Abstract
Rice blast is one of the most destructive diseases affecting rice worldwide. The adoption of host resistance has proven to be the most economical and effective approach to control rice blast. In recent years, sequence-specific nucleases (SSNs) have been demonstrated to be powerful tools for the improvement of crops via gene-specific genome editing, and CRISPR/Cas9 is thought to be the most effective SSN. Here, we report the improvement of rice blast resistance by engineering a CRISPR/Cas9 SSN (C-ERF922) targeting the OsERF922 gene in rice. Twenty-one C-ERF922-induced mutant plants (42.0%) were identified from 50 T0 transgenic plants. Sanger sequencing revealed that these plants harbored various insertion or deletion (InDel) mutations at the target site. We showed that all of the C-ERF922-induced allele mutations were transmitted to subsequent generations. Mutant plants harboring the desired gene modification but not containing the transferred DNA were obtained by segregation in the T1 and T2 generations. Six T2 homozygous mutant lines were further examined for a blast resistance phenotype and agronomic traits, such as plant height, flag leaf length and width, number of productive panicles, panicle length, number of grains per panicle, seed setting percentage and thousand seed weight. The results revealed that the number of blast lesions formed following pathogen infection was significantly decreased in all 6 mutant lines compared with wild-type plants at both the seedling and tillering stages. Furthermore, there were no significant differences between any of the 6 T2 mutant lines and the wild-type plants with regard to the agronomic traits tested. We also simultaneously targeted multiple sites within OsERF922 by using Cas9/Multi-target-sgRNAs (C-ERF922S1S2 and C-ERF922S1S2S3) to obtain plants harboring mutations at two or three sites. Our results indicate that gene modification via CRISPR/Cas9 is a useful approach for enhancing blast resistance in rice.
Introduction
Rice (Oryza sativa L.) is one of the most important food crops in the world, feeding nearly 50% of the world’s population. Rice blast, caused by the filamentous ascomycete fungus Magnaporthe oryzae, is one of the most destructive diseases affecting rice in all rice-growing countries and often causes serious damage to global rice production [1, 2]. Enhancing the resistance of rice to M. oryzae has been shown to be the most economical and effective approach for controlling rice blast [3, 4].
Over the course of evolution, plants have evolved sophisticated mechanisms to resist pathogen infection. In plant cells, surface-localized pattern recognition receptors (PRRs) rapidly perceive pathogen-associated molecular patterns (PAMPs) [5] and activate a battery of defense mechanisms [6]. PAMP-triggered immunity (PTI) is considered a conserved and ancient form of plant immunity that acts as the first line of inducible defense to various pathogens [6, 7]. The plant hormones abscisic acid, salicylic acid, jasmonic acid and ethylene play important roles in this defense response [8–10].
Plant ethylene responsive factors (ERF), a subfamily of the APETELA2/ethylene response factor (AP2/ERF) transcription factor superfamily in plants, are involved in the modulation of multiple stress tolerance and have been implicated in multiple responses to abiotic and biotic stresses [11–13]. For example, expression of the rice ERF genes OsBIERF1, OsBIERF3 and OsBIERF4 are not only induced by M. oryzae infection, but also up-regulated by salt, cold, drought and wounding [14]. Likewise, the overexpression of BrERF11 enhanced disease resistance to Ralstonia solanacearum in tobacco [15]. However, overexpression of PpERF3b in tobacco contributes to susceptibility to disease caused by Pseudomonas syringae pv. Tabaci [16]. This is in line with the finding that silencing StERF3 in potato produced enhanced foliage resistance to Phytophthora infestans [17]. Similarly, knockdown of expression of the rice ERF gene OsERF922 by RNA interference (RNAi) enhanced rice resistance to M. oryzae, indicating that OsERF922 acts as a negative regulator of blast resistance in rice [18].
In conventional rice breeding, it takes approximately a decade to pyramid multiple blast resistance genes into a rice variety via crossing and backcrossing, while the high pathogenic variability in M. oryzae often leads to the rapid break down of resistant cultivars [19]. Thus, enhancement of PTI for the development of broad-spectrum resistance has been suggested as an effective approach for breeding varieties resistant to rice blast [20–22]. Breeding strategy using RNAi-based down-regulation of rice transcription factor expression has been demonstrated to be an alternative approach for enhancing rice resistance to blast [18, 23–25]. However, expression of RNAi transgenes varies in different transgenic lines; a large number of transgenic plants are required to identify candidates in which the transgene is highly expressed over multiple generations. In addition, rice plants derived by RNAi methods are usually regarded as transgenic and are subjected to rigorous regulatory processes [26].
In recent years, sequence-specific nucleases (SSNs), including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) 9 (CRISPR/Cas9) have been demonstrated to be useful tools for plant genome editing [27, 28]. In particular, CRISPR/Cas9 has been demonstrated to be the most effective SSN thus far and has been used for genome editing in major crops such as rice [29–41], maize [42–45], wheat [39, 46, 47], sorghum [30], tomato [48, 49], soybean [50–52] and potato [53].
The gene-specific DNA double-strand breaks (DSBs) caused by the SSNs are repaired primarily by the high-fidelity homologous recombination (HR) or error-prone non-homologous end joining (NHEJ) pathways [54]. NHEJ often introduces small insertion or deletion (InDel) mutations at the cut site that lead to the loss of gene function. Compared with RNAi technology, SSN-based genome editing can achieve complete knockout without incorporating exogenous DNA. To date, successful examples of ZFN- and TALEN-based improvements of agronomically important traits in major crops have been reported [47, 55–60]. Here, we report the improvement of rice blast resistance via CRISPR/Cas9-targeted knockout of the ERF transcription factor gene OsERF922 in Kuiku131, a japonica rice variety widely cultivated in northern China.
Materials and Methods
Plant growth
The japonica rice variety Kuiku131 and all transgenic plants were grown in a net house and greenhouse at 28–35°C in Beijing, or fields in the experimental station under normal growth conditions in Sanya. The experimental station is specialized for genetically modified crops planting permitted by Chinese Ministry of Agriculture. For blast inoculation at the seedling stage, rice seeds were grown in 60 × 30 × 5 cm plastic seedling-nursing trays supplemented with a mineral nutrient solution in a greenhouse maintained at approximately 28–35°C under natural sunlight [61]. Briefly, moistened seeds of wild-type rice and rice mutants (T2 progeny) were sown in rows (15 seed per row) in triplicate trays.
Vector construction
The C-ERF922-expressing vector (pC-ERF922) was constructed according to a method described by Ma et al. [38]. Briefly, the target site (Fig 1A) sequence-containing primers E922-FS2/E922-RS2 (S1 Table) were synthesized by Sangon Biotech (http://www.sangon.com/) and combined by annealing. Then, the target site sequence-containing chimeric primers were cloned into the sgRNA expression cassette pYLsgRNA-U6a at a BsaI site [38]. The integrated sgRNA expression cassette was then amplified by nested PCR [38] using the primers U-F/E922-RS2 and E922-FS2/gR-R (for the first round) and the site-specific primers Pps-GGL and Pgs-GGR (for the second round) (S1 Table). Subsequently, the amplicons containing ERF922-S2-sgRNA with different BsaI-cutting sites were cloned into the CRISPR/Cas9 Multi-targeting vector pYLCRISPR/Cas9Pubi-H at a BsaI site [38]; the resultant construct pC-ERF922 contained a Cas9p expression cassette (Pubi::NLS::Cas9p::NLS::Tnos) and a hygromycin resistance cassette (2×P35S::HPT::T35STnos) (Fig 1B).
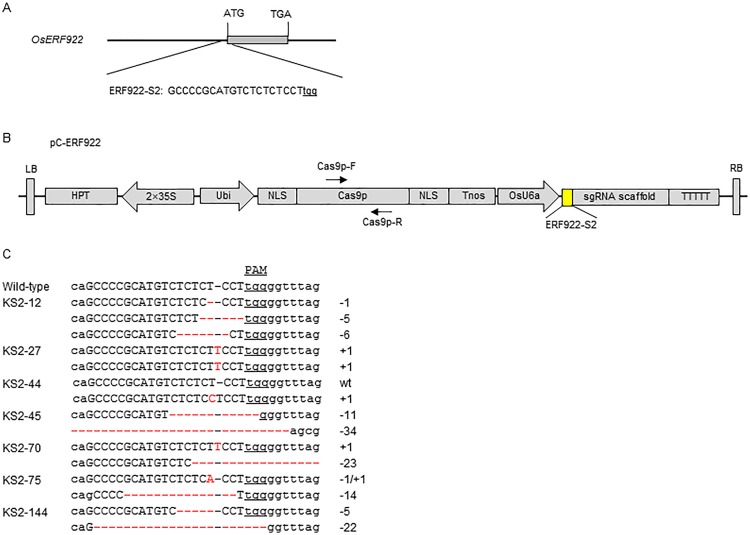
Fig 1. CRISPR/Cas9-induced OsERF922 gene modification in rice.
(A) Schematic of OsERF922 gene structure and the C-ERF922 target site (ERF922-S2). OsERF922 contains a single exon indicated by gray rectangles. The translation initiation codon (ATG) and termination codon (TGA) are shown. The target site nucleotides are shown in capital letters, and the protospacer adjacent motif (PAM) site is underlined. (B) Schematic diagram of the pC-ERF922 construct for expressing the CRISPR/Cas9 protein C-ERF922. The positions and orientations of the primers Cas9p-F and Cas9p-R, which were used to screen Cas9-free mutants, are indicated by small arrows. The expression of Cas9 is driven by the maize ubiquitin promoter (Ubi); the expression of the sgRNA scaffold is driven by the rice U6a small nuclear RNA promoter (OsU6a); the expression of hygromycin (HPT) is driven by 2 CaMV35S promoters (2 × 35S). NLS: nuclear localization signal; Tnos: gene terminator; LB and RB: left border and right border, respectively. (C) Nucleotide sequences at the target site in the 7 T0 mutant rice plants. The recovered mutated alleles are shown below the wild-type sequence. The target site nucleotides are indicated as black capital letters and black dashes. The PAM site is underlined. The red dashes indicate the deleted nucleotides. The red capital letters indicate the inserted nucleotides. The numbers on the right indicate the type of mutation and the number of nucleotides involved. “−” and “+” indicate the deletion and insertion of the indicated number of nucleotides, respectively; “−/+” indicates the simultaneous deletion and insertion of the indicated number of nucleotide.
The C-ERF922S1S2- and C-ERF922S1S2S3-expressing vectors (pC-ERF922S1S2 and pC-ERF922S1S2S3) were constructed according to the method described by Ma et al. [38]. Briefly, the target site-containing sequence primers E922-FS1/E922-RS1, E922-FS2/E922-RS2 and E922-FS3/E922-RS3 (S1 Table, Fig 1A and S1 Fig) were combined by annealing, and then the target site sequence-containing chimeric primers were cloned into the sgRNA expression cassettes pYLsgRNA-U3, pYLsgRNA-U6a and pYLsgRNA-U6b at a BsaI site [38], respectively. The integrated sgRNA expression cassettes were then amplified by nested PCR [38] using U-F/Reverse adapter primers (E922-RS1, E922-RS2 and E922-RS3) and Forward adapter primers (E922-FS1, E922-FS2 and E922-FS3)/gR-R for the first round, and the corresponding site-specific primers Pps-GGL/Pgs-GG2, Pps-GG2/Pgs-GGR and Pps-GGL/Pgs-GG2, Pps-GG2/Pgs-GG3, Pps-GG3/Pgs-GGR for the second round (S1 Table). Subsequently, the two-target-sgRNAs (ERF922-S1-sgRNA and ERF922-S2-sgRNA) and three-target-sgRNAs (ERF922-S1-sgRNA, ERF922-S2-sgRNA and ERF922-S3-sgRNA) expression cassettes were ligated into the pYLCRISPR/Cas9Pubi-H vector [38], resulting in the C-ERF922S1S2- and C-ERF922S1S2S3-expressing constructs pC-ERF922S1S2 and and C-ERF922S1S2S3-expressing constructs pC-ERF922S1S2 and pC-ERF922S1S2S3 (S2 Fig).
Rice transformation
The Cas9/sgRNA-expressing binary vectors (pC-ERF922, pC-ERF922S1S2 and pC-ERF922S1S2S3) were transformed into an Agrobacterium tumefaciens strain EHA105 by electroporation. Agrobacterium-mediated transformation of the embryogenic calli derived from the japonica rice variety Kuiku131 was performed according to Hiei et al. [62]. Briefly, hygromycin-containing medium was used to select hygromycin-resistant calli, and then the hygromycin-resistant calli were transferred onto regeneration medium for the regeneration of transgenic plants. After 2–3 months of cultivation, transgenic seedlings were transferred to a field during the rice growing season.
Protoplast assay
Kuiku131 seeds were sterilized with 0.2% potassium permanganate solution for 24 h and then imbibed in water at 37°C for 24 h before germination. Seedlings were grown in cylindrical glass bottles lined with wet toilet paper under a regime of 12 h light (150–200 μmol m-2 s-1)/12 h dark at 26°C in an incubator for 10–14 days before protoplast isolation. The protoplast isolation and transformation were performed following protocols published by Zhang et al. [63] and Shan et al. [64], respectively.
Healthy and fresh rice stems and sheaths from 50 rice plants were used. A bundle of rice plants were cut together into fine strips approximately 0.5 mm in length using sharp razors. The fine strips were immediately transferred into an enzyme solution (1.5% Cellulase RS, 0.75% Macerozyme R-10, 0.6 M mannitol, 10 mM MES at pH 5.7, 10 mM CaCl2 and 0.1% BSA). After 5 h digestion with gentle shaking (60–80 rpm) in the dark, an equal volume of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl and 2 mM MES at pH 5.7) was added, followed by light shaking for 10 sec. The protoplasts were released by filtering through 40 μm nylon meshes into round bottom tubes and were washed 2–3 times using W5 solution. The pellets were collected by centrifugation at 250 g for 3 min. After washing, the pellets were resuspended in MMG solution (0.4 M mannitol, 15 mM MgCl2 and 4 mM MES at pH 5.7) at a concentration of 2 × 106 cells mL-1, as calculated using a hematocytometer.
Protoplast transformation was carried out in a poly-ethylene glycol (PEG) solution [40% (W/V) PEG 4000, 0.2 M mannitol and 0.1 M CaCl2]. For one sample, 20 μg of plasmid DNA was mixed with 200 μL protoplasts (approximately 4 × 105 cells) and 220 μl freshly prepared PEG solution, and the mixture was incubated at room temperature for 20 min in the dark. After incubation, 880 μL of W5 solution was added slowly, and the protoplast cells were harvested by centrifugation at 250 g for 3 min. The protoplast cells were resuspended gently in 2 mL WI solution (0.5 M mannitol, 20 mM KCl and 4 mM MES at pH 5.7) and cultured in 6-well plates in darkness at room temperature for 48 h.
The genomic DNA was extracted from protoplast cells transformed with pC-ERF922 plasmid by the SDS method [65]. The protoplast genomic DNA was subjected to PCR with the gene-specific primer pair E922-KF/E922-KR (S1 Table) to amplify DNA fragments across the target site. Then, the PCR amplicons were cloned into the pEASY-Blunt vector (TransGen Biotech, Beijing, China), and a total of 48 randomly selected colonies were further characterized by Sanger DNA sequencing.
Identification of mutant transgenic plants
Genomic DNA was extracted from individual transgenic plants using SDS extraction according to Dellaporta et al. [65]. All transgenic hygromycin-resistant T0 plants were characterized by PCR using the Cas9-specific primers Cas9p-F/Cas9p-R (Fig 1B and S1 Table). Subsequently, all PCR-positive plants were subjected to PCR using the gene-specific primer pairs E922-KF/E922-KR and E922P-2F/E922-KR2 (S1 Table) to amplify DNA fragments across the target sites (ERF922-S2, ERF922-S1, ERF922-S2 and ERF922-S3, respectively); the PCR amplicons were then directly sequenced. The sequencing chromatograms with superimposed peaks of bi-allelic and heterozygous mutations were decoded by the Degenerate Sequence Decoding (DSD) method [66]-based web tool DSDecode (http://dsdecode.scgene.com/) [67].
Pathogen inoculation
To evaluate the resistance to M. oryzae at the seedling stage, the inoculation of rice blast fungus M. oryzae was performed according to a method described by Wang et al. [20]. Briefly, 3–4 week-old wild-type and homozygous mutant plants were inoculated by spraying with conidial suspensions (2 × 105 conidia mL-1, 0.02% Tween 20) of M. oryzae isolate 06-47-6. The inoculated plants were grown in an ENCONAIR phytotron at 26°C (95% humidity) in the dark for 24 h, then were grown under conditions of a 16 h/8 h light/dark cycle with 95% humidity. Disease severity was evaluated according to the method described by Fukuoka et al. [68] at 7 d post inoculation (dpi). The area of the lesions was determined for the third leaves of 10 plants of each line. The experiments were repeated three times.
To confirm the presence of disease, all seedlings of wild-type and homozygous mutant lines were transplanted to the field and injection-inoculated at the tillering stage according to a method described by Ma et al. [69]. Briefly, when rice plants grow to 5–6 tillers, conidial suspensions (2 × 104 conidia mL-1) of M. oryzae were injected into the rice stem with a syringe until the suspension emerged from the heart-leaf. The injection sites on the stems were approximately 10 cm from the top of the tillers. Five tillers were injected for each plant. Disease severity was evaluated according to method described by Kobayashi et al. [70] at 7 dpi. The length of the lesions was assessed on the inoculated leaves of five tillers for each line. The experiments were repeated three times.
Agronomic trait characterization
Wild-type and homozygous T2 mutant lines were grown in the field under normal growth conditions in Beijing. Agronomic traits were characterized by measuring plant height, flag leaf length and width, the number of productive panicles, panicle length, the number of grains per panicle, the seed setting rate, and thousand seed weight after the rice had reached maturity. Five plants were investigated for each line.
Results
CRISPR/Cas9 design and the assessment of gene-editing activity
To design a CRISPR/Cas9 (C-ERF922) targeting the OsERF922 gene in rice, a 20-bp nucleotide sequence containing the initiation codon of the open reading frame of OsERF922 was chosen as the target site (ERF922-S2) (Fig 1A). The predicted Cas9 cleavage site in the coding region of the gene was seven base-pairs downstream from the ATG initiation codon. The binary plasmid pC-ERF922 (Fig 1B) was then constructed based on the CRISPR/Cas9 vector described by Ma et al. [38]. To test the gene-editing efficacy of C-ERF922, rice protoplasts were transformed with pC-ERF922, and genomic DNA was extracted to amplify the DNA fragment containing the target site. PCR amplicons generated with the primers E922-KF and E922-KR (S1 Table) were cloned into the pEASY-Blunt vector to isolate the colonies for sequencing. Three mutants (6.3%, S3 Fig) were recovered from 48 randomly selected colonies. Sequencing revealed that the mutation in colony C1 was a single nucleotide substitution; in colony C2 was a 5-bp deletion; and in colony C3 was a 30-bp insertion (S3 Fig). These observations showed that the C-ERF922-expression construct in pC-ERF922 exhibits gene-editing activity in rice protoplasts and can be used for creating mutant rice plants.
Recovery of rice plants with mutations in OsERF922
The pC-ERF922 construct was used to transform the rice variety Kuiku131 by Agrobacterium-mediated transformation, with the goal of enhancing its blast resistance by gene-specific editing. We obtained 50 positive transgenic (T0) plants and analyzed the target site in 21 of the plants (S4 Fig). Direct Sanger-sequencing of the target-containing amplicons followed by decoding via the DSD method [66, 67] showed that among the 21 plants, there were 16 bi-allelic mutations, 3 homozygous mutations, 1 heterozygous mutation, and 1 chimeric mutation (Table 1). Based on allele mutation types, more than half (64.3%, 27/42) of the mutations were nucleotide deletions, 23.8% (10/42) of the mutations were nucleotide insertions, and 11.9% (5/42) of the mutations were simultaneous nucleotide deletions and insertions (Table 1). As for the deletion mutations, the majority (63.0%, 17/27) were short (≤ 10 bp) deletions and the other 37.0% (10/27) were longer deletions ranging from 11 bp to 34 bp (S4 Fig); as for the insertion mutations, 90.0% (9/10) were 1 bp insertions (S4 Fig).
Table 1. Ratios of mutant genotype and mutation type at the target site (ERF922-S2) in T0 mutant plants.
| Mutant genotype ratios (%) a | Mutation type ratios (%) b | |||||
|---|---|---|---|---|---|---|
| Chimera* | Bi-allele | Homozygote | Heterozygote | Deletion | Insertion | Deletion and insertion |
| 4.8(1/21) | 76.1 (16/21) | 14.3(3/21) | 4.8 (1/21) | 64.3 (27/42) | 23.8 (10/42) | 11.9 (5/42) |
a Based on the number of each mutant genotype out of the total number of all mutant genotypes at the target site.
b Based on the number of each allele mutation type out of the total number of all allele mutation types at the target site.
* Refers to a plant with at least three distinct alleles detected at the target site.
Transmission of C-ERF922-induced mutations from the T0 to the T1 and T2 generations
To determine whether and how the C-ERF922-induced mutations were transmitted to the next generation, 4 bi-allelic (KS2-45, 70, 75, 144), 1 chimeric (KS2-12), 1 homozygous (KS2-27) and 1 heterozygous (KS2-44) T0 mutant plants (Fig 1C) were self-pollinated, and their progenies were genotyped at the target site. A total of 120 T1 plants derived from the T0 mutant plants were genotyped by PCR and DNA sequencing (Table 2). We found that all allelic mutations in the T0 mutant plants were transmitted to the T1 generation with a transmission rate of 100% (Table 2). In theory, allelic mutations in the bi-allelic T0 mutant plants should segregate to T1 plants following Mendelian genetic law (1xx:2xy:1yy). As expected, homozygous genotypes were detected in all T1 populations derived from the T0 mutant plants, even when the T1 segregation pattern of progeny from the chimeric T0 mutant plant KS2-12 was more diverse and less predictable. For all bi-allelic T0 mutant plants with the exception of KS2-45, the segregation ratio of [homozygote (xx): bi-allele mutants (xy): homozygote (yy)] in the T1 populations fit a 1:2:1 ratio shown to be statistically reliable in a chi-square test of the T1 plants (Table 2). For example, the bi-allelic T0 mutant plant KS2-70 harbors two mutations [a 23-bp deletion (-23) and a 1-bp insertion (+1)]; its T1 progenies segregated in a ratio of [11(-23): 20(-23, +1): 10(+1)], matching the (1xx: 2xy: 1yy) ratio well (Table 2).
Table 2. Segregation and types of C-ERF922-induced mutations in the target gene and their transmission to subsequent generations.
| Mutation transmission in the T1 or T2 generations | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mutant plants a | Genotype | Mutation type b | No. of plants tested | Wt | Bi-allele | Homozygote | Heterozygote | Transmission ratio (%) c | No. of T-DNA-free plants d |
| T0 generation | |||||||||
| KS2-12 | Chimera* | -1, -5, -6 | 8 | 0 | 2(-1, -5), 2(-1, -6) | 4(-1) | 0 | 100.0 | 0 |
| KS2-27 | Homozygote | +1 | 6 | 0 | 0 | 6(+1) | 0 | 100.0 | 1 |
| KS2-44 | Heterozygote | +1, wt | 3 | 0 | 0 | 2(+1) | 1(+1, wt) | 100.0 | 0 |
| KS2-45 | Bi-allele | -11, -34 | 37 | 0 | 26(-11, -34) | 3(-11), 8(-34) | 0 | 100.0 | 3 |
| KS2-70 | Bi-allele | -23, +1 | 41 | 0 | 20(-23, +1) | 11(-23), 10(+1) | 0 | 100.0e | 6 |
| KS2-75 | Bi-allele | -14, -1/+1 | 14 | 0 | 5(-14, -1/+1) | 4(-14), 5(-1/+1) | 0 | 100.0e | 0 |
| KS2-144 | Bi-allele | -5, -22 | 11 | 0 | 5(-5, -22) | 3(-5), 3(-22) | 0 | 100.0e | 0 |
| T1 generation | |||||||||
| KS2-27-1 | Homozygote | +1 | 30 | 0 | 0 | 30(+1) | 0 | 100.0 | 7 |
| KS2-44-1 | Heterozygote | +1, wt | 40 | 12 | 4(-1/+1, -1), 2(-1, +1) | 14(+1) | 8(+1, wt) | 70.0 | 0 |
| KS2-45-1 | Bi-allele | -11, -34 | 57 | 0 | 28(-11, -34) | 10(-11), 19(-34) | 0 | 100.0e | 9 |
| KS2-45-2 | Homozygote | -11 | 30 | 0 | 0 | 30(-11) | 0 | 100.0 | 0 |
| KS2-45-6 | Homozygote# | -34 | 30 | 0 | 0 | 30(-34) | 0 | 100.0 | 30 |
a KS2-27-1 and KS2-44-1 were progenies of KS2-27 and KS2-44, respectively; KS2-45-1, KS2-45-2 and KS2-45-6 were progenies of KS2-45.
b “−” indicates the deletion of the indicated number of nucleotides; “+” indicates the insertion of the indicated number of nucleotides; “−/+” indicates the simultaneous deletion and insertion of the indicated number of nucleotides at the same site.
c Based on the number of plants carrying the observed mutation out of the total number of plants tested.
d Mutant plants not containing DNA from the pC-ERF922 construct.
e Segregation of the bi-allele lines conforms to a Mendelian 1: 2: 1 ratio according to the χ2 test (P > 0.5).
* Refers to a plant with at least three distinct alleles detected at the target site
# T-DNA-free homozygote.
The transmission of mutations from T1 mutant plants to the T2 generation was further investigated using 3 homozygous (KS2-27-1, KS2-45-2 and KS2-45-6), 1 bi-allelic (KS2-45-1) and 1 heterozygous (KS2-44-1) T1 mutant plants (Table 2). Similarly, all allelic mutations in the T1 mutant plants were transmitted to the T2 generation, and the transmission rates ranged from 70% to 100%; the segregation pattern of T2 plants derived from the bi-allelic T1 mutant plant KS2-45-1 also followed Mendelian genetic law (Table 2). In addition, all T1 plants derived from the homozygous T0 mutant plant (KS2-27) and T2 plants derived from homozygous T1 mutant plants (KS2-27-1, KS2-45-2 and KS2-45-6) were homozygous for the same mutations (Table 2), indicating that the mutations in these homozygous mutant lines were stably transmitted to the next generation as expected. Interestingly, several new allelic mutations [(-1/+1, -1) and 2(-1, +1)] were detected among the T2 progeny of KS2-44-1 (Table 2 and S5 Fig), revealing that C-ERF922 activity was sustainable in the heterozygous T1 mutant plant. Together, these results clearly demonstrate that CRISPR/Cas9-induced gene mutations can be stably transmitted to subsequent generations.
Selection of T-DNA-free mutant rice lines
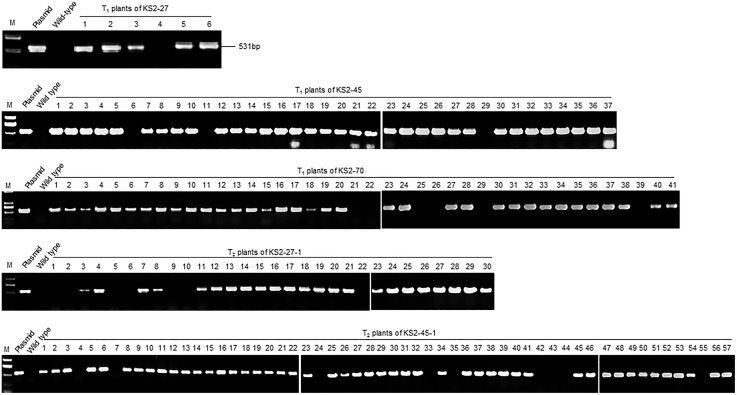
To investigate the possibility of obtaining rice lines harboring the desired modifications in OsERF922 but without transferred DNA (T-DNA) of the construct pC-ERF922, we designed the Cas9 gene-specific PCR primers Cas9p-F and Cas9p-R (Fig 1B and S1 Table) and performed PCR assays of the T1 and T2 plants. All 120 T1 plants were subjected to PCR assays, and 10 (8.3%) T1 plants failed to generate a Cas9-specific 531-bp amplicon from the transferred pC-ERF922 construct (Fig 2 and Table 2). Similarly, the PCR assay also failed to detect the pC-ERF922 construct in 16 out of 157 (10.2%) T2 plants derived from the 4 T1 mutant plants (KS2-27-1, KS2-44-1, KS2-45-1 and KS2-45-2) (Fig 2 and Table 2). Notably, all 30 T2 plants derived from the T1 mutant plant KS2-45-6 failed to generate the Cas9-specific amplicon (Table 2) because the KS2-45-6 plant was a Cas9-free homozygous mutant harboring the desired OsERF922 modifications. These results indicate that T-DNA-free plants carrying the desired gene modifications can be acquired through genetic segregation.
Fig 2. PCR-based identification of T-DNA-free rice mutant plants.
PCR products amplified from the progenies of KS2-27, KS2-45, KS2-70, KS2-27-1 and KS2-45-1 genomic DNA using the primers Cas9p-F and Cas9p-R. The numbers above the gel image refer to individual offspring of KS2-27, KS2-45, KS2-70, KS2-27-1 and KS2-45-1, respectively. M: DNA molecular weight marker; Plasmid: pC-ERF922; Wild-type: genomic DNA from Kuiku131.
Resistance to M. oryzae was enhanced in C-ERF922-induced rice mutants
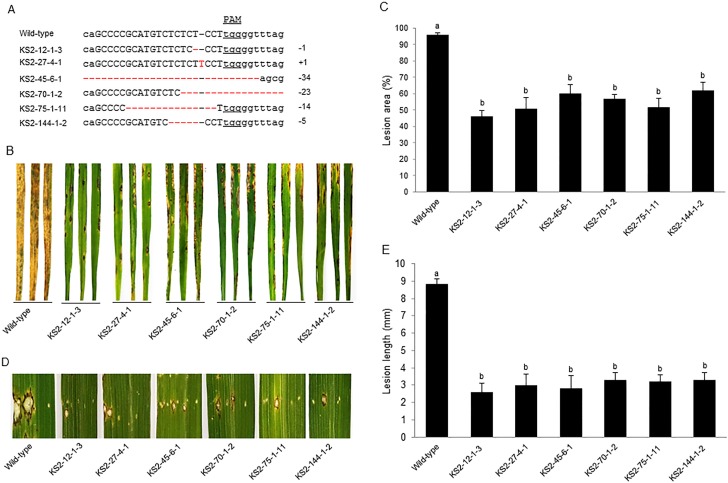
To characterize the blast resistance phenotype of the rice mutants, 6 homozygous mutant T2 lines (Fig 3A) with different types of allelic mutations were inoculated with the fungal pathogen M. oryzae isolate 06-47-6 at the seedling stage. The leaves of wild-type plants nearly died due to pathogen infection, likely because the pathogenicity of isolate 06-47-6 was very strong, and the wild-type variety was highly susceptible (Fig 3B). Nevertheless, the lesion areas formed by pathogen infection were significantly decreased in all mutant rice lines compared with wild-type plants (Fig 3B). The differences were further evaluated by quantification of the lesion areas and significance analysis using Student’s t-test (Fig 3C), which indicated that the mutant rice lines enhanced rice blast resistance. Similarly, lesion lengths formed by pathogen infection were also decreased in the mutant rice lines compared with the wild-type plants at the tillering stage (Fig 3D), and significant difference analysis of quantitative lesion length revealed that all mutant rice lines were significantly different from wild-type plants (Fig 3E). These results indicated that C-ERF922-induced frame shifts in the OsERF922 gene enhanced resistance to M. oryzae in the rice mutants because OsERF922 negatively regulates the blast resistance of rice [18].
Fig 3. Identification of blast resistance in homozygous mutant rice lines.
(A) Nucleotide sequences of the target site in the 6 homozygous T2 mutant rice lines used for pathogen inoculation. The recovered mutated alleles are shown below the wild-type sequence. The target site nucleotides are indicated using black capital letters and black dashes. The PAM site nucleotides are underlined. Red dashes indicate the deleted nucleotides. Red capital letters indicate the inserted nucleotides. The numbers on the right indicate the type of mutation and the number of nucleotides involved. “−” and “+” indicate the deletion and insertion of the indicated number of nucleotides, respectively. (B) The blast resistance phenotypes of the mutant rice lines and wild-type plants at the seedling stage. Leaves were detached from the inoculated plants at 7 dpi for photography. The experiments were repeated three times with similar results. (C) Histograms showing the average area of lesions formed on the third leaves of 10 plants for each line. The values marked with different letters are significantly different (P < 0.01, Student’s t-test). (D) Blast resistance phenotypes of the mutant rice lines and the wild-type plants at the tillering stage. Leaves were detached from the inoculated leaves of plants at 7 dpi for photography. The experiments were repeated three times with similar results. (E) Histograms showing the average length of lesions formed on the inoculated leaves of five tillerings for each line. The values marked with different letters are significantly different (P < 0.01, Student’s t-test).
The main agronomic traits were not altered in C-ERF922-induced rice mutants
To determine whether mutations in the OsERF922 gene affect agronomic traits, we characterized all 6 homozygous T2 mutant lines (Fig 3A) by measuring their plant height, flag leaf length and width, the number of productive panicles, panicle length, the number of grains per panicle, seed setting rate, and thousand seed weight. Student’s t-test revealed that none of the 6 T2 mutant lines differed significantly from wild-type plants under normal growth conditions with regard to the agronomic traits investigated (Table 3).
Table 3. Analysis of the agronomic traits of 6 homozygous T2 mutant lines.
| Mutant lines | Plant height (cm) | Flag leaf length (cm) | Flag leaf width (mm) | No. of productive panicles | Panicle length (cm) | No. of grainsper panicle | Seed setting rate (%) | Thousand seed weight (g) |
|---|---|---|---|---|---|---|---|---|
| WT | 59.8±1.9a | 26.2±3.2a | 13.6±0.5a | 9.0±1.0a | 12.4±1.0a | 69.6±3.2a | 90.0±1.1a | 26.3±0.3a |
| KS2-12-1-3 | 60.1±1.5a | 26.8±3.3a | 13.4±0.5a | 9.2±1.3a | 12.9±1.4a | 71.2±2.8a | 90.2±1.5a | 26.2±0.3a |
| KS2-27-4-1 | 59.8±2.4a | 27.2±2.9a | 13.6±1.1a | 9.4±1.5a | 12.6±0.7a | 69.2±3.0a | 89.0±0.8a | 26.5±0.3a |
| KS2-45-6-1 | 60.8±1.6a | 25.4±1.9a | 13.8±0.8a | 9.2±1.4a | 12.7±1.6a | 67.8±3.1a | 89.1±1.9a | 26.1±0.1a |
| KS2-70-1-2 | 60.0±2.1a | 25.8±1.6a | 13.6±1.2a | 9.4±1.6a | 13.0±0.5a | 68.8±3.7a | 88.4±0.5a | 26.2±0.2a |
| KS2-75-1-11 | 59.2±1.9a | 26.4±3.6a | 13.8±0.9a | 9.2±1.5a | 12.3±0.9a | 71.8±2.8a | 89.4±0.9a | 26.4±0.3a |
| KS2-144-1-2 | 59.6±1.7a | 25.2±3.3a | 13.6±1.3a | 9.4±1.7a | 12.5±0.5a | 69.0±2.4a | 89.8±1.1a | 26.3±0.2a |
The results are shown for five mutant plants of each mutant line and are represented as the mean ± SE. The values marked with the same letter (a) are non-significantly different (P < 0.05, Student’s t-test).
The mutagenic frequency and mutagenic frequency of homozygous plants were increased by targeting multiple sites within OsERF922
To examine whether the mutagenic frequency could be increased by targeting multiple sites within one gene, we designed Cas9/two-target-sgRNAs (C-ERF922S1S2) and Cas9/three-target-sgRNAs (C-ERF922S1S2S3) to target two sites (ERF922-S1 and ERF922-S2, Fig 1A and S1 Fig) and three sites (ERF922-S1, ERF922-S2 and ERF922-S3; Fig 1A and S1 Fig) within the OsERF922 gene in rice, respectively. Higher overall mutagenic frequencies and mutagenic frequencies of homozygous plants are of value because they facilitate the generation of homozygous mutants in the T0 generation for crop improvement. The C-ERF922S1S2- and C-ERF922S1S2S3-expressing constructs (pC-ERF922S1S2 and pC-ERF922S1S2S3, S2 Fig) were generated based on CRISPR/Cas9 multi-targeting vector described by Ma et al. [38] and were used to transform the rice variety Kuiku131 by Agrobacterium-mediated transformation. We obtained 30 positive transgenic (T0) plants for the analysis of mutations for each transformation and detected 21 (70.0%) and 27 (90.0%) mutant plants, respectively (Table 4, S2 and S3 Tables). Direct Sanger-sequencing of the target-containing amplicons followed by decoding with the DSD method [66, 67] showed that among the C-ERF922S1S2-induced mutant plants were 19 (63.3%) plants harboring mutations at both target sites (Table 4 and S2 Table). Furthermore, 47.6% (10/21, Table 5) of the mutants were homozygotes. In addition, among C-ERF922S1S2S3-induced mutant plants, all 27 (90.0%) plants harbored mutations at all three target sites (Table 4 and S3 Table), and 40.7% (11/27, Table 5) of the mutants were homozygotes. These results demonstrated that the mutagenic frequencies increased when targeting more sites within one gene, and Cas9/two-target-sgRNAs resulted in the highest mutagenic frequency in homozygotes (Tables 1 and 5).
Table 4. Targeting multiple sites in rice using CRISPR/Cas9 and the number of plants with mutations at single, double and triple target sites.
| No. of plants harboring mutations at target sites | |||||
|---|---|---|---|---|---|
| Transformant | No. of tested plants | ERF922-S1 | ERF922-S2 | ERF922-S3 | All* |
| pC-ERF922S1S2 | 30 | 21(70.0%) | 19(63.3%) | 19(63.3%) | |
| pC-ERF922S1S2S3 | 30 | 27(90.0%) | 27(90.0%) | 27(90.0%) | 27(90.0%) |
* Based on the number of plants with mutations for all combinations of target sites.
Table 5. Targeting multiple sites in rice using CRISPR/Cas9 and the mutant genotype ratios at the target sites.
| ERF922-S1 (%) # | ERF922-S2 (%) # | ERF922-S3 (%) # | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Transformant | Hetero | Bi | Homo | Hetero | Bi | Homo | Hetero | Bi | Homo | All Homo * |
| pC-ERF922S1S2 | 14.3(3/21) | 28.6(6/21) | 57.1(12/21) | 15.8(3/19) | 26.3(5/19) | 57.9(11/19) | 47.6(10/21) | |||
| pC-ERF922S1S2S3 | 3.8(1/27) | 37.0(10/27) | 59.3(16/27) | 3.8(1/27) | 44.4(12/27) | 51.8(14/27) | 3.8(1/27) | 48.1(13/27) | 48.1(13/27) | 40.7(11/27) |
# Based on the number of mutant genotypes out of the total number of mutant genotypes at each the target site.
* Based on the number of homozygote genotypes out of the total number mutant genotypes at all target sites. Hetero: Heterozygote; Bi: Bi-allele; Homo: Homozygote.
Discussion
Genome editing using SSNs provides an opportunity for crop improvement. Thus far, SSNs have been used to improve a variety of important crops, such as rice [56, 60], wheat [47], maize [55], soybean [58] and potato [59], by creating specific gene knockouts. However, examples of crop improvement via the creation of novel genotypes, agronomic traits or disease resistance remain limited. The first example was using ZFNs to target the maize IPK1 gene, which resulted in reduced levels of phytate—an anti-nutritional component of feed grains—and reduced phosphate pollution in waste streams from cattle-feeding operations [55]. In rice, the disease-susceptibility gene and the sucrose efflux transporter gene OsSWEET14, which aids in pathogen survival and virulence, was mutated by TALENs to produce disease-resistant rice with normal phenotypes [56]; moreover, using TALENs to target the OsBADH2 gene produced a generation of fragrant rice that contain 2-acetyl-1-pyrroline (2AP), a major fragrance compound [60]. In addition, knocking out all three MILDEW-RESISTANCE LOCUS (MLO) alleles in bread wheat using one pair of TALENs resulted in the creation of stable mutant lines exhibiting broad-spectrum resistance to powdery mildew [47]. In the present study, we used C-ERF922, C-ERF922S1S2 and C-ERF922S1S2S3 to knockout OsERF922 and achieved 42.0%, 70.0% and 90.0% recovery of C-ERF922-, C-ERF922S1S2- and C-ERF922S1S2S3-induced mutant plants, respectively, in T0 transgenic plants; all of the allele mutations were transmitted to the T1 and T2 generations. We obtained more than 20 mutant plants that harbor the desired modification in OsERF922 but not containing the transgene, which was eliminated via segregation in the T1 and T2 generations. Inoculation with M. oryzae revealed that blast resistance in the T2 homozygous mutant lines tested was significantly enhanced compared with that of wild-type plants at both the seedling and tillering stages. In addition, we showed that there was no significant difference between T2 homozygous mutant lines and wild-type plants with respect to the agronomic traits, such as plant height, flag leaf length and width, the number of productive panicles, panicle length, the number of grains per panicle, seed setting rate, and thousand seed weight. This study provides a successful example of improving rice blast resistance using CRISPR/Cas9 technology.
The C-ERF922-induced mutagenic frequency of T0 plants in this study was 42.0%, similar to those previously reported for CRISPR/Cas9-induced mutations in rice [31, 35, 38, 39]. In addition, the genotypes of these T0 mutant plants were primarily bi-allelic (76.1%) and homozygotic (14.3%) (Table 1), which was also similar to previous observations [29–33, 35–38, 71]. This phenomenon is in stark contrast to that of TALEN-induced mutant genotypes, wherein the heterozygous mutants were more frequent than bi-allele mutants in T0 plants [60, 64, 72, 73]. This difference might be caused by the different target site cleavage efficiencies of CRISPR/Cas9 and TALENs.
Multiple mutations were detected at the target site in the T0 mutant plant KS2-12 (Fig 1C). The presence of chimeric mutations in a single mutant plant may result from delayed cleavage in the primary embryogenic cell. This phenomenon has been reported in rice [29, 34, 35, 38], Arabidopsis [74], wheat [47], tomato [49] and maize [42]. In addition, the segregation ratios observed for the T1 plants derived from KS2-12 were not Mendelian, probably because the chimeric mutations were restricted to somatic cells that did not participate in the production of gametes. Furthermore, the segregation ratios found in T1 plants obtained from the bi-allelic mutant plant KS2-45 did not conform to a Mendelian ratio, but the T2 plants derived from the same bi-allelic mutant plant did (Table 2), indicating that the number of T1 plants was smaller or that homozygous mutation (11-bp deletion) may be induced through detrimental mutations caused by T-DNA insertion, which has less chance of survival compared with the other types of mutants. Notably, several novel mutations were detected in the T2 offspring of KS2-44-1 (S5 Fig). This could be explained by the fact that KS2-44-1 was a heterozygote in which the C-ERF922 construct remained active and continually cleaved the target site in T2 plants, resulting in new mutations. However, we did not detect new mutations in the T1 offspring of KS2-44, probably due to the existence of only 3 T1 plants (Table 2).
Both CRISPR/Cas9 and TALENs are effective tools for gene modification; however, each has specific advantages and limitations. Compared with TALENs, CRISPR/Cas9-expressing vectors are much easier to construct and can be competed in just two or three days [38, 75], whereas the construction of TALENs typically requires over seven days [76–78]. Furthermore, CRISPR/Cas9 induces a much higher mutation rate than TALENs. For example, the frequency of CRISPR/Cas9-targeted mutagenesis ranged from 21.1% to 66.7% (average 44.4%) for 11 rice genes [35]. Likewise, Ma and colleagues recently reported that the average mutation rate was 85.4% for CRISPR/Cas9-based editing of 46 target sites in rice [38]. However, the frequency of TALEN-targeted mutagenesis ranged from 0–30% overall [47, 64, 73]. Nevertheless, the requirement for a PAM (-NGG) sequence and the possibility of off-target effects are limitations of CRISPR/Cas9 system. For example, previous studies have demonstrated that off-target effects were common at the level of one nucleotide mismatch in plant species [37, 50], fish [79] and human cells [80, 81]. In contrast, off-target effects were extremely rare in the event of one nucleotide mismatch for the TALENs-editing system [82, 83]. The present study indicates that the CRISPR/Cas9 system is indeed a powerful tool for crop improvement via site-specific genome editing.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Dr. Richard A Wilson and two anonymous reviewers for their comments and suggestions for improvement of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by a grant from the Major Science and Technology Project to Create New Crop Cultivars using Gene Transfer Technology (grant number 2014ZX0801001B.
References
- 1.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012; 13(4): 414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu W, Liu J, Triplett L, Leach JE, Wang GL. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 2014; 52: 213–241. 10.1146/annurev-phyto-102313-045926 [DOI] [PubMed] [Google Scholar]
- 3.Boyd LA, Ridout C, O'Sullivan DM, Leach JE, Leung H. Plant-pathogen interactions: disease resistance in modern agriculture. Trends Genet. 2013; 29(4): 233–240. 10.1016/j.tig.2012.10.011 [DOI] [PubMed] [Google Scholar]
- 4.Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Asfaliza R, et al. Blast resistance in rice: a review of conventional breeding to molecular approaches. Mol Biol Rep. 2013; 40(3): 2369–2388. 10.1007/s11033-012-2318-0 [DOI] [PubMed] [Google Scholar]
- 5.Postel S, Kemmerling B. Plant systems for recognition of pathogen-associated molecular patterns. Semin Cell Dev Biol. 2009; 20(9): 1025–1031. 10.1016/j.semcdb.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 6.Jones JD, Dangl JL. The plant immune system. Nature. 2006; 444(7117): 323–329. [DOI] [PubMed] [Google Scholar]
- 7.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006; 124(4): 803–814. [DOI] [PubMed] [Google Scholar]
- 8.Rojo E, Solano R, Sánchez-Serrano JJ. Interactions between signaling compounds involved in plant defense. J Plant Growth Regul. 2003; 22(1): 82–98. [Google Scholar]
- 9.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005; 43: 205–227. [DOI] [PubMed] [Google Scholar]
- 10.De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant-Microbe Interact. 2005; 18(9): 923–937. [DOI] [PubMed] [Google Scholar]
- 11.Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, et al. The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta. 2007; 225(3): 575–588. [DOI] [PubMed] [Google Scholar]
- 12.Vogel MO, Moore M, Konig K, Pecher P, Alsharafa K, Lee J, et al. Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant cell. 2014; 26(3): 1151–1165. 10.1105/tpc.113.121061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller M, Munne-Bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015; 169(1): 32–41. 10.1104/pp.15.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y, Song F, Goodman RM, Zheng Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J Plant Physiol. 2006; 163(11): 1167–1178. [DOI] [PubMed] [Google Scholar]
- 15.Lai Y, Dang F, Lin J, Yu L, Shi Y, Xiao Y, et al. Overexpression of a Chinese cabbage BrERF11 transcription factor enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Physiol Biochem. 2013; 62: 70–78. 10.1016/j.plaphy.2012.10.010 [DOI] [PubMed] [Google Scholar]
- 16.Sherif S, El-Sharkawy I, Paliyath G, Jayasankar S. PpERF3b, a transcriptional repressor from peach, contributes to disease susceptibility and side branching in EAR-dependent and -independent fashions. Plant Cell Rep. 2013; 32(7): 1111–1124. 10.1007/s00299-013-1405-6 [DOI] [PubMed] [Google Scholar]
- 17.Tian Z, He Q, Wang H, Liu Y, Zhang Y, Shao F, et al. The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant cell Physiol. 2015; 56(5): 992–1005. 10.1093/pcp/pcv025 [DOI] [PubMed] [Google Scholar]
- 18.Liu DF, Chen XJ, Liu JQ, Ye JC, Guo ZJ. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot. 2012; 63(10): 3899–3912. 10.1093/jxb/ers079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia Y. Marker assisted selection for the control of rice blast disease. Pestic Outlook. 2003; 14(4): 150–152. [Google Scholar]
- 20.Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, et al. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007; 65(6): 799–815. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Guo Z. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice. Int J Mol Sci. 2008; 9(12): 2601–2613. 10.3390/ijms9122601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bundo M, Coca M. Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol J. 2015. November 18 10.1111/pbi.12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang CJ, Shimono M, Maeda S, Inoue H, Mori M, Hasegawa M, et al. Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant-Microbe Interact. 2009; 22(7): 820–829. 10.1094/MPMI-22-7-0820 [DOI] [PubMed] [Google Scholar]
- 24.Yara A, Yaeno T, Hasegawa M, Seto H, Montillet JL, Kusumi K, et al. Disease resistance against Magnaporthe grisea is enhanced in transgenic rice with suppression of omega-3 fatty acid desaturases. Plant Cell Physiol. 2007; 48(9): 1263–1274. [DOI] [PubMed] [Google Scholar]
- 25.Yara A, Yaeno T, Hasegawa M, Seto H, Seo S, Kusumi K, et al. Resistance to Magnaporthe grisea in transgenic rice with suppressed expression of genes encoding allene oxide cyclase and phytodienoic acid reductase. Biochem Biophys Res Commun. 2008; 376(3): 460–465. 10.1016/j.bbrc.2008.08.157 [DOI] [PubMed] [Google Scholar]
- 26.McGinnis KM. RNAi for functional genomics in plants. Brief Funct Genomics. 2010; 9(2): 111–117. 10.1093/bfgp/elp052 [DOI] [PubMed] [Google Scholar]
- 27.Baltes NJ, Voytas DF. Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 2015; 33(2): 120–131. 10.1016/j.tibtech.2014.11.008 [DOI] [PubMed] [Google Scholar]
- 28.Weeks DP, Spalding MH, Yang B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J. 2016; 14(2): 483–495. 10.1111/pbi.12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013; 23(10): 1229–1232. 10.1038/cr.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013; 41(20): e188 10.1093/nar/gkt780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, et al. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013; 23(10): 1233–1236. 10.1038/cr.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotech. 2013; 31(8): 686–688. [DOI] [PubMed] [Google Scholar]
- 33.Xu R, Li H, Qin R, Wang L, Li L, Wei P, et al. Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice. 2014; 7(1): 5 10.1186/s12284-014-0005-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, et al. Generation of inheritable and "transgene clean" targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep. 2015; 5: 11491 10.1038/srep11491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014; 12(6): 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]
- 36.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014; 42(17): 10903–10914. 10.1093/nar/gku806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Endo M, Mikami M, Toki S. Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 2015; 56(1): 41–47. 10.1093/pcp/pcu154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015; 8(8): 1274–1284. 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 39.Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014; 9(10): 2395–2410. 10.1038/nprot.2014.157 [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Shen L, Fu Y, Yan C, Wang K. A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genomics. 2015; 42(12): 703–706. 10.1016/j.jgg.2015.09.011 [DOI] [PubMed] [Google Scholar]
- 41.Ikeda T, Tanaka W, Mikami M, Endo M, Hirano HY. Generation of artificial drooping leaf mutants by CRISPR-Cas9 technology in rice. Genes Genet Syst. 2016; 90(4): 231–235. 10.1266/ggs.15-00030 [DOI] [PubMed] [Google Scholar]
- 42.Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014; 41(2): 63–68. 10.1016/j.jgg.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 43.Feng C, Yuan J, Wang R, Liu Y, Birchler JA, Han F. Efficient targeted genome modification in maize using CRISPR/Cas9 system. J Genet Genomics. 2016; 43(1): 37–43. 10.1016/j.jgg.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 44.Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, et al. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J Genet Genomics. 2016; 43(1): 25–36. 10.1016/j.jgg.2015.10.006 [DOI] [PubMed] [Google Scholar]
- 45.Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015; 169(2): 931–945. 10.1104/pp.15.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3. 2013; 3(12): 2233–2238. 10.1534/g3.113.008847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014; 32(9): 947–951. 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- 48.Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014; 166(3): 1292–1297. 10.1104/pp.114.247577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun. 2015; 467(1): 76–82. 10.1016/j.bbrc.2015.09.117 [DOI] [PubMed] [Google Scholar]
- 50.Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC biotechnol. 2015; 15: 16 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, et al. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol. 2016; 217: 90–97. 10.1016/j.jbiotec.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, et al. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015; 169(2): 960–970. 10.1104/pp.15.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015; 34(9): 1473–1476. 10.1007/s00299-015-1816-7 [DOI] [PubMed] [Google Scholar]
- 54.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011; 45: 247–271. 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- 55.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009; 459(7245): 437–441. 10.1038/nature07992 [DOI] [PubMed] [Google Scholar]
- 56.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012; 30(5): 390–392. 10.1038/nbt.2199 [DOI] [PubMed] [Google Scholar]
- 57.Wendt T, Holm PB, Starker CG, Christian M, Voytas DF, Brinch-Pedersen H, et al. TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol Biol. 2013; 83(3): 279–285. 10.1007/s11103-013-0078-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, et al. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J. 2014; 12(7): 934–940. 10.1111/pbi.12201 [DOI] [PubMed] [Google Scholar]
- 59.Clasen BM, Stoddard TJ, Luo S, Demorest ZL, Li J, Cedrone F, et al. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J. 2016; 14: 169–176. 10.1111/pbi.12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shan Q, Zhang Y, Chen K, Zhang K, Gao C. Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol J. 2015; 13: 791–800. 10.1111/pbi.12312 [DOI] [PubMed] [Google Scholar]
- 61.Li W, Lei C, Cheng Z, Jia Y, Huang D, Wang J, et al. Identification of SSR markers for a broad-spectrum blast resistance gene Pi20(t) for marker-assisted breeding. Mol Breeding. 2008; 22(1): 141–149. [Google Scholar]
- 62.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994; 6(2): 271–282. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011; 7(1): 30 10.1186/1746-4811-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, et al. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant. 2013; 6(4): 1365–1368. 10.1093/mp/sss162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II3. Plant Mol Biol Rep. 1983; 1(4): 19–21. [Google Scholar]
- 66.Ma X, Chen L, Zhu Q, Chen Y, Liu YG. Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol Plant. 2015; 8(8): 1285–1287. 10.1016/j.molp.2015.02.012 [DOI] [PubMed] [Google Scholar]
- 67.Liu W, Xie X, Ma X, Li J, Chen J, Liu YG. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant. 2015; 8(9): 1431–1433. 10.1016/j.molp.2015.05.009 [DOI] [PubMed] [Google Scholar]
- 68.Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009; 325(5943): 998–1001. 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- 69.Ma J, Lei C, Xu X, Hao K, Wang J, Cheng Z, et al. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance toleaf and neck blast in rice. Mol Plant-Microbe Interact. 2015; 28(5): 558–568. 10.1094/MPMI-11-14-0367-R [DOI] [PubMed] [Google Scholar]
- 70.Kobayashi N, Yanoria MJT, Fukuta Y. Differential varieties bred at IRRI and virulence analysis of blast isolates from the Philippines. JIRCAS Working Rep. 2007; 53: 17–30. [Google Scholar]
- 71.Xie K, Zhang J, Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014; 7(5): 923–926. 10.1093/mp/ssu009 [DOI] [PubMed] [Google Scholar]
- 72.Wang M, Liu Y, Zhang C, Liu J, Liu X, Wang L, et al. Gene editing by co-transformation of TALEN and chimeric RNA/DNA oligonucleotides on the rice OsEPSPS gene and the inheritance of mutations. Plos One. 2015; 10(4): e0122755 10.1371/journal.pone.0122755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H, Gou F, Zhang J, Liu W, Li Q, Mao Y, et al. TALEN-mediated targeted mutagenesis produces a large variety of heritable mutations in rice. Plant Biotechnol J. 2016; 14: 186–194. 10.1111/pbi.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA. 2014; 111(12): 4632–4637. 10.1073/pnas.1400822111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowder LG, Zhang D, Baltes NJ, Paul JW III, Tang X, Zheng X, et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015; 169(2): 971–985. 10.1104/pp.15.00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011; 39(12): e82 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011; 29(8): 699–700. 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- 78.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011; 39(14): 6315–6325. 10.1093/nar/gkr188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013; 140(24): 4982–4987. 10.1242/dev.099085 [DOI] [PubMed] [Google Scholar]
- 80.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013; 31(9): 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin YN, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014; 42(11): 7473–7485. 10.1093/nar/gku402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011; 39(21): 9283–9293. 10.1093/nar/gkr597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015; 33(2): 175–178. 10.1038/nbt.3127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.