated lin-23(e1883) mutant adult. (C) Mock RNAi-treated lin-35(rr33) mutant adult. (D) cdc-25.2 RNAi-treated lin-35(rr33) mutant adult. Left, the anterior side. Scale bars, 50 μm. (E) Average numbers of intestinal nuclei after RNAi depletion of mock, cdc-25.1, and cdc-25.2 in lin-23(e1883) mutants (n = 33, 15, and 38, respectively), and in lin-35(rr33) mutants (n = 33, 54, and 25, respectively). * p = 0.025 and 0.7 in lin-23(e1883) and lin-35(rr33), respectively. ** p < 0.001.
Intestinal hyper-nucleation in place of endoreduplication was also reported in lin-35(rr33) early larvae.46 Because LIN-35 is a C. elegans homolog of the mammalian transcriptional repressor, Retinoblastoma (Rb), this hyper-nucleation phenotype could be a consequence of a failure to repress transcription of a cell-cycle regulator gene. We assumed that cdc-25.2 could also be transcriptionally repressed either directly or indirectly by LIN-35 during early larval stages. Therefore, if cdc-25.2 was not properly repressed in the lin-35 mutants, extra intestinal nuclear divisions might be induced instead of endoreduplication. To test this possibility, we examined whether the intestinal hyper-nucleation of lin-35(rr33) mutants was suppressed by cdc-25.2 RNAi. Indeed, we found that RNAi against cdc-25.2, but not cdc-25.1, suppressed the intestinal hyper-nucleation of the lin-35 mutants (Fig. 4C-E).
To test whether the level of cdc-25.2 mRNA is upregulated in lin-35 mutants when additional nuclear divisions occur, we quantified cdc-25.2 mRNA levels in synchronized wild-type and lin-35(rr33) mutant populations during the late L1 to early L2 stages by qRT-PCR, while simultaneously observing their intestinal nuclear divisions (Fig. 5). To minimize delay or fluctuation of development in lin-35 mutants as compared to wild type, the populations were synchronized and cultured at 15°C, the lowest permissive temperature, and progression of development was carefully monitored through observation of intestinal nuclear divisions and larval molting. We found that both wild type and lin-35(rr33) mutants started L1 nuclear divisions at 25 hours of cultivation and molted between 25 and 27 hours of cultivation, indicating that there was no significant difference in the developmental rate between wild type and lin-35 mutants at 15°C, as previously described (Fig. 5A and C).46 We then observed additional nuclear divisions in lin-35 mutants, but not in wild type, at 27 and 29 hours of cultivation (Fig. 5A). These additional nuclear divisions in lin-35 mutants were clearly identified by observing int2 nuclei at 27 and 29 hours of cultivation (Fig. 5B). Notably, we found that the level of cdc-25.2 mRNA was upregulated during the 27-to-29-hour cultivation window in lin-35 mutants, but not in wild type, as assessed by qRT-PCR quantification (Fig. 5C). These results indicate that cdc-25.2 mRNA was temporarily upregulated in lin-35 mutants when additional nuclear divisions occurred after the L1-to-L2 molting. These results also suggest that cdc-25.2 is negatively regulated by lin-35 at the transcriptional level during the early L2 stage, possibly to promote transition of cell cycle mode from nuclear division to endoreduplication.
Figure 5.
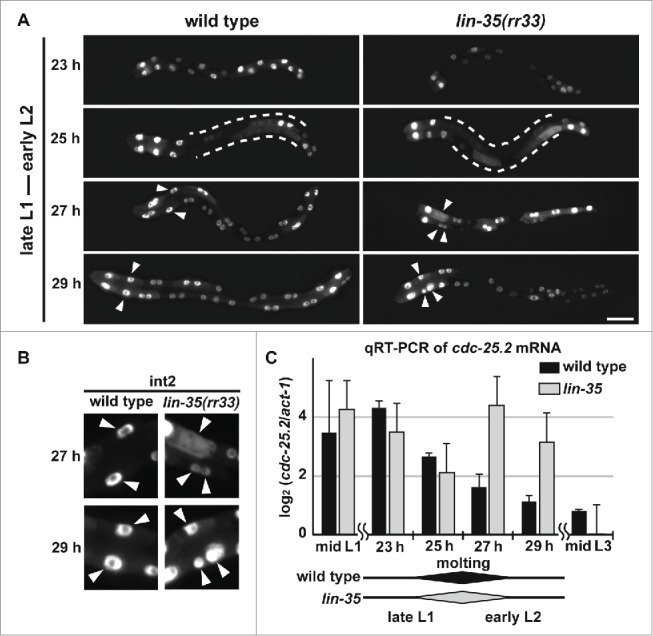
cdc-25.2 mRNA levels were negatively regulated by LIN-35. (A) GFP-marked intestinal nuclei in wild type and lin-35(rr33) mutants during early larval development. Hours (h) indicate the feeding period of synchronized L1 larvae in culture plates at 15°C. Dotted lines indicate regions of intestine in which GFP signals temporarily dispersed, and therefore intestinal nuclei were not clearly identified. Arrowheads indicate nuclei of int2 cells. Left, the anterior side. Scale bar, 25 μm. (B) Enlarged images of int2 nuclei. Excessive numbers of int2 nuclei were observed in 27 h- and 29 h-old lin-35(rr33) mutant early larvae (arrowheads). In contrast, the number of int2 nuclei was consistently 2 in wild-type larvae. (C) A time course of cdc-25.2 mRNA levels during early larval development in wild type and lin-35(rr33) mutants. The cdc-25.2 mRNA level at each time point in wild type and lin-35(rr33) mutants was measured 3 times by qRT-PCR, averaged, normalized to that of act-1 mRNA, and shown as the log2 value. Rhomboid shapes at the bottom indicate the duration of L1-to-L2 molting observed in wild type and lin-35(rr33) mutant larval samples.
Discussion
The intestinal development of C. elegans is an excellent model system to study cell division cycles because it consists of 3 distinct periods that employ different division modes (Figs. 1I and 6). The first is the period of cell divisions during embryogenesis. The second is the period of binucleations that occur in the int3-7 cells (occasionally also in the int8-9 cells) at the L1 larval stage, which produces 30–34 nuclei in the 20 cells. The third is the period of endoreduplications that occur before each larval molting and increase DNA content. In this study, we demonstrated that CDC-25.2 serves an essential role in intestinal development in the later stage of the first period and through the second period. First, we showed that CDC-25.2 is required for intestinal divisions only after the 16E cell stage. Homozygous progeny produced from cdc-25.2 heterozygous mothers (M+ Z−) and rare homozygous progeny produced from cdc-25.2 homozygous mothers (M− Z−) generated the same 16 intestinal nuclei (Table 1), indicating that there was no contribution of maternal cdc-25.2 product to the intestinal divisions before the 16E cell stage. In addition, the cdc-25.1(gf) mutation that causes intestinal hyperplasia did not completely suppress the intestinal division defects of the cdc-25.2 mutants (Fig. 3). Furthermore, cdc-25.1 RNAi was less suppressive than cdc-25.2 RNAi on intestinal hyper-nucleation in lin-23(e1883) and lin-35(rr33) mutants (Fig. 4). These results and studies by others strongly suggest that, although they may overlap, the functional periods of CDC-25.1 and CDC-25.2 are different. That is, CDC-25.1 functions earlier than CDC-25.2, probably up to the 16E cell stage, in embryonic intestinal cell divisions (Fig. 6).19,47 Because neither cdc-25.3 nor cdc-25.4 deletion mutants showed any obvious intestinal division defects (Table 1), CDC-25.2 appears to be the only member of the family that regulates intestinal divisions after the 16E cell stage to accomplish the rest of the embryonic cell divisions as well as the entire postembryonic nuclear divisions (Fig. 6).
Figure 6.
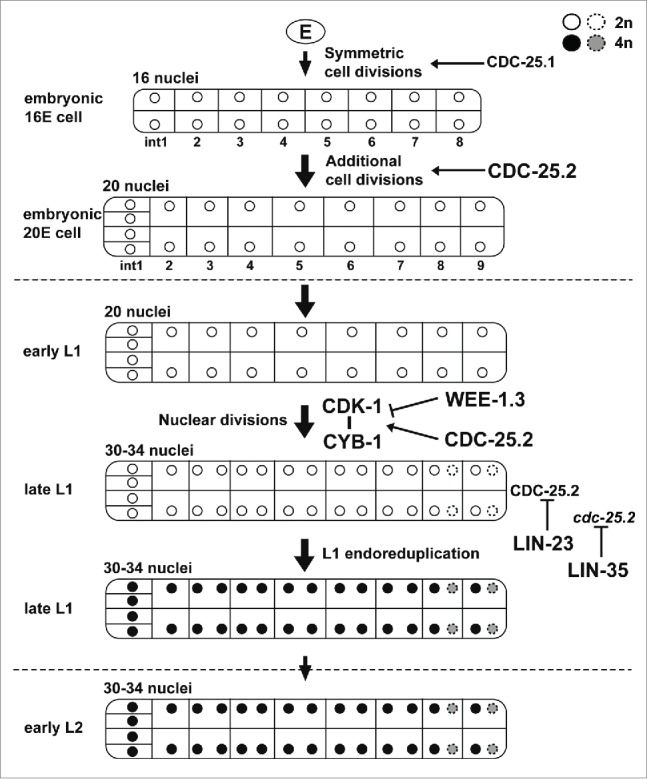
A proposed model for the function and regulation of CDC-25.2 during C. elegans intestinal development. Tubes and internal compartments indicate the whole intestine and intestinal cells, respectively. Circles indicate intestinal nuclei and the gray scale indicates different DNA contents of the nuclei after endoreduplications (white, 2n; black and gray, 4n). Broken circles indicate intestinal nuclei that occasionally divided during nuclear divisions. Vertical arrows indicate the progression of intestinal divisions. Broken horizontal lines indicate the borders of developmental stages. Horizontal arrows indicate that respective intestinal divisions are positively regulated by CDC-25.1 or CDC-25.2. The horizontal T-bar indicates the inhibition of the CDK-1/CYB-1 complex by WEE-1.3. The vertical T-bars indicate the inhibition of CDC-25.2 protein and cdc-25.2 mRNA by LIN-23 and LIN-35, respectively.
In the wild type, after completion of binucleations in the L1 stage, although the number of intestinal nuclei (30–34) does not increase any further, all the intestinal nuclei double their DNA content via endoreduplication before each larval molting.10 It was previously reported that endoreduplication at the end of the L1 stage was replaced by extra nuclear divisions in lin-23(e1883) mutants, which resulted in intestinal hyper-nucleation.45 Since it was also reported that LIN-23 may negatively regulate embryonic intestinal cell divisions through ubiquitin-mediated degradation of CDC-25.1,19,47 we hypothesized that LIN-23 might be required during the L1 stage to restrict intestinal nuclear divisions through inactivation of CDC-25.2. Indeed, we found that the intestinal hyper-nucleation of lin-23(e1883) mutants was almost completely suppressed by cdc-25.2 RNAi (Fig. 4). This result suggests that LIN-23 negatively regulates CDC-25.2 at the end of the L1 stage to successfully shift the intestinal division mode from binucleation to endoreduplication (Fig. 6). However, whether LIN-23 inactivates CDC-25.2 directly or indirectly remains to be elucidated. Extra nuclear divisions in place of endoreduplication was also observed in lin-35(rr33) mutants at the early L2 stage.46 We found that the intestinal hyper-nucleation of lin-35(rr33) mutants was also almost completely suppressed by cdc-25.2 RNAi (Fig. 4), and that the level of cdc-25.2 mRNA was transiently upregulated after the L1-to-L2 molting in the lin-35 mutants (Fig. 5C). These results indicate that the level of cdc-25.2 mRNA needs to be repressed in a timely manner by LIN-35 in order to accomplish L1 endoreduplication (Fig. 6). However, precisely how this process occurs is not yet clear. Taken together, our results indicate that cdc-25.2 activity (CDC-25.2 protein and cdc-25.2 mRNA) needs to be negatively regulated by both LIN-23 and LIN-35 at the end of the L1 stage or at the beginning of the L2 stage to achieve successful transition from the nuclear division to L1-stage endoreduplication (Fig. 6).
We previously reported that an absence of CDC-25.2 during oogenesis interfered with normal oocyte maturation and caused the endomitotic oocyte (Emo) phenotype.21 In this study, we found that, although additional 4 cell divisions during embryogenesis and binucleations at the L1 stage were totally abrogated, intestinal endoreduplication was not affected in the cdc-25.2 mutants; int2 nuclei had the same DNA levels in wild type and cdc-25.2 mutants (Table 2). In fact, inactivation of CDC-25.2 appeared to be essential for initiating and accomplishing intestinal endoreduplication. These observations suggest that, under certain circumstances, inactivation of CDC-25.2 causes endomitosis or endoreduplication instead of merely arresting the cell cycle. Endoreduplication is a specialized cell cycle in which the M phase is bypassed, but the other 3 phases, G1, S, and G2, continue to occur (reviewed in ref. 48). It would be intriguing to understand the mechanism by which inactivation of CDC-25.2 induces endoreduplication instead of cell cycle arrest.
To better understand the regulatory mechanism of intestinal divisions in C. elegans, we tested possible involvement of other conserved cell-cycle regulators by performing systematic RNAi analysis. First, among the genes of wee-1 family, RNAi depletion of wee-1.3 partially suppressed the cdc-25.2 intestinal phenotype. As wee-1.3 RNAi also suppressed the cdc-25.2 Emo phenotype during oogenesis,21 this suppression in the intestine indicates that the counteractive interaction between CDC-25.2 and WEE-1.3 is conserved between the organs in C. elegans. To identify the specific CDK and cyclin that function during C. elegans intestinal divisions, we screened the available C. elegans orthologs via intestine-specific RNAi analysis using the OLB11 strain. Among the cdk genes, RNAi depletion of cdk-1, cdk-2, and cdk-4, and among the cyclin genes, RNAi depletion of cyb-1, cyd-1, cye-1, and cyl-1 reduced the number of intestinal nuclei in OLB11 (Table 4). Furthermore, RNAi depletion of cdk-1, cyb-1, and cyd-1 did not affect relative DNA-staining intensity of int2 nuclei at the adult stage, nor did the cdc-25.2(ok597) mutation (Table 2), suggesting that these 3 genes are most likely not involved in the regulation of endoreduplication. However, in a previous study, it was reported that intestinal development in 2 cyd-1 mutants, he112 and he116, was arrested at the 16E cell stage with 2C DNA content, indicating that both binucleations and endoreduplications were abrogated in the cyd-1 mutants.42 Therefore, our results on cyd-1 RNAi might not be accurate. The simplest explanation for this discrepancy is that our cyd-1 RNAi treatment in OLB11 did not completely inactivate the gene. The OLB11 strain contains an rde-1 transgene that is driven by an intestine-specific elt-2 promoter in the rde-1 mutant background, which restores RNAi activity specifically in the intestine (Table S1).41 However, because elt-2 begins to be expressed at the embryonic 2E cell stage, maternal gene products that are already loaded before the 2E cell stage are not effectively inactivated by RNAi in OLB11.33 Therefore, the effectiveness of RNAi depletion in OLB11 differs among genes depending on their timing of expression during development. This difference in RNAi effectiveness may at least partially explain why intestine-specific RNAi phenotypes of cdk and cyclin homolog genes in OLB11 were weaker than those seen in previous studies. Nevertheless, our RNAi results and those of previous studies37,42 are consistent with the view that CDK-1 is the most likely candidate CDK that is required for intestinal nuclear divisions but not for endoreduplications. Because cyb-1 RNAi displayed the phenotype that was most similar to that of cdk-1 RNAi among the cyclin genes in OLB11, and because CYB-1 and CYB-3 were previously shown to associate with CDK-1,40 we assume that CYB-1 is one of the most likely candidates for the cyclin that works in concert with CDK-1 during intestinal nuclear divisions. Because cyb-3 RNAi in OLB11 caused embryonic lethality, we could not determine whether CYB-3 also functions after the 16E cell stage, possibly together with CDK-1. Taken together, based on our intestine-specific RNAi analysis and previous studies, we hypothesize that, after the 16E cell stage, CDC-25.2 positively and WEE-1.3 negatively regulates CDK-1, which forms a complex with CYB-1, to control intestinal nuclear divisions (Fig. 6). On the other hand, RNAi depletion of cdk-2, cdk-4, and cye-1 in OLB11 not only reduced the number of intestinal nuclei, but also significantly reduced the relative DNA content of int2 nuclei (Table 2). These results suggest that CDK-2, CDK-4, CYE-1, and also possibly CYD-1 (see above) are required not only for nuclear divisions but also for endoreduplications in the intestine. Previous studies on cye-1 mutations, which revealed defects in both binucleation and endoreduplication,43,44 support this role of CYE-1.
In this study, we demonstrated that CDC-25.2 and its regulation are essential for the progression of diversified intestinal divisions including embryonic cell divisions, postembryonic nuclear divisions, and endoreduplications. Nevertheless, the biological significance of CDC-25.2′s role in intestinal development is still elusive, notably because the intestine of cdc-25.2 mutants, which contains only 16 nuclei instead of 30–34 nuclei, seems to function normally, at least under standard growth conditions. Further analysis is required to better understand the biological importance of this specialized developmental process, in which CDC-25.2 is involved.
Materials and methods
Strains
C. elegans strains used in this study are listed in a separate Table (Table S1). Worms were cultured and handled at 20°C using standard procedures.49
Construction and microscopic observation of transgenic animals
To visualize intestinal nuclei, An elt-2::gfp transgene, rrIs01[Pelt-2::GFP], was introduced into some strains by genetic crossing (Table S1). To express cdc-25.2 under the control of the intestine-specific elt-2 promoter, a full-length cdc-25.2 cDNA was inserted into the pOLB1872 plasmid between the BamHI and SmaI restriction sites, which was then microinjected into recipient worms with the pRF4 injection marker plasmid as previously described.50 The generated transgenic worms containing the heritable extrachromosomal array, kkuEx03[Pelt-2::cdc-25.2::mCherry], were then crossed with wild-type and cdc-25.2(ok597) heterozygous rrIs01[Pelt-2::GFP] transgenic worms for overexpression and rescue experiments, respectively (Table S1). For staging, newly hatched larvae were first synchronized at the L1 stage in the absence of food, then cultured with food until they reached the desired developmental stage. To examine the number of intestinal nuclei, the staged worms were transferred to M9 buffer on a poly-l-lysine-coated glass slide, covered with a coverslip, and observed under a fluorescence microscope (Zeiss Axioskop 2). Images were recorded using an Orca-ERG digital camera (Hamamatsu) with Openlab 5 software (Improvision).
Immunohistochemistry and confocal microscopy
Embryonic specimens were prepared as previously described51 and immunostained with rabbit polyclonal anti-GFP primary antibody (Invitrogen A11122, 1:400) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) secondary antibody (Invitrogen A11008, 1:200). Images were recorded using a FluoView FV1000 confocal microscope (Olympus).
In situ hybridization
The C. elegans cDNA clone yk472b2 was used to generate RNA probes for cdc-25.2. Digoxigenin (DIG) labeling of RNA probes was performed according to the manufacturer's instructions (Roche, DIG application manual for in situ hybridization; http://www.roche-applied-science.com/PROD_INF/MANUALS/InSitu/InSi_toc.htm). Larvae were synchronized, fixed, and hybridized with sense or antisense RNA probe as previously described.34 Fixed specimens were hybridized with 1:500 diluted cdc-25.2 sense or antisense RNA probe at 60°C for 16 hours. In situ hybridization images were taken using an AxioCam MRc 5 digital camera and processed with AxioVision software (Zeiss).
DNA quantification
DNA staining of intact worms was carried out as previously described.10,52 Fluorescence images were taken with an Orca-ERG digital camera (Hamamatsu) using the same exposure conditions in all cases. Thereafter, the fluorescence intensities of Hoechst 33342-stained intestinal nuclei were measured using Openlab 5 software (Improvision). The area of each intestinal nucleus was set as region of interest (ROI), and the average fluorescence intensity of each ROI (iav) was measured. Average background intensity (bav) was also measured and subtracted from the iav of each intestinal nucleus (iav-bav). The DNA content of each intestinal nucleus was calculated as (iav-bav) × area of ROI. This value was normalized to that of wild-type int2 for quantitative comparison.
RNA interference
RNAi depletion of wee-1 family genes in the wild-type and cdc-25.2(ok597) worms was performed by the feeding RNAi method,53 with the E. coli strain HT115(DE3) that harbored L4440-derived vectors in which a C. elegans genomic insert, F35H8.7 (wee-1.1) or Y53C12A.1 (wee-1.3), was inserted. Gravid adult hermaphrodites were transferred to each feeding RNAi plate, and the resulting progeny were examined for their intestinal phenotypes when they became adults. RNAi depletion of cdc-25.1 and cdc-25.2 in lin-23(e1883) and lin-35(rr33) mutant worms was performed by the soaking RNAi method:54 dsRNA was transcribed in vitro from corresponding yk EST clones (Table S2) and dissolved in soaking buffer. Synchronized L1 larvae were soaked in the dsRNA solution and incubated for 48 hours at 20°C. Then, the soaked worms were placed onto OP50-seeded NGM plates and grown until they reached the L4 or young adult stage, at which time their intestinal phenotypes were observed. For RNAi depletion of cell-cycle regulator genes in N2 and OLB11 worms, dsRNA of each gene prepared by in vitro transcription was microinjected into the gonads of young adult hermaphrodites, and their progeny were examined for the resulting intestinal phenotypes when they became adults. cDNA inserts of corresponding yk EST clones (Table S2) were PCR-amplified using the T7 primer and the CMo422 primer pair (Table S3) as DNA templates for in vitro transcription. Those DNA templates for which yk clones were not available were PCR-amplified from an N2 cDNA pool using gene-specific primers (Table S3).
Quantitative real-time RT-PCR
To quantify mRNA levels of cdc-25.2, total RNA was prepared from synchronized wild-type N2 and lin-35(rr33) mutant worms at respective developmental stages. To collect mid-L1 and mid-L3 stage worms, synchronized L1 worms were fed for 10 hours and 30 hours at 20°C, respectively, before harvesting. To collect late-L1 to early-L2 stage worms, synchronized L1 worms were fed for 23, 25, 27, and 29 hours at 15°C. Real-time measurement of cdc-25.2 mRNA levels was performed as previously described.21 The measured values were normalized to that of act-1 mRNA and presented as log2 values for quantitative comparisons.
Statistical analysis
P values were calculated by Student's t-test, and most of the data were obtained from 3 independent experiments with multiple sample numbers (n), which are described in the figure legends.
Supplementary Material
Abbreviations
- CDC
cell division cycle
- CDK
cyclin-dependent kinase
- 16E
16 great-great-granddaughter cells of the founder blastomere E
- L1 to L4
the first to the fourth larval stage
- M
maternally loaded
- Z
zygotically expressed
- Emo
endomitotic oocyte
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Olaf Bossinger (Düsseldorf, Germany) for the OLB11 strain and pOLB1872 plasmid, Dr. Koki Noguchi (Mishima, Japan) for helpful advice on in situ hybridization, Dr. Junho Lee (Seoul, Korea) for the feeding RNAi clones, and Dr. Yuji Kohara (Mishima, Japan) for the yk EST clones. The VC402 strain was provided by the C. elegans Reverse Genetics Core Facility at UBC, which is part of the International C. elegans Gene Knockout Consortium. Some strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Republic of Korea [grant numbers 2010-0011182 and NRF-2013R1A1A2009090 to Y.-H.S., 2010-0009509 and NRF-2013R1A1A2009820 to I.K.]. This research was also supported by the 2014 KU Brain Pool Program of Konkuk University to I.K. and by the Hi Seoul Science Fellowship from the Seoul Scholarship Foundation to Y.-U.L. We also thank CureBio for supporting this project.
References
- [1].Sharrock WJ. Yolk proteins of Caenorhabditis elegans. Dev Biol 1983; 96:182-188; PMID:6337890; http://dx.doi.org/ 10.1016/0012-1606(83)90321-4 [DOI] [PubMed] [Google Scholar]
- [2].Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. Inducible antibacterial defense system in C. elegans. Curr Biol 2002; 12:1209-1214; PMID:12176330; http://dx.doi.org/ 10.1016/S0960-9822(02)00928-4 [DOI] [PubMed] [Google Scholar]
- [3].An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 2003; 17:1882-1893; PMID:12869585; http://dx.doi.org/ 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 2003; 115:489-502; PMID:14622602; http://dx.doi.org/ 10.1016/S0092-8674(03)00889-4 [DOI] [PubMed] [Google Scholar]
- [5].Deppe U, Schierenberg E, Cole T, Krieg C, Schmitt D, Yoder B, von Ehrenstein G. Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA 1978; 75:376-380; PMID:272653; http://dx.doi.org/ 10.1073/pnas.75.1.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983; 100:64-119; PMID:6684600; http://dx.doi.org/ 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- [7].Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol 1999; 216:114-134; PMID:10588867; http://dx.doi.org/ 10.1006/dbio.1999.9471 [DOI] [PubMed] [Google Scholar]
- [8].Schroeder DF, McGhee JD. Anterior-posterior patterning within the Caenorhabditis elegans endoderm. Development 1998; 125:4877-4887; PMID:9811572 [DOI] [PubMed] [Google Scholar]
- [9].Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 1977; 56:110-156; PMID:838129; http://dx.doi.org/ 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- [10].Hedgecock EM, White JG. Polyploid tissues in the nematode Caenorhabditis elegans. Dev Biol 1985; 107:128-133; PMID:2578115; http://dx.doi.org/ 10.1016/0012-1606(85)90381-1 [DOI] [PubMed] [Google Scholar]
- [11].Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature 1979; 279:428-430; PMID:16068179; http://dx.doi.org/ 10.1038/279428a0 [DOI] [PubMed] [Google Scholar]
- [12].Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 1987; 49:559-567; PMID:3032459; http://dx.doi.org/ 10.1016/0092-8674(87)90458-2 [DOI] [PubMed] [Google Scholar]
- [13].Ashcroft NR, Srayko M, Kosinski ME, Mains PE, Golden A. RNA-mediated interference of a cdc25 homolog in Caenorhabditis elegans results in defects in the embryonic cortical membrane, meiosis, and mitosis. Dev Biol 1999; 206:15-32; PMID:9918692; http://dx.doi.org/ 10.1006/dbio.1998.9135 [DOI] [PubMed] [Google Scholar]
- [14].Ashcroft NR, Golden A. CDC-25.1 regulates germline proliferation in Caenorhabditis elegans. Genesis 2002; 33:1-7; PMID:12001064; http://dx.doi.org/ 10.1002/gene.10083 [DOI] [PubMed] [Google Scholar]
- [15].Clucas C, Cabello J, Büssing I, Schnabel R, Johnstone IL. Oncogenic potential of a C. elegans cdc25 gene is demonstrated by a gain-of-function allele. EMBO J 2002; 21:665-674; PMID:11847114; http://dx.doi.org/ 10.1093/emboj/21.4.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kostić I, Roy R. Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans. Development 2002; 129:2155-2165; PMID:11959825 [DOI] [PubMed] [Google Scholar]
- [17].Kim J, Lee AR, Kawasaki I, Strome S, Shim YH. A mutation of cdc-25.1 causes defects in germ cells but not in somatic tissues in C. elegans. Mol Cells 2009; 28:43-48; PMID:19533027; http://dx.doi.org/ 10.1007/s10059-009-0098-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yoon S, Kawasaki I, Shim YH. CDC-25.1 controls the rate of germline mitotic cell cycle by counteracting WEE-1.3 and by positively regulating CDK-1 in Caenorhabditis elegans. Cell Cycle 2012; 11:1354-1363; PMID:22421141; http://dx.doi.org/ 10.4161/cc.19755 [DOI] [PubMed] [Google Scholar]
- [19].Hebeisen M. Roy R. CDC-25.1 stability is regulated by distinct domains to restrict cell division during embryogenesis in C. elegans. Development 2008; 135:1259-1269; PMID:18287204; http://dx.doi.org/ 10.1242/dev.014969 [DOI] [PubMed] [Google Scholar]
- [20].Ashcroft NR, Kosinski ME, Wickramasinghe D, Donovan PJ, Golden A. The four cdc25 genes from the nematode Caenorhabditis elegans. Gene 1998; 214:59-66; PMID:9651482; http://dx.doi.org/ 10.1016/S0378-1119(98)00228-5 [DOI] [PubMed] [Google Scholar]
- [21].Kim J, Kawasaki I, Shim YH. cdc-25.2, a C. elegans ortholog of cdc25, is required to promote oocyte maturation. J Cell Sci 2010; 123:993-1000; PMID:20200231; http://dx.doi.org/ 10.1242/jcs.060442 [DOI] [PubMed] [Google Scholar]
- [22].Yan B, Memar N, Gallinger J, Conradt B. Coordination of cell proliferation and cell fate determination by CES-1 snail. PLoS Genet 2013; 9: e1003884; PMID:24204299; http://dx.doi.org/ 10.1371/journal.pgen.1003884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Edgar BA, O'Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell 1989; 57:177-187; PMID:2702688; http://dx.doi.org/ 10.1016/0092-8674(89)90183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sadhu K, Reed SI, Richardson H, Russell P. Human homolog of fission yeast cdc25 mitotic inducer is predominantly expressed in G2. Proc Natl Acad Sci USA 1990; 87:5139-5143; PMID:2195549; http://dx.doi.org/ 10.1073/pnas.87.13.5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nagata A, Igarashi M, Jinno S, Suto K, Okayama H. An additional homolog of the fission yeast cdc25+ gene occurs in humans and is highly expressed in some cancer cells. New Biol 1991; 3:959-968; PMID:1662986 [PubMed] [Google Scholar]
- [26].Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 1992; 69:977-988; PMID:1606618; http://dx.doi.org/ 10.1016/0092-8674(92)90616-K [DOI] [PubMed] [Google Scholar]
- [27].Edgar BA, O'Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell 1990; 62:469-480; PMID:2199063; http://dx.doi.org/ 10.1016/0092-8674(90)90012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Courtot C, Fankhauser C, Simanis V, Lehner CF. The Drosophila cdc25 homolog twine is required for meiosis. Development 1992; 116:405-416; PMID:1286615 [DOI] [PubMed] [Google Scholar]
- [29].Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J 1994; 13:1549-1556; PMID:8156993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gabrielli BG, De Souza CP, Tonks ID, Clark JM, Hayward NK, Ellem KA. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J Cell Sci 1996; 109:1081-1093; PMID:8743955 [DOI] [PubMed] [Google Scholar]
- [31].Lammer C, Wagerer S, Saffrich R, Mertens D, Ansorge W, Hoffmann I. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J Cell Sci 1998; 111:2445-2453; PMID:9683638 [DOI] [PubMed] [Google Scholar]
- [32].Ferguson AM, White LS, Donovan PJ, Piwnica-Worms H. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol Cell Biol 2005; 25:2853-2860; PMID:15767688; http://dx.doi.org/ 10.1128/MCB.25.7.2853-2860.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fukushige T, Hendzel MJ, Bazett-Jones DP, McGhee JD. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc Natl Acad Sci USA 1999; 96:11883-11888; PMID:10518545; http://dx.doi.org/ 10.1073/pnas.96.21.11883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Motohashi T, Tabara H, Kohara Y Protocols for large scale in situ hybridization on C. elegans larvae. WormBook, ed. The C. elegans Research Community. WormBook 2006; doi/10.1895/wormbook.1.103.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wood WB, Laufer JS, Strome S. Developmental determinants in embryos of Caenorhabditis elegans. J Nematol 1982; 14:267-273; PMID:19295708. [PMC free article] [PubMed] [Google Scholar]
- [36].Wilson MA, Hoch RV, Ashcroft NR, Kosinski ME, Golden A. A Caenorhabditis elegans wee1 homolog is expressed in a temporally and spatially restricted pattern during embryonic development. Biochim Biophys Acta 1999; 1445:99-109; PMID:10209262; http://dx.doi.org/ 10.1016/S0167-4781(99)00027-5 [DOI] [PubMed] [Google Scholar]
- [37].Boxem M, Srinivasan DG, van den Heuvel S. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development 1999; 126:2227-2239; PMID:10207147 [DOI] [PubMed] [Google Scholar]
- [38].Park M, Krause MW. Regulation of postembryonic G1 cell cycle progression in Caenorhabditis elegans by a cyclin D/CDK-like complex. Development 1999; 126:4849-4860; PMID:10518501 [DOI] [PubMed] [Google Scholar]
- [39].Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev 2002; 16:2135-2146; PMID:12183367; http://dx.doi.org/ 10.1101/gad.999002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].van der Voet M, Lorson MA, Srinivasan DG, Bennett KL, van den Heuvel S. C. elegans mitotic cyclins have distinct as well as overlapping functions in chromosome segregation. Cell Cycle 2009; 8:4091-4102; PMID:19829076; http://dx.doi.org/ 10.4161/cc.8.24.10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pilipiuk J, Lefebvre C, Wiesenfahrt T, Legouis R, Bossinger O. Increased IP3/Ca2+ signaling compensates depletion of LET-413/DLG-1 in C. elegans epithelial junction assembly. Dev Biol 2009; 327:34-47; PMID:19109941; http://dx.doi.org/ 10.1016/j.ydbio.2008.11.025 [DOI] [PubMed] [Google Scholar]
- [42].Boxem M, van den Heuvel S. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development 2001; 128:4349-4359; PMID:11684669 [DOI] [PubMed] [Google Scholar]
- [43].Fay DS, Han M. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development 2000; 127:4049-4060; PMID:10952902 [DOI] [PubMed] [Google Scholar]
- [44].Grishok A, Sharp PA. Negative regulation of nuclear divisions in Caenorhabditis elegans by retinoblastoma and RNA interference-related genes. Proc Natl Acad Sci USA 2005; 102:17360-17365; PMID:16287966; http://dx.doi.org/ 10.1073/pnas.0508989102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kipreos ET, Gohel SP, Hedgecock EM. The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development 2000; 127:5071-5082; PMID:11060233 [DOI] [PubMed] [Google Scholar]
- [46].Ouellet J, Roy R. The lin-35/Rb and RNAi pathways cooperate to regulate a key cell cycle transition in C. elegans. BMC Dev Biol 2007; 7:38; PMID:17466069; http://dx.doi.org/ 10.1186/1471-213X-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Segref A, Cabello J, Clucas C, Schnabel R, Johnstone IL. Fate specification and tissue-specific cell cycle control of the Caenorhabditis elegans intestine. Mol Biol Cell 2010; 21:725-738; PMID:20053685; http://dx.doi.org/ 10.1091/mbc.E09-04-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kipreos ET. C. elegans cell cycles: invariance and stem cell divisions. Nat Rev Mol Cell Biol 2005; 6:766-776; PMID:16314866; http://dx.doi.org/ 10.1038/nrm1738 [DOI] [PubMed] [Google Scholar]
- [49].Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77:71-94; PMID:4366476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 1991; 10:3959-3970; PMID:1935914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].van Fürden D, Johnson K, Segbert C, Bossinger O. The C. elegans ezrin-radixin-moesin protein ERM-1 is necessary for apical junction remodelling and tubulogenesis in the intestine. Dev Biol 2004; 272:262-276; PMID:15242805; http://dx.doi.org/ 10.1016/j.ydbio.2004.05.012 [DOI] [PubMed] [Google Scholar]
- [52].Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 1998; 94:635-645; PMID:9741628 [DOI] [PubMed] [Google Scholar]
- [53].Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2000; 2:RESEARCH0002-RESEARCH0002.10; PMID:11178279; http://dx.doi.org/ 10.1186/gb-2000-2-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol 2001; 11:171-176; PMID:11231151; http://dx.doi.org/ 10.1016/S0960-9822(01)00052-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.