Diversity in neuronal types provides us with the ability to form complex thoughts and functions. The molecular mechanisms used to generate neuronal subtype diversity in the nervous system are still only beginning to be understood. As directed differentiation of region-specific neurons from embryonic stem cells, induced pluripotent stem cells, and fibroblasts follows developmental principles, understanding the mechanisms used to generate neuronal subtypes will be fundamental to cell replacement therapies. A terminal selector model has emerged for how unique neuronal subtypes acquire their terminal identities. In this model, a transcription factor referred to as a “terminal selector” promotes the identity of a neuron by binding to a common cis-element in the promoters of multiple neuron subtype-specific genes.1 A recent study shows that the highly conserved HMG transcription factor gene sox-2, well known for its role in pluripotency, goes beyond stem cell biology to function as a terminal selector that cooperates with 2 different transcription cofactors to distinguish between 2 postmitotic neuron identities.2
C. elegans adult hermaphrodites have 302 neurons which fall into 118 different classes. There are 3 classes of olfactory neurons: AWA, AWB, and AWC, which exist as bilateral pairs across the left-right axis. Each of the neuron classes has differential gene expression, unique synaptic targets, and senses different odors. In addition, the pair of AWC neurons are asymmetrically differentiated into 2 subtypes, AWCON and AWCOFF, with distinct odorant receptor expression and sensory functions.3 A forward genetic screen identified a missense mutation ky707 in the highly conserved HMG DNA-binding domain of the transcription factor gene sox-2 that results in a cell identity transformation of AWB olfactory neurons into AWCON olfactory neurons at molecular, morphological, and functional levels.2 This mutant phenotype is similar to a previously reported mutation in the LIM homeodomain gene lim-4.4 AWB and AWCON neurons connect with different sets of sensory neurons and interneurons for mediating repulsion and attractive behaviors, respectively. Functional transformation of AWB to AWCON suggests that ectopic AWCON neurons transformed from AWB are recruited to the same neural circuits as native AWCON in mediating attractive behaviors (Fig. 1).
Figure 1.
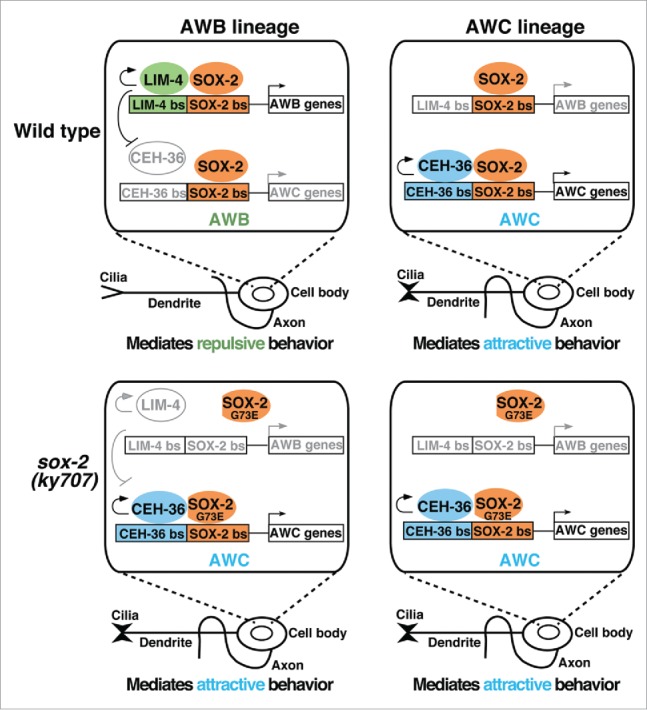
SOX-2 partners with different transcription factors to specify AWB and AWC identities. Upper: SOX-2 cooperates with LIM-4 or CEH-36 to specify AWB or AWC identity, respectively. Lower: The sox-2(ky707) mutation affects binding of SOX-2G73E to the LIM-4/SOX-2 target site but not the CEH-36/SOX-2 site, allowing CEH-36/SOX-2G73E cooperation to specify AWC identity in the AWB lineage. Colored/black, active; gray, inactive.
In sox-2 null mutants, the expression of terminal identity genes of both AWB and AWC are abolished, indicating that sox-2 plays an essential role in the specification of AWB and AWC neurons.2 Similarly, mutations in the LIM homeodomain gene lim-4 or the Otx-type homeobox gene ceh-36 also abolish terminal identity gene expression in AWB or AWC neurons, respectively.1 Together, these results indicate that SOX-2 and LIM-4 are terminal selectors in AWB neurons, while SOX-2 and CEH-36 are terminal selectors in AWC neurons. Analagous to vertebrate Sox2 partnering with different transcription factors for distinct cell specification,5 C. elegans SOX-2 cooperates with LIM-4 in terminal AWB neuron differentiation and with CEH-36 in terminal AWC neuron differentiation. These transcription factors bind to respective adjacent target sites of LIM-4/SOX-2 and CEH-36/SOX-2 to specify terminal AWB and AWC cell identities, respectively (Fig. 1).2 In AWB neurons, lim-4 represses ceh-36 expression, thereby preventing the execution of the alternative AWC identity.6 Gene dosage experiments further suggest a competition model in which LIM-4 competes with CEH-36 for cooperation with SOX-2 in AWB neurons, with LIM-4 being the preferred partner of SOX-2, thereby inhibiting SOX-2/CEH-36-mediated AWC induction (Fig. 1).2 In sox-2(ky707) mutants, the cooperation between mutant SOX-2G73E and LIM-4 is much reduced compared with the interaction between wild-type SOX-2 and LIM-4, due to decreased DNA binding of mutant SOX-2G73E to the LIM-4/SOX-2 adjacent target site. However, the cooperation between mutant SOX-2G73E and CEH-36 is not affected, since mutant SOX-2G73E is as efficient at binding the CEH-36/SOX-2 adjacent target site as wild-type SOX-2.2 This allows mutant SOX-2G73E to pair up with CEH-36 in AWB neurons, resulting in a cell identity transformation from AWB to AWCON in sox-2(ky707) mutants (Fig. 1).2
These findings reveal that a single amino acid change can result in a cell identity transformation at molecular, morphological, and functional levels. They also demonstrate that cooperative DNA binding is important for determining terminal identities of neurons, and support the notion that cell identities are determined both by activating a particular identity and suppressing an alternative identity. Interestingly, ectopic AWCON neurons in sox-2(ky707) mutants are specified largely independently of the genetic pathway specifying native AWCON neurons.2 This indicates that ectopic AWCON and native AWCON are specified with overlapped but distinct mechanisms. Another recent study has shown that sox-2 is also important for terminal differentiation of glutamatergic and cholinergic neurons in C. elegans, although it does not play a role in embryonic neurogenesis.7 Although vertebrate Sox2 has been implicated in generation of GABAergic neurons and retinal ganglion cells, whether Sox2 directly controls terminal differentiation of GABAneurons or RGCs is not known. Therefore, recent findings in C. elegans may provide mechanistic insight into the potential role of Sox2 in terminal neuronal differentiation in vertebrates.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Hobert O. WormBook 2010:1-24; PMID:20891032; http://dx.doi.org/ 10.1895/wormbook.1.12.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alqadah A, et al.. EMBO J 2015; 34:2574-89; PMID:26341465; http://dx.doi.org/ 10.15252/embj.201592188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hsieh YW, et al.. Genesis 2014; 52:544-54; PMID:24478264; http://dx.doi.org/ 10.1002/dvg.22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sagasti A, et al.. Genes Dev 1999; 13:1794-806; PMID:10421632; http://dx.doi.org/ 10.1101/gad.13.14.1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kamachi Y, Kondoh H. Development 2013; 140:4129-44; PMID:24086078; http://dx.doi.org/ 10.1242/dev.091793 [DOI] [PubMed] [Google Scholar]
- [6].Kim K, et al.. Development 2010; 137:963-74; PMID:20150279; http://dx.doi.org/ 10.1242/dev.044719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vidal B, et al.. Development 2015; 142:2464-77; PMID:26153233; http://dx.doi.org/ 10.1242/dev.125740 [DOI] [PMC free article] [PubMed] [Google Scholar]


