Abstract
Cutaneous and mucosal leishmaniasis is widely distributed in Central and South America. Leishmania of the Viannia subgenus are the most frequent species infecting humans. L. (V.) braziliensis, L. (V.) panamensis are also responsible for metastatic mucosal leishmaniasis. Conventional or real time PCR is a more sensitive diagnostic test than microscopy, but the cost and requirement for infrastructure and trained personnel makes it impractical in most endemic regions. Primary health systems need a sensitive and specific point of care (POC) diagnostic tool. We developed a novel POC molecular diagnostic test for cutaneous leishmaniasis caused by Leishmania (Viannia) spp. Parasite DNA was amplified using isothermal Recombinase Polymerase Amplification (RPA) with primers and probes that targeted the kinetoplast DNA. The amplification product was detected by naked eye with a lateral flow (LF) immunochromatographic strip. The RPA-LF had an analytical sensitivity equivalent to 0.1 parasites per reaction. The test amplified the principal L. Viannia species from multiple countries: L. (V.) braziliensis (n = 33), L. (V.) guyanensis (n = 17), L. (V.) panamensis (n = 9). The less common L. (V.) lainsoni, L. (V.) shawi, and L. (V.) naiffi were also amplified. No amplification was observed in parasites of the L. (Leishmania) subgenus. In a small number of clinical samples (n = 13) we found 100% agreement between PCR and RPA-LF. The high analytical sensitivity and clinical validation indicate the test could improve the efficiency of diagnosis, especially in chronic lesions with submicroscopic parasite burdens. Field implementation of the RPA-LF test could contribute to management and control of cutaneous and mucosal leishmaniasis.
Author Summary
Cutaneous leishmaniasis is a parasitic disease transmitted by the bite of sandflies that produces skin ulcers. The severe, disfiguring form of the disease is characterized by parasite dissemination to the mucosa of the nose and palate. Current diagnosis is based on microscopy which has low sensitivity in chronic cases. Molecular methods (PCR) that detect parasite DNA are highly sensitive but costs and personnel training make it impossible to implement it in resource-limited areas. We developed a novel molecular method (RPA-LF) that could be applied in the field because it does not require sophisticated equipment. It is also very sensitive and specific to detect the principal Leishmania species that produce cutaneous leishmaniasis in Latin America. Future field implementation of RPA-LF could have a positive impact on disease management and control.
Introduction
Dermal and mucosal leishmaniasis are widely distributed in Central and South America, affecting an estimated 190,000–300,000 people annually [1]. Many different Leishmania species grouped under the subgenera Leishmania or Viannia can produce dermal leishmaniasis. Epidemiologically, Viannia is the most relevant subgenus in this region since it is highly prevalent and also responsible for metastatic mucosal leishmaniasis (L. (V.) braziliensis, L. (V.) panamensis, L. (V.) guyanensis), the severe form of tegumentary disease [2,3].
Microscopy is still the most common diagnostic method used in endemic regions but its sensitivity is not ideal and markedly affected by the experience of the microscopist [4]. Furthermore, the sensitivity of this method tends to decrease with disease chronicity, which is characterized by a low number of amastigotes in the lesions [4]. Serological tests were used in the past and the identification of new antigens and formats for serodiagnosis of American cutaneous leishmaniasis is still considered [5,6]. However, in general, they have proven to be of limited value due to the variable immune responses of patients and no clear distinction between current disease and past infections or exposure.
Conventional or quantitative PCR from dermal or mucosal samples have high diagnostic sensitivity (≈87–98%) and specificity (≥84%) [7]. This molecular method is currently the gold standard in leishmaniasis reference centers or tertiary care facilities. However, the need for expensive equipment, trained personnel, and relatively complex laboratory facilities are beyond the capability of the typical health infrastructure in endemic areas.
Therefore, there is a clear need to provide primary health systems with diagnostic tools that are simple, easy to use and have good sensitivity and specificity. To address this critical gap, we developed a novel-point-of-care molecular test to diagnose dermal and mucosal leishmaniasis produced by Leishmania Viannia spp. We designed primers and probes that targeted the kinetoplast DNA minicircles, similar to the strategy we used previously to detect L. infantum chagasi [8]. Leishmania DNA was amplified using isothermal Recombinase Polymerase Amplification (RPA), a method originally described by Piepenburg et al.[9]. The amplification product was detected in a lateral flow immunochromatographic strip (LF) which is read with the naked eye. Its analytical sensitivity and specificity indicated that it could be used as a point-of-care diagnostic test for dermal and mucosal leishmaniasis in endemic areas of Latin America.
Materials and Methods
Ethical statement
The current study was approved by the Office of Sponsored Programs of the University of Texas Medical Branch. The activities of NAMRU-6 were conducted in compliance with all applicable federal and international regulations governing the protection of human subjects. This study was approved by the Institutional Review Board of the U.S. Naval Medical Research Unit 6 (NMRCD.2007.0018), and administratively approved by the Madre de Dios Regional Health Directorate in Peru. CIDEIM provided DNA samples stored in its cryobank and consent forms from patients for multiple uses were obtained. The IRB of UTMB waived ethical approval for this study based on the utilization of DNA from de-identified patients.
Design of primers and probe
The primer sets for Leishmania Viannia are 30–35 nucleotides long and target conserved sequences identified by computational alignment of L. Viannia kDNA minicircle sequences reported in GenBank. Primers were designed with 40–60% GC content, few direct/inverted repeats, and absence of long homopolymer tracts. We focused principally on conserved regions and to a lesser extent on regions with moderate variability, obtaining a 120 bp RPA amplicon in agarose gels. To enable detection by lateral flow, the reverse primer was biotinylated at the 5’ end. We designed a 45bp conserved internal probe (Biosearch technologies -Petaluma, CA) that included FAM (5’-carboxy fluorescein amidite) at the 5’ end, an internal dSpacer and a SpacerC3 in the 3’ end, as suggested by the manufacturer (TwistDx). Forward Primer: Fw- GATGAAAATGTACTCCCCGACATGCCTCTG. Reverse Primer: Rev-bio-CTAATTGTGCACGGGGAGGCCAAAAATAGCGA. Internal Probe: The probe contains a 5’-fluorescein group (FAM), an internal (THF)-tetrahydrofuran residue, and a C3 spacer block at the 3’ end. Probe-FAM-GTAGGGGNGTTCTGCGAAAACCGAAAAATG[THF]CATACAGAAACCCCG[C3-spacer].
Parasite DNA isolation
Promastigote suspensions of reference strains or clinical strains thawed from cryopreserved stocks or absorbed in Whatman FTA filter paper (Sigma-Aldrich) were subjected to 95°C for 2 minutes in a dry bath to lyse the parasites. DNA purification for the majority of the samples was carried out using the DNeasy Blood & Tissue Kit (Qiagen) following the recommendations of the vendor and adjusted to10 ng/μL. In addition, a small number of clinical samples (n = 8) obtained from ulcers of Peruvian patients suspected of having cutaneous leishmaniasis were absorbed in Whatman FTA filter paper. The filter papers (n = 2 of 6 mm diameter) were placed in direct contact with the ulcer allowing lymph and cells to be absorbed by the papers. Once dried, each patient sample was packed individually using sealed plastic bags which were labeled and transported to the central lab at room temperature. Two 3mm diameter filter papers were punched from the original samples, washed 3 times with 200 μL of FTA buffer (Whatman, GE Healthcare Life Sciences) followed by 3 washes with TE buffer pH 8 (Sigma). The papers were then suspended in water and heated at 95°C for 30 minutes; 2.5 μL of the solution were used to run the RPA-LF test.
RPA reaction and lateral flow reading
The amplification mixture was comprised of: 1) forward primer, 2) biotinylated reverse primer, 3) FAM-labeled probe (stocks-5μM), 4) magnesium acetate, and 5) the rehydrated cocktail (Twist amp nfo RPA kit -TwistDx, UK). Parasite DNA (5–25ng/μL) was immediately added to the mixture and subjected to amplification at 45°C for 30 minutes using a dry bath. The RPA product was diluted 1:25 in the dipstick assay buffer and 30 μL were placed in a 1.5 Eppendorf tube or 96-well microplate. The bottom tip of the lateral flow strip was then immersed in the sample (GenLine HybriDetect, Milenia Biotec, Germany) making the amplification product run upwards by capillarity. Parasite amplification was confirmed with the naked eye after 5 minutes by the appearance of the test band in the lower part of the strip. This band is produced when anti-biotin antibodies immobilize the amplified DNA which contains the biotinylated primers. The gold particles, which are covered with mouse anti-FAM antibodies, bind to the probe labeled with FAM making the test band visible. The reaction was validated by the appearance of the control band in the upper part of the strip. This band appears upon the immobilization of excess free-gold particles (which are covered with mouse antibodies) by means of anti-mouse antibodies.
Quantitative PCR
The RPA-LF sensitivity was compared with SYBRgreen real-time PCR using the primers described by Pita-Pereira et al. [10].
Leishmania samples
The analytical evaluations of RPA-LF were carried out using known concentrations of DNA (10ng/μL). We evaluated banked strains of L. braziliensis from Brazil (n = 15), Colombia (n = 5), and Peru (n = 13); L. guyanensis from Brazil (n = 11) and Colombia (n = 6); L. panamensis from Colombia (n = 7), Nicaragua (n = 1), and Panama (n = 1); L. lainsoni from Brazil (n = 3) and Peru (n = 7); and L. shawi (n = 2) and L. naiffi (n = 6) from Brazil. Also, we evaluated DNA purified from lesion biopsies of patients from Peru who were infected with L. braziliensis (n = 9) and L. guyanensis (n = 4), as well as non-leishmanial (PCR-negative) skin lesions (n = 5).
Results
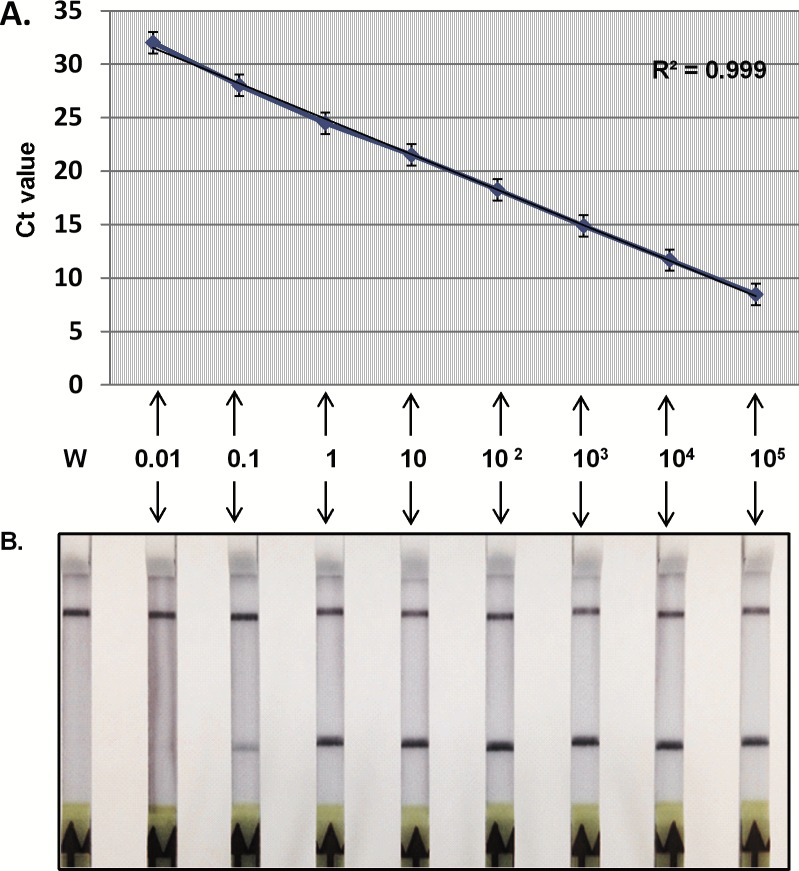
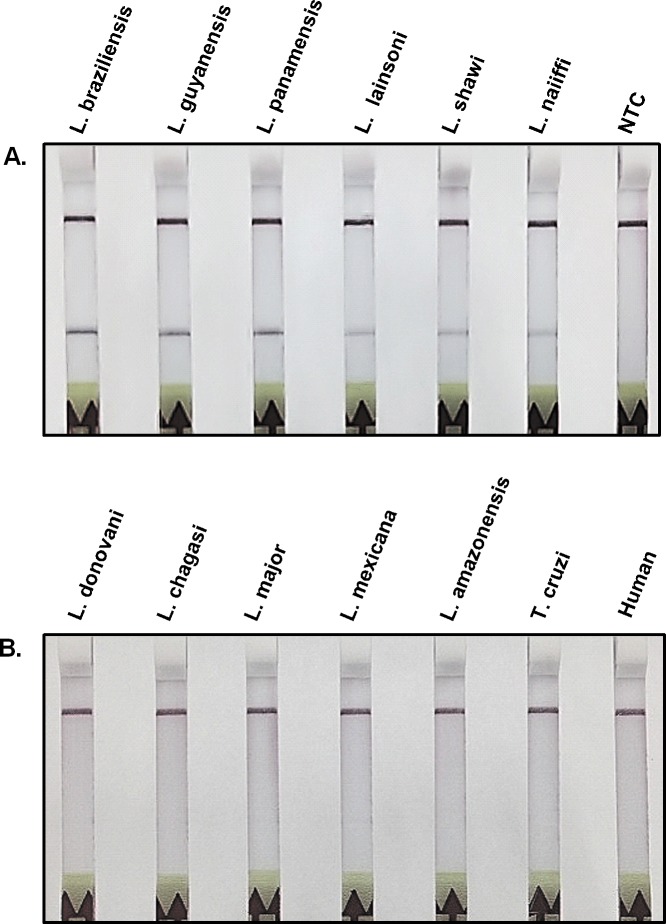
The RPA-LF amplified Leishmania DNA with an analytical sensitivity equivalent to 0.1 parasite per reaction, which corresponded to aCt value of 28 in the real-time PCR used as the gold standard (Fig 1). The capacity of RPA-LF to detect the most relevant species of the subgenus Viannia was initially determined by the amplification of a small number of banked strains of Leishmania Viannia spp: L. braziliensis, L. panamensis, L. guyanensis, L. lainsoni, L. shawi and L. naiffi. The specificity was confirmed by the lack of amplification of L. donovani, L. chagasi, L. mexicana, L. amazonensis, L. major, Trypanosoma cruzi and human DNA (Fig 2).
Fig 1. Sensitivity of RPA-LF to detect L. viannia spp. compared with real-time PCR (the current gold standard).
Ten-fold serial dilutions of parasite DNA, extracted with Qiagen DNeasy blood and tissue kit, were amplified by qPCR (SYBRgreen) (A) or RPA-LF (B). W = water. The control band is the upper band, while the test band is the lower band.
Fig 2. Specificity of RPA-LF to amplify species of the Viannia subgenus.
A) The most relevant L. Viannia species (L. braziliensis, L. guyanensis, L. panamensis) produced stronger bands in the lateral flow strip than other less common species of this subgenus. B) Species of the Leishmania subgenus, Trypanosoma cruzi, and human DNA were not amplified by the RPA-LF test. NTC = no template control.
We further evaluated panels of strains from different species within the Viannia subgenus isolated in endemic areas of Brazil, Colombia, and Peru. Fifteen out of 15 L. braziliensis strains from Brazil, 6/6 strains from Colombia, and 12/12 from Peru, isolated from humans or dogs from different geographical areas, were amplified by RPA-LF (Table 1). The test also demonstrated good sensitivity to detect several L. guyanensis strains obtained from endemic regions of Brazil (11/11) and Colombia (6/6) (Table 1). Similarly, L. panamensis strains originally isolated from patients of Colombia (7/7), Nicaragua (1/1), and Panama (1/1) were readily amplified by RPA-LF. A small group of L. Viannia species known to occasionally infect humans were also evaluated by RPA-LF. Two Brazilian strains of L. shawi, a species closely related to L. guyanensis, produced strong bands indicating that the primers efficiently amplified this parasite species. However, 5/6 strains of Leishmania naiffi, usually found in mammals of the Amazon region and less frequently in other parts of South America, were amplified less efficiently than other Viannia species and generated weaker bands (Table 1). In the case of L. lainsoni, a parasite found in wild mammals and sporadically infecting humans, RPA-LF produced a weak yet clearly detectable band in 3/3 strains from Brazil and 6/7 from Peru. One L. naiffi-L. lainsoni hybrid from Brazil was also detected by RPA/LF. Collectively, these results indicated that the test is capable of detecting all the epidemiologically relevant species of the Viannia subgenus. We developed an interactive map that depicts the geographical distribution of Leishmania species evaluated by RPA-LF (http://www.scribblemaps.com/maps/view/Leish_Viannia/9-18-15).
Table 1. RPA-LF detection of Leishmania Viannia species isolated from different countries in Latin America.
| Leishmania spp. | Country | Region1 | WHO code | RPA-LF | Map code2 |
|---|---|---|---|---|---|
| L. braziliensis | Brazil | Pará | MHOM/BR/1975/M2903 | + | 1 |
| Ceará | MHOM/BR/1987/H-210 | + | 2 | ||
| Amazonas | MHOM/BR/1988/IM3482 | + | 3 | ||
| Ceará | MCAN/BR/1990/C35 | + | 4 | ||
| Ceará | MCAN/BR/1991/C51 | + | 5 | ||
| Amazonas | MHOM/BR/1994/IM3946 | + | 6 | ||
| Espírito Santo | MHOM/BR/1994/HAD-1 | + | 7 | ||
| Bahia | MHOM/BR/1996/SBS | + | 8 | ||
| Bahia | MHOM/BR/2001/LTCP13183 | + | 9 | ||
| Acre | MHOM/BR/2002/NMT-RBO037 | + | 10 | ||
| Bahia | MHOM/BR/2001/NMT-LTCP14369-P | + | 11 | ||
| Rio de Janeiro | MHOM/BR/2008/NC | + | 12 | ||
| Pernambuco | MHOM/BR/2010/MMS | + | 13 | ||
| Santa Catarina | MHOM/BR/2006/LSC128 | + | 14 | ||
| Santa Catarina | MHOM/BR/2006/LSC185 | + | 15 | ||
| Colombia | Caqueta | MHOM/CO/87/1270 | + | 16 | |
| Nariño | MHOM/CO/85/2388 | + | 17 | ||
| Putumayo | MHOM/CO/82/L71 | + | 18 | ||
| Caqueta | MHOM/CO/88/1403 | + | 19 | ||
| Meta | MHOM/CO/85/1110 | + | 20 | ||
| Nariño | MHOM/CO/97/3144 | + | 21 | ||
| Peru | Cusco | MHOM/PE/14/LDP-0053 | + | 22 | |
| Loreto | MHOM/PE/14/LDP-0057 | + | 23 | ||
| Junín | MHOM/PE/14/LDP-0060 | + | 24 | ||
| Junín | MHOM/PE/14/LDP-0065 | + | 25 | ||
| Cusco | MHOM/PE/14/LDP-0067 | + | 26 | ||
| Junín | MHOM/PE/14/LDP-0073 | + | 27 | ||
| Cusco | MHOM/PE/14/LDP-0075 | w | 28 | ||
| Madre de Dios | MHOM/PE/13/LDP-2036 | + | 29 | ||
| Madre de Dios | MHOM/PE/13/LDP-2039 | + | 30 | ||
| Madre de Dios | MHOM/PE/13/LDP-2059 | + | 31 | ||
| Madre de Dios | MHOM/PE/14/LDP-2074 | + | 32 | ||
| WHO | MHOM/PE/84/LTB300 | + | 33 | ||
| L. guyanensis | Brazil | Amazonas | MHOM/BR/1997/NMT-MAO 210P | + | 34 |
| Amazonas | MHOM/BR/1997/NMT-MAO 212P | + | 35 | ||
| Amazonas | MHOM/BR/1997/NMT-MAO 237P | + | 36 | ||
| Amazonas | MHOM/BR/1997/NMT-MAO 246P | + | 37 | ||
| Amazonas | MHOM/BR/1997/NMT-MAO 292P | + | 38 | ||
| Amazonas | MHOM/BR/1997/NMT-MAO 307P | + | 39 | ||
| Amazonas | MHOM/BR/1997/NMT-MAO 317P | + | 40 | ||
| Amazonas | MHOM/BR/1997/NMT-MAO 325P | + | 41 | ||
| Amazonas | MHOM/BR/2007/031-LOP | + | 42 | ||
| Amazonas | MHOM/BR/2007/033-MECM | + | 43 | ||
| WHO | MHOM/BR/75/M4147 | + | 44 | ||
| Colombia | Caqueta | MHOM/CO/83/1028 | + | 45 | |
| Putumayo | MHOM/CO/82/L76 | + | 46 | ||
| Putumayo | MHOM/CO/82/L75 | + | 47 | ||
| Caqueta | MHOM/CO/88/1390 | + | 48 | ||
| Tolima | MHOM/CO/2008/A197 | + | 49 | ||
| Putumayo | MHOM/CO/83/1011 | + | 50 | ||
| L. panamensis | Nicaragua | Chontales | MHOM/NI/1988/XD45 | + | 51 |
| Colombia | Putumayo | MHOM/CO/92/1735 | + | 52 | |
| Valle | MHOM/CO/84/1048 | + | 53 | ||
| Nariño | MHOM/CO/85/2476 | + | 54 | ||
| Nariño | MHOM/CO/85/2472 | + | 55 | ||
| Cauca | MHOM/CO/86/1180 | + | 56 | ||
| Narino | MHOM/CO/83/2017 | + | 57 | ||
| Cauca | MHOM/CO/95/1989 | + | 58 | ||
| Panama | WHO | MHOM/PA/71/LS94 | + | 59 | |
| L. lainsoni | Brazil | Pará | MHOM/BR/1981/M6426 | + | 60 |
| Rondônia | MCOE/BR/1983/IM1367 | + | 61 | ||
| Pará | MCUN/BR/1983/IM1721 | + | 62 | ||
| Peru | Amazonas | MHOM/PE/14/LDP-0061 | w | 63 | |
| Loreto | MHOM/PE/13/LDP-1021 | + | 64 | ||
| Madre de Dios | MHOM/PE/15/LDP-2138 | + | 65 | ||
| Madre de Dios | MHOM/PE/15/LDP-2169 | w | 66 | ||
| Madre de Dios | MHOM/PE/15/LDP-2236 | + | 67 | ||
| Madre de Dios | MHOM/PE/15/LDP-2242 | - | 68 | ||
| Huanuco | MHOM/PE/88/BAB1730 | + | 69 | ||
| L. shawi | Brazil | Pará | IWHI/BR/1985/IM2322 | + | 70 |
| Pará | MCEB/BR/1984/M8408 | + | 71 | ||
| L. naiffi | Brazil | Pará | ISQU/BR/1985/IM2264 | + | 72 |
| Pará | MDAS/BR/1987/IM3280 | - | 73 | ||
| Pará | MDAS/BR/1979/M5533 | + | 74 | ||
| Amazonas | MHOM/BR/1991/IM3740 | + | 75 | ||
| Pará | MHOM/BR/2011/S50 | + | 76 | ||
| Pará | MHOM/BR/2011/58-AMS | w | 77 | ||
| L. naiffi/ lainsoni | Acre | MHOM/BR/2002/NMT-RBO004 | + | 78 |
+ indicates positive reading; W = indicates weak band
1Region: Brazil = State; Colombia = Department; Peru = Region
2Link to the interactive map: http://www.scribblemaps.com/maps/view/Leish_Viannia/9-18-15
In a small number of clinical samples we found that RPA-LF has excellent agreement with PCR as determined in DNA samples from patients of Peru infected with L. braziliensis or L. guyanensis (Table 2). All 9 of the samples from clinical lesions due to L. (V.) braziliensis, and all 4 of the samples from clinical lesions due to L. (V.) guyanensis were positive by RPA-LF. The samples from negative controls were uniformly negative by RPA-LF. The high sensitivity and specificity identified with these limited number of samples warrants large-scale field testing to determine the diagnostic sensitivity of the RPA-LF.
Table 2. Agreement between RPA-LF and PCR to amplify Leishmania Viannia DNA purified from lesions of cutaneous leishmaniasis patients from Peru.
| Leishmania spp. | Country | Region | WHO Code | qPCR | RPA-LF | Map Code1 |
|---|---|---|---|---|---|---|
| L. braziliensis | Peru | Ucayali | MHOM/PE/2012/LDP-0005-Bx | L.b. | + | 79 |
| Cusco | MHOM/PE/2012/LDP-0011-Bx | L.b. | + | 80 | ||
| Junín | MHOM/PE/2012/LDP-0012-Bx | L.b. | + | 81 | ||
| Madre de Dios | MHOM/PE/2013/LDP-0034-Bx | L.b. | + | 82 | ||
| Junín | MHOM/PE/2014/LDP-0052-Bx | L.b. | + | 83 | ||
| Junín | MHOM/PE/2014/LDP-0052-FP | L.b. | + | 84 | ||
| Loreto | MHOM/PE/2014/LDP-0057-Bx | L.b. | + | 85 | ||
| Loreto | MHOM/PE/2014/LDP-0057-FP | L.b. | + | 86 | ||
| Loreto | MHOM/PE/2014/LDP-0057-L | L.b. | + | 87 | ||
| L. guyanensis | Huánuco | MHOM/PE/2012/LDP-0007-Bx | L.g. | + | 88 | |
| San Martin | MHOM/PE/2012/LDP-0014-Bx | L.g. | + | 89 | ||
| San Martin | MHOM/PE/2013/LDP-0041-Bx | L.g. | + | 90 | ||
| San Martin | MHOM/PE/2013/LDP-0041-FP | L.g. | + | 91 | ||
| Negative Controls | San Martin | MHOM/PE/2012/LDP-0017-Bx | - | - | ||
| Junín | MHOM/PE/2012/LDP-0030-Bx | - | - | |||
| Ucayali | MHOM/PE/2013/LDP-0042-Bx | - | - | |||
| Cusco | MHOM/PE/2013/LDP-0043-Bx | - | - | |||
| Iquitos | MHOM/PE/2015/LDP-0083-Bx | - | - |
+ indicates positive reading
L.b. = L. braziliensis as determined by qPCR; L.g. = L. guyanensis as determined by qPCR.
1Link to the interactive map: http://www.scribblemaps.com/maps/view/Leish_Viannia/9-18-15
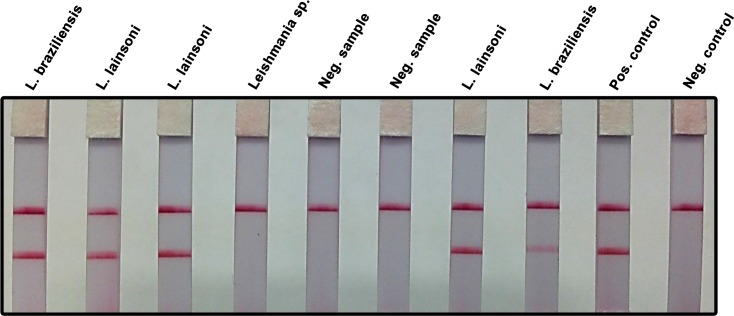
A potential limitation for field application of molecular diagnostic methods is the need for equipment such as vortex and high speed centrifuge to purify DNA from the clinical sample. We evaluated a DNA extraction method based on brief washing, elution and 95°C heating of the sample absorbed in filter paper (details in Materials & Methods). Using this approach we successfully amplified by RPA-LF all the samples of patients infected with Leishmania Viannia spp. as confirmed by real time PCR (Fig 3). This result indicated that RPA-LF could be implemented in basic diagnostic settings.
Fig 3. RPA-LF amplification of clinical samples using a simplified DNA extraction method.
Samples from patients suspected of having cutaneous leishmaniasis were obtained by pressing Whatman FTA filter paper (two 6 mm diameter discs) over the dermal lesions. Two, 3 mm diameter papers were cut from the original samples using a punch, washed thrice with FTA washing reagent and twice with TE buffer pH 8. A 2.5 μL aliquot was amplified by RPA and subsequently read using a lateral flow strip. Patients infected with Leishmania Viannia spp. (L. lainsoni, L. braziliensis) were readily detected. The test did not amplify a strain originally labeled as Leishmania sp. in NAMRU-6, Peru. Presumably, it did not belong to the Viannia subgenus as confirmed by real-time PCR at UTMB. There was agreement between NAMRU-6 and UTMB labs with regard to the negative clinical samples. L. lainsoni was used as positive control; the negative control was run without template.
Discussion
We developed a field-applicable molecular diagnostic test that distinguishes between the subgenera Viannia and Leishmania by selectively detecting strains of the Viannia subgenus. Our primers and probes were designed to target the kinetoplast DNA minicircles due to the high copy number (≈ 10,000) of this circular network of genomic mitochondrial DNA [11]. This remarkable number of copies provides a comparative advantage over other parasite targets with regard to test sensitivity. We targeted the Viannia subgenus because it encompasses the most relevant species causing cutaneous leishmaniasis in Latin America. The evaluation of the RPA-LF test included strains from Brazil, Colombia, and Peru, in which the recently reported incidence was 26,008, 17,420, and 6,405 cases/year, respectively [1]. The number of patients requiring diagnosis in these countries could be even greater since it was estimated that under-reporting varied between 2.8 and 4.6 fold [1].
The discrimination between Viannia and Leishmania subgenera is clinically relevant because in Latin America these infections may be treated differently [12]. Also, infection with L. braziliensis, L. panamensis, and less frequently L. guyanensis require prolonged patient follow up due to the risk of mucosal metastasis after apparent successful treatment [13,14].Leishmania (V.) shawi was readily detected by the RPA-LF test. Early studies suggested that L. shawi was not frequently reported in humans and seemed to be of low prevalence in nature [15,16]. However, more recent studies in Northeastern Brazil found that 6.5% (5/77) of isolates were identified as L. shawi and that some of them could be considered hybrids with L. braziliensis [17].The RPA-LF was less efficient at amplifying L. naiffi, a species found in armadillos and occasionally infecting humans in different countries of South America [18,19]. Therefore, further test optimization would be necessary for epidemiological studies aimed at this particular species.
During the development phase, we detected variability in distinct batches of the lateral flow strips (Milenia Biotec, Germany) regarding increased background that led to the appearance of faint test bands in the negative controls. The problem was resolved by using higher dilutions of the amplification product (1:100–1:200). Each laboratory should standardize and select the lateral flow strips that best suits its needs. There are different commercial options of immunochromatographic strips for lateral flow reading. They are offered in containers with multiple strips (Milenia, Biotec), individual cards (Abingdon Health, UK), or cassettes (UStar, China) that are putatively less prone to contamination.
Scrapings or brushings of cutaneous lesions absorbed in filter paper were shown to be amenable to molecular diagnosis using PCR [20]. We have already shown that RPA-LF could use this preservation-transportation method to amplify Leishmania DNA from the blood of dogs infected with L. chagasi [8].The test was capable of amplifying DNA equivalent to 0.1 parasites in the reaction mix, which was comparable to the detection limit of our qPCR. Preliminary results using a small number of samples from lesions suggested that RPA-LF can efficiently detect parasite DNA in the presence of host DNA with high sensitivity and specificity. Nevertheless, the diagnostic sensitivity will have to be evaluated under field conditions in a larger number of patients. It is well established that parasite burdens tend to be highly variable and that parasites are more difficult to detect in chronic lesions [4]. Therefore, it will be particularly important to evaluate the diagnostic sensitivity of the RPA-LF in chronic lesions with >3 months of evolution.
A significant advantage of the RPA-LF is that samples can be rapidly processed, without the need of sophisticated equipment, outside of a traditional laboratory (e.g. at a house, school, or community center). Furthermore, initial evaluations strongly suggested that DNA extraction could be accomplished efficiently using a method that does not require equipment other than a boiling bath, giving additional support to the feasibility of adapting RPA-LF to the POC. RPA-LF is a less complex test than other isothermal amplification methods. RPA-LF results would be available in approximately one hour and the patients could initiate treatment if tested positive. Compared to a PCR reference test, this approach should enable earlier initiation of treatment, significantly increasing compliance and treatment efficacy. The need for delivering samples to a central reference lab, that leads to delayed therapeutic decisions and increased risk of patient loss, would be avoided. Importantly, the implementation of the field-applicable RPA-LF could replace or repurpose the need for experienced microscopists (and microscopes). It will improve the efficiency to diagnose leishmaniasis of short evolution time and, more importantly, in chronic lesions with parasite burdens below the microscopy threshold. The RPA-LF test may well fill the need for a field-applicable test, which is critical to cutaneous and mucosal leishmaniasis management.
Acknowledgments
We thank Maryori Vidarte for reconfirmation of the species identification of Colombian strains of Leishmania recovered the cryobank of the Centro Internacional de Entrenamiento e Investigaciones Médicas-CIDEIM.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The views expressed in this article are those of the authors only and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
Several authors of this manuscript are employees of the U.S. Government. This work was prepared as part of their duties. Title 17 U.S.C. § 105 provides that 'Copyright protection under this title is not available for any work of the United States Government.' Title 17 U.S.C. § 101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties.
Data Availability
All the relevant data are within the paper and link to an interactive map.
Funding Statement
This study was partially funded by the Armed Forces Health Surveillance Center, Global Emerging Infections Surveillance and Response System, the AFHSC/GEIS of the U.S. Department of Defense and the Congresionally Directed Medical Research Programs (CDMRP) Award W81XWH-14-2-0196; Log # PR130282. Support was also received from the Institute for Translational Sciences at the University of Texas Medical Branch (UTMB), which is supported in part by a Clinical and Translational Science Award (UL1 TR001439) from the National Center for Advancing Translational Sciences, National Institutes of Health; MTT Award (UL1TR000071). Partial support was also provided by the Center for Tropical Diseases from UTMB. AGL is sponsored by a training grant of the Fogarty International Center of the US National Institutes of Health (2D43 TW007393). The participation of OLF in this work was supported by a doctoral scholarship awarded by the Departamento Administrativo de Ciencia, Tecnología e Innovación-COLCIENCIAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, et al. (2012) Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weigle K, Saravia NG (1996) Natural history, clinical evolution, and the host-parasite interaction in New World cutaneous leishmaniasis. Clinics in dermatology 14: 433–450. [DOI] [PubMed] [Google Scholar]
- 3.Blum J, Lockwood DN, Visser L, Harms G, Bailey MS, et al. (2012) Local or systemic treatment for New World cutaneous leishmaniasis? Re-evaluating the evidence for the risk of mucosal leishmaniasis. Int Health 4: 153–163. 10.1016/j.inhe.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 4.Weigle KA, de Davalos M, Heredia P, Molineros R, Saravia NG, et al. (1987) Diagnosis of cutaneous and mucocutaneous leishmaniasis in Colombia: a comparison of seven methods. Am J Trop Med Hyg 36: 489–496. [DOI] [PubMed] [Google Scholar]
- 5.Barroso-Freitas AP, Passos SR, Mouta-Confort E, Madeira MF, Schubach AO, et al. (2009) Accuracy of an ELISA and indirect immunofluorescence for the laboratory diagnosis of American tegumentary leishmaniasis. Trans R Soc Trop Med Hyg 103: 383–389. 10.1016/j.trstmh.2008.12.019 [DOI] [PubMed] [Google Scholar]
- 6.Romero LI, Paz HM, Ortega-Barria E, Bayard V, Hochberg LP, et al. (2004) Evaluation of serological assays based on a novel excreted antigen preparation for the diagnosis of Cutaneous Leishmaniasis in Panama. J Microbiol Methods 57: 391–397. [DOI] [PubMed] [Google Scholar]
- 7.Adams ER, Gomez MA, Scheske L, Rios R, Marquez R, et al. (2014) Sensitive diagnosis of cutaneous leishmaniasis by lesion swab sampling coupled to qPCR. Parasitology 141: 1891–1897. 10.1017/S0031182014001280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellanos-Gonzalez A, Saldarriaga OA, Tartaglino L, Gacek R, Temple E, et al. (2015) A Novel Molecular Test to Diagnose Canine Visceral Leishmaniasis at the Point of Care. The American journal of tropical medicine and hygiene: 15–0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piepenburg O, Williams CH, Stemple DL, Armes NA (2006) DNA detection using recombination proteins. PLoS Biol 4: e204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pita-Pereira D, Lins R, Oliveira MP, Lima RB, Pereira BA, et al. (2012) SYBR Green-based real-time PCR targeting kinetoplast DNA can be used to discriminate between the main etiologic agents of Brazilian cutaneous and visceral leishmaniases. Parasit Vectors 5: 15 10.1186/1756-3305-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris E, Kropp G, Belli A, Rodriguez B, Agabian N (1998) Single-step multiplex PCR assay for characterization of New World Leishmania complexes. Journal of Clinical Microbiology 36: 1989–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Organization WH (2010) Control of the leishmaniases. World Health Organization technical report series: xii. [PubMed]
- 13.Arevalo J, Ramirez L, Adaui V, Zimic M, Tulliano G, et al. (2007) Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J Infect Dis 195: 1846–1851. [DOI] [PubMed] [Google Scholar]
- 14.Llanos-Cuentas A, Tulliano G, Araujo-Castillo R, Miranda-Verastegui C, Santamaria-Castrellon G, et al. (2008) Clinical and parasite species risk factors for pentavalent antimonial treatment failure in cutaneous leishmaniasis in Peru. Clin Infect Dis 46: 223–231. 10.1086/524042 [DOI] [PubMed] [Google Scholar]
- 15.Lainson R, Braga R, De Souza A, Povoa M, Ishikawa E, et al. (1989) Leishmania (Viannia) shawi sp. n., a parasite of monkeys, sloths and procyonids in Amazonian Brazil. Annales de parasitologie humaine et comparée 64: 200 [DOI] [PubMed] [Google Scholar]
- 16.Shaw J, Ishikawa E, Lainson R, Braga R, Silveira F (1990) Cutaneous leishmaniasis of man due to Leishmania (Viannia) shawi Lainson, de Souza, Povoa, Ishikawa & Silveira, in Para State, Brazil. Annales de parasitologie humaine et comparée 66: 243–246. [DOI] [PubMed] [Google Scholar]
- 17.Brito ME, Andrade MS, Mendonca MG, Silva CJ, Almeida EL, et al. (2009) Species diversity of Leishmania (Viannia) parasites circulating in an endemic area for cutaneous leishmaniasis located in the Atlantic rainforest region of northeastern Brazil. Trop Med Int Health 14: 1278–1286. 10.1111/j.1365-3156.2009.02361.x [DOI] [PubMed] [Google Scholar]
- 18.Naiff RD, Freitas RA, Naiff MF, Arias JR, Barrett TV, et al. (1991) Epidemiological and nosological aspects of Leishmania naiffi Lainson & Shaw, 1989. Mem Inst Oswaldo Cruz 86: 317–321. [DOI] [PubMed] [Google Scholar]
- 19.Pratlong F, Deniau M, Darie H, Eichenlaub S, Proll S, et al. (2002) Human cutaneous leishmaniasis caused by Leishmania naiffi is wide-spread in South America. Ann Trop Med Parasitol 96: 781–785. [DOI] [PubMed] [Google Scholar]
- 20.Veland N, Boggild AK, Valencia C, Valencia BM, Llanos-Cuentas A, et al. (2012) Leishmania (Viannia) species identification on clinical samples from cutaneous leishmaniasis patients in Peru: assessment of a molecular stepwise approach. Journal of clinical microbiology 50: 495–498. 10.1128/JCM.05061-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the relevant data are within the paper and link to an interactive map.