ABSTRACT
Exposure to stressful life events during pregnancy exerts profound effects on neurodevelopment and increases the risk for several neurodevelopmental disorders including major depression. The mechanisms underlying the consequences of gestational stress are complex and remain to be elucidated. This study investigated the effects of gestational stress on depressive-like behavior and epigenetic modifications in young adult offspring. Gestational stress was induced by a combination of restraint and 24-hour light disturbance to pregnant dams throughout gestation. Depressive-like and anxiety-like behaviors of young adult offspring were examined. The expression and promoter methylation of brain derived neurotrophic factor (BDNF) were measured using RT-qPCR, Western blot, methylated DNA immunoprecipitation (MeDIP) and chromatin immunoprecipitation (ChIP). In addition, the expressions of histone deacetylases (HDACs) and acetylated histone H3 lysine 14 (AcH3K14) were also analyzed. Our results show that offspring from gestational stress dams exhibited depressive-like and anxiety-like behaviors. Biochemically, stress-offspring showed decreased expression of BDNF, increased expression of DNMT1, HDAC1, and HDAC2, and decreased expression of AcH3K14 in the hippocampus as compared to non-stress offspring. Data from MeDIP and ChIP assays revealed an increased methylation as well as decreased binding of AcH3K14 on specific BDNF promoters. Pearson analyses indicated that epigenetic changes induced by gestational stress were correlated with depressive-like and anxiety-like behaviors. These data suggest that gestational stress may be a suitable model for understanding the behavioral and molecular epigenetic changes observed in patients with depression.
KEYWORDS: Anxiety, BDNF, depression, epigenetics, gestational stress, HDAC, histone deacetylation, promoter methylation
Introduction
Major depression disorder is a heterogeneous disorder with a wide spectrum of symptoms. The etiology of major depression is complex and not well understood. Accumulated evidence suggests that exposure to stressful life events during pregnancy exerts profound effects on neurodevelopment and increases the risk for several neurodevelopmental disorders including major depression, bipolar disorder, schizophrenia, and autism.1-6 Recent studies suggest that epigenetic modifications of DNA and chromatin structure induced by environmental factors, including stress, may contribute to the complex phenotypes of neuropsychiatric disorders.7-12 For example, patients with psychosis exhibit an increase in brain DNA methyltransferases (DNMTs) and ten-eleven-translocation hydroxylases (TETs),13-17 leading to downregulation of candidate genes through promoter CpG residue methylation/hydroxymethylation. A study of postmortem hippocampus from suicide victims with childhood abuse history showed increased DNA methylation on the promoter region of the glucocorticoid-receptor gene.18 Differential methylation from genome-wide DNA methylation analyses suggests an epigenetic mechanism associated with major depression disorders.19 Furthermore, hippocampal DNMT inhibition displayed antidepressant effects in rats.20,21
Current theories regarding the pathogenesis of symptoms present in depression have depended largely on animal models. Because of the complex nature of depression, various animal models have been developed using different paradigms including genetic engineering, brain damage, and environmental manipulations in different genetic background rodents.22,23 Depressive-like rodent models can be divided largely into 2 groups: 1) acute stress models including the forced swim test (FST), tail suspension test (TST) and helplessness which offer rapid phenotyping and tests of antidepressant action; 2) chronic stress models including chronic mild stress models developed by applying physical stresses over couple of weeks and psychosocial stress models such as social defeat which offer a platform to investigate the neuroplasticity associated with chronic stress and drug actions. The molecular insights of depressive-like animal models cover a variety of systems including adrenergic, dopaminergic, GABAergic, serotonergic, glutamatergic, and mediators in the immune system, such as cytokines, and neurotrophins, such as brain derived neurotrophic factors (BDNF), and modifications of chromatin, such as histone methylation. Diverse molecular characterizations of rodent models have provided different capacities for exploring the pathophysiology of depression and its new therapeutics.24
At the present time, a significant focus in research on major depression has been the interplay between genetic and environmental factors. Epigenetics refers to the state of DNA CpG methylation/demethylation and chromatin structure, which controls gene transcription or silencing by facilitating or blocking transcription machinery access. The development of depression cannot be studied only in the post-mortem brain of patients and requires the use of animal models. In order to study epigenetic mechanisms involved in the pathogenesis of major depression, we measured depressive-like and anxiety-like behaviors exhibited by young adult offspring of dams exposed to restraint stress combined with 24-hour constant light disturbance throughout gestation. Gestational stress is a paradigm widely used for modeling psychiatric disorders.
BDNF is a significant target gene of depression.25,26 Reduced expression of BDNF and increased promoter methylation of BDNF-exons-iv and -ix have been reported in the brain and blood of patients with depression.27-33
The mice we used in this study were Kunming species mice. Kunming mice are the most widely used outbreed colony in China. The molecular genetic profiles of the species and the extent of genetic differentiation among populations are still unclear. Because Kunming mice were originated from Swiss mice, they may share a similar genetic background. These mice show: high disease resistance, good adaptive capacity, high breeding coefficient, and good survival rate.34
In this study we analyzed: 1) if gestational stress induces depressive-like and anxiety-like behavior in offspring and 2) if such behavior changes are associated with epigenetic alterations of BDNF. The goal of the present study was to increase the understanding of epigenetic mechanisms underlying development of depression by recapitulating interactions between genes and the environment in animal models.
Results
Gestational stress leads to depressive-like and anxiety-like behaviors in offspring
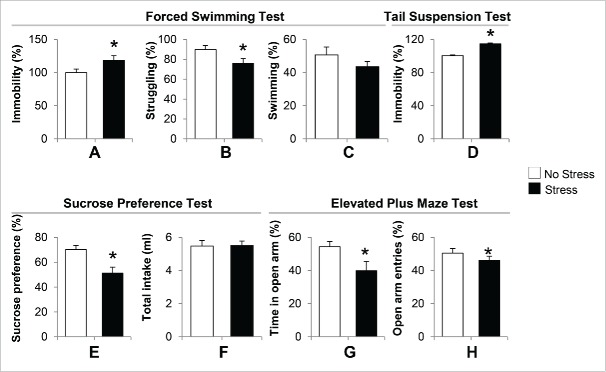
We measured depressive-like and anxiety-like behaviors in offspring born to gestational-stress dams. We chose offspring at the age of postnatal day 40 (PND40) for all behavior and biochemical experiments because the most common time of onset of depression in humans is between the ages of 20 and 30 y old. Mice at PND40 are approximately equivalent to human adolescents or young adults. Fig. 1A to 1C depict the duration of immobility, swimming and struggling between stress and non-stress offspring. During 5 min of a forced swimming test, immobility behavior in the water tank was significantly increased in gestational-stress offspring compared to non-stress offspring (Fig. 1A). In addition, gestational-stress offspring displayed significant reduction in struggling time (Fig. 1B) compared to non-stress offspring. However, there was no marked difference in the duration of swimming between the 2 groups (Fig. 1C). We next applied the tail suspension test, another standard test for measuring depressive behavior in mice. As shown in Fig. 1D, a significant increase in immobility during the tail suspension test was observed in gestational-stress offspring compared to non-stress offspring during 5 min of test. Furthermore, we examined the sucrose preference test to see if gestational stress induces anhedonic behavior in offspring. Fig. 1E shows that gestational-stress offspring exhibited a significantly reduced preference to sucrose solution compared to non-stress offspring. In terms of total liquid intake, there was no difference between the 2 groups (Fig. 1F). This result suggests that gestational stress induces anhedonic-like behavior in offspring. In addition, we used the elevated plus maze test to measure if gestational-stress offspring developed anxiety behaviors. As shown in Fig. 1G-H, a significant difference was detected in both the time spent and number of entries in open arm between the 2 groups of offspring during a 10-minute test. Our data indicate that the paradigm of a combination of restraint and light stress can induce depressive-like and anxiety-like behaviors in young adult offspring.
Figure 1.
Gestational stress induced depressive-like and anxiety-like behaviors in young adult offspring. In the forced swimming test, immobility (A), swimming (B) and struggling (C) behaviors were analyzed during a 5-minute test. In the tail suspension test, immobility (D) was recorded during a 5-minute test. In the sucrose consumption test, the sucrose preference (E) was calculated as percentage of consumed sucrose solution over total liquid intake (F) for 24. h In the elevated plus maze test, percentage of open arm entries (G) and percentage of time spent in open arms (H) were scored for a 10-minute test. Data are presented as mean ± SEM; n = 10 for each group; *P < 0 .05 (Student t-test) vs. no-stress offspring.
Gestational stress decreases BDNF transcripts and protein expression in offspring hippocampus
In order to probe possible mechanisms by which gestational stress induces depressive-like and anxiety-like behaviors, next, we evaluated BDNF mRNA and protein expression in the hippocampus. Mouse BDNF consists of nine exon-driven transcripts producing at least nine BDNF splice variants.35 Detailed mRNA analysis of whole hippocampus revealed changes in four (BDNF-i, -iv, -vi, and -ix) out of seven BDNF (-i, -ii, -iii, -iv, -v, -vi, and -ix) transcripts measured (Fig. 2A). BDNF-vii and -viii were excluded from the measurement because of very low expression in the hippocampal region. Immunoblot data normalized by β-actin show a marked decrease (about 40%) in the protein level of BDNF in the hippocampus of gestational-stress offspring compared to non-stress offspring (Fig. 2B). Downregulated BDNF protein is correlated with decreased expression of individual transcripts in gestational-stress offspring such as BDNF-i, -iv, -vi, and -ix, but not of other variants.
Figure 2.
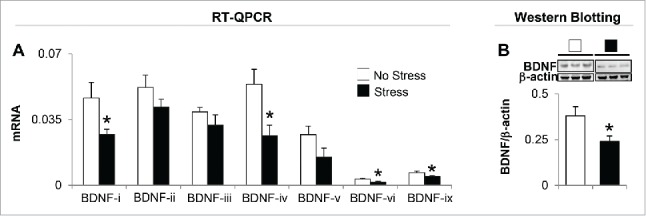
Gestational stress changes the expression of brain-derived neurotrophic factor (BDNF) transcripts in the hippocampus of young adult offspring. Among seven BDNF variants tested, BDNF transcripts (-i, -iv, -vi, and -ix) (A) and protein expression (B) in the hippocampus of gestational-stress offspring are significantly decreased compared to the non-stress offspring. Data are presented as mean ± SEM of 10 mice for each group. *P < 0.05 (one-way ANOVA followed by Bonferroni test vs. the corresponding value for non-stress offspring).
Gestational stress induces increased expression of DNMT1 and DNA methylation on BDNF promoters
We then measured DNMT1 mRNA levels in the hippocampus of offspring born from both non-stress and stress dams. As shown in Fig. 3A, there is a marked increase in the expression of DNMT1 mRNA in the gestational stress offspring. Similar to mRNA, the protein level of DNMT1 in the hippocampus of gestational-stress offspring also significantly increased as compared to non-stress offspring (Fig. 3B). To examine whether decreased BDNF transcripts are due to epigenetic regulation, especially promoter methylation induced by gestational stress, we used methylated DNA immunoprecipitation (MeDIP) with specific 5-methylcytosine (5mC) antibody to measure the enrichment of the most important epigenetic mark, 5mC, on the BDNF variants such as BDNF-i, -iv, -vi, and -ix, which are decreased by stress in the hippocampus. As shown in Fig. 3C, high levels of 5mC were found at BDNF-i, -iv, -vi, and -ix regulatory regions in gestational-stress offspring compared to non-stress offspring. These findings suggest that gestational stress leads to CpG methylation on specific BDNF promoters. Consistent with reported findings, the enrichment of 5mC at BDNF-i, -iv, -vi, and -ix promoters was negatively correlated with the levels of corresponding BDNF transcripts (Fig. 4) when analyzed using Pearson correlation, suggesting an epigenetic mechanism by which promoter methylation may be responsible for the downregulation of hippocampal BDNF in gestational-stress offspring. We also checked the specificity of methylation induced by gestational stress in the same brain region using GAPDH as a control gene. As expected, GAPDH failed to show enrichment of 5mC in gestational-stress offspring, indicating that CpG methylation induced by gestational stress is gene specific: GAPDH [5mC enrichment on promoter (%)]: non-stress, 0.036 ± 0.004, n = 8; stress, 0.041 ± 0.01, n = 8 (P = 0.9, Student t-test).
Figure 3.
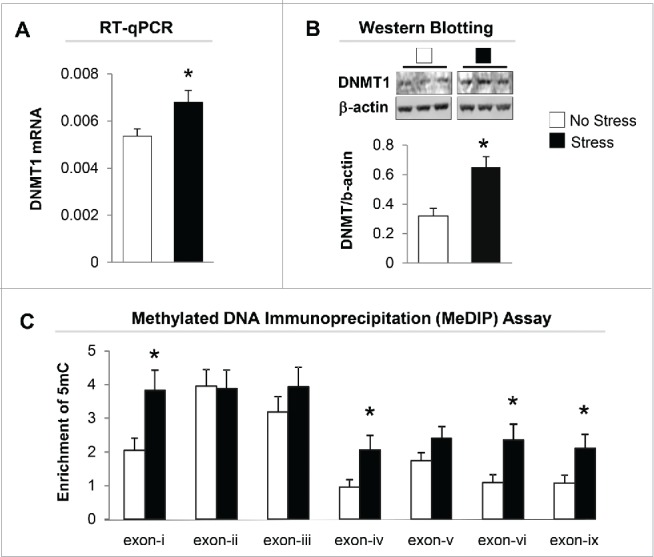
Gestational stress significantly increases expression of DNMT1 mRNA (A) and protein (B) in the hippocampus of gestational-stress offspring. The representative immunoblots show a major band of approximately 190 kDa for DNMT1. All values are means ± SEM of 8 mice for each group. * P < 0.05 (Student t-test) vs. the corresponding control values. (C) Gestational stress significantly increases the levels of 5-methylcytosine (5mC) on promoter regions (-i, -iv, -vi and -ix) in the hippocampus of offspring compared to non-stress offspring. Data are presented as mean ± SEM of 10 mice for each group. * P < 0.05 (one-way ANOVA followed by Bonferroni test vs. the corresponding values for non-stress offspring).
Figure 4.
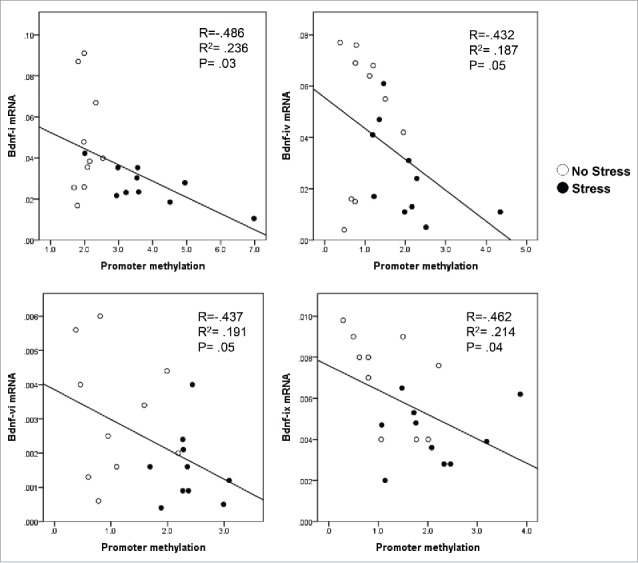
The enrichment of 5mC (promoter methylation) on exons of BDNF-i (A), -iv (B), -vi (C), and -ix (D) is negatively correlated with the corresponding transcripts by Pearson correlation analysis.
Gestational stress changes the expression of histone deacetylases in offspring hippocampus
Chromatin modification is an important epigenetic mechanism in regulating transcription. To probe whether gestational stress leads to chromatin modifications that consequently affect BDNF expression, we measured the expression of histone deacetylases (HDAC), especially HDAC1, HDAC2, and HDAC3 in the hippocampus, because these enzymes are key components in gene regulation. As shown in Fig. 5, the data from both RT-qPCR and immunoblotting show that the mRNA and protein expression levels of HDAC1 and HDAC2 but not HDAC3 increased significantly in stress offspring as compared to non-stress offspring. This suggests that deacetylation occurs on histone H3 tails, which may be associated with downregulation of BDNF.
Figure 5.
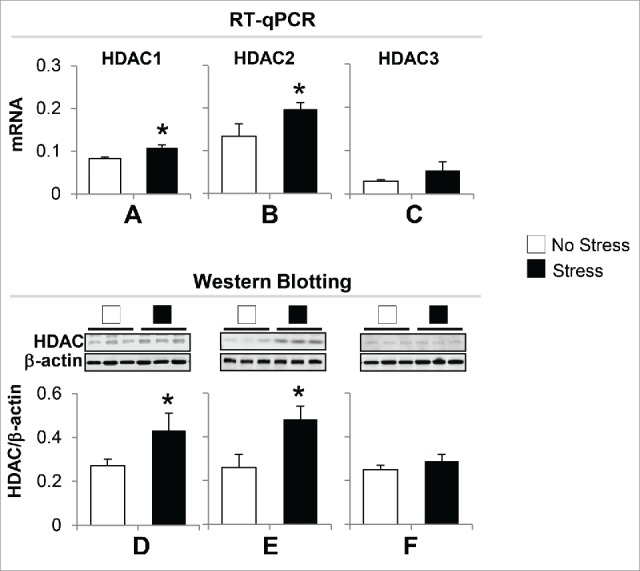
Gestational stress induces alteration of hippocampal HDACs expression in offspring. (A) The mRNA of HDAC1 and HDAC2 but not HDAC3 are significantly increased in the hippocampus of gestational-stress offspring as compared to non-stress offspring. (B) Immunoblot data normalized by β-actin protein levels show a marked increase in the protein levels of HDAC1 and HDAC2 but not HDAC3 in the hippocampus of gestational-stress mice compared to non-stress offspring. Data are presented as mean ± SEM of 10 mice for each group. *P < 0.05 (one-way ANOVA followed by Bonferroni test vs. the corresponding values for non-stress offspring).
Gestational stress decreases levels of Acetyl Histone H3 Lysine 14 (AcH3K14) and its binding on BDNF promoters in offspring hippocampus
The significant decrease of BDNF-i, -iv, -vi, and -xi expression and increased HDACs observed in the hippocampus of gestational-stress offspring prompted us to investigate whether the downregulation of BDNF and depressive-like behavior phenotypes are also the consequence of post-translational histone modification. We first measured the protein level of AcH3K14 with specific antibody. As shown in Fig. 6, significant decrease in AcH3K14 expression was observed in the hippocampus of gestational-stress offspring compared to non-stress offspring. To examine if such a decrease is involved in regulating BDNF expression, we assayed and compared the levels of AcH3K14 at BDNF-i, -iv, -vi, and -xi promoter regions in whole hippocampus using chromatin immunoprecipitation (ChIP). As shown in Fig. 7, a strong (more than 40%) decrease in AcH3K14 occurred at BDNF-i, -iv -vi, and -xi promoters in gestational-stress offspring compared to non-stress offspring.
Figure 6.
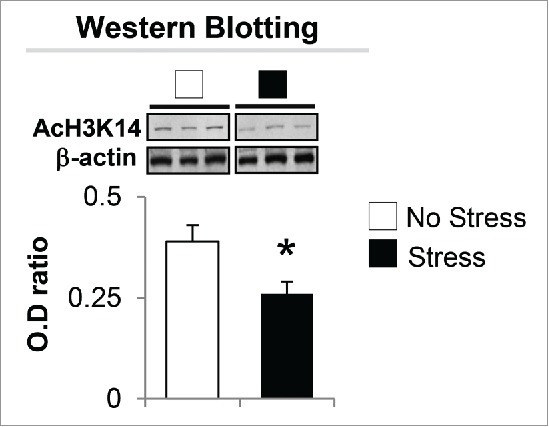
Immunoblot analysis shows a decrease in the protein level of AcH3K14 in the hippocampus of gestational-stress offspring. Data are presented as mean ± SEM of 10 mice for each group. * P < 0.05 (Student t-test) vs. non-stress offspring.
Figure 7.
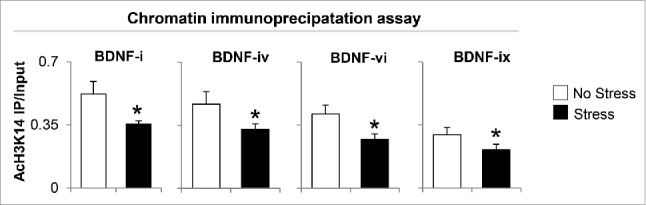
Gestational stress causes decrease of AcH3K14 binding to BDNF exons (-i, -iv, -vi and -ix) in the hippocampus compared to non-stress offspring. Data are presented as mean ± SEM of 10 mice for each group. * P < 0.05 (One-way ANOVA followed by Bonferroni test vs. corresponding values for non-stress offspring for ChIP assay). IP: immunoprecipitation.
Correlation
To examine potential relationships among the protein levels of BDNF, HDAC1, HDAC2, and AcH3K14 and the behavioral data such as immobility (forced swim test), sucrose preference and percentage of time spent in open arm, we performed Pearson correlation analyses. As shown in Table 1, there is a significant negative correlation between levels of BDNF and HDAC1 (r = -0.44, P=0.05), indicating that decreased BDNF was associated with increased HDAC1. In addition, the level of BDNF was positively correlated with sucrose preference (r = 0.65, P = 0.03) while levels of HDAC1 and HDAC2 demonstrated a significant inverse correlation with sucrose preference (r = −0.64, P = 0.003 and r = −0.45, P = 0.05). However, no significant correlations were found among biochemical measurements with immobility and percentage of time spent in open arm observed in this study. The data imply an important role for BDNF, HDAC1, and HDAC2 in regulating motivational behavior.
Table 1.
Correlation analysis for selected biological and behavioral measures.
| HDAC1 Level | HDAC2 Level | AcH3 Level | FST Immobility | Sucrose Preference | % of Time in Open Arm | |
|---|---|---|---|---|---|---|
| BDNF Level | r = −0.44 P = 0.05 | r = −0.21P = 0.37 | r = 0.22P = 0.35 | r = −0.09P = 0.68 | r = 0.65P = 0.003 | r = 0.38P=0.09 |
| HDAC1 Level | r = 0.31P = 0.19 | r = −0.26P = 0.27 | r = −0.03P = 0.90 | r = −0.64P = 0.003 | r = −0.14P = 0.57 | |
| HDAC2 Level | r = −0.27p = .25 | r = 0.26p = 0.25 | r = −0.45p = 0.05 | r = −0.36p = 0.12 | ||
| AcH3 Level | r = −0.42p = .07 | r = 0.43p = 0.06 | r = 0.07p = 0.77 |
Discussion
The goal of this study was to determine whether gestational stress induces depressive-like and anxiety-like behavior phenotypes in young adult offspring and whether such behavioral deficits are accompanied by differences in the expression of epigenetic-related biomarkers as previously found in the brains of patients with depression.25,27-33,36 To reach this goal, we stressed pregnant dams with a combination of stressors: restraint stress with 24-h constant light disturbance throughout the gestational period. Light disturbance has been reported to induce anxiety-like and depressive-like responses in both rodents and humans.37 To test the consequence of such stress on young adult offspring, we used standard paradigms widely used for animal models of human psychiatric disorders, i.e., forced swim test, tail suspension test, sucrose preference test and elevated plus maze. We found that exposure to the stressors above during gestation lead to depressive-like and anxiety-like behavior phenotypes in young adult offspring mice including increased immobility, decreased struggling, reduced sucrose preference and decreased percentage of time and number of entries in open arms. The behavior abnormalities observed in multiple paradigms suggest depressive and anxiety traits in young adult offspring born to gestational-stress dams, offering a valid model for depression and anxiety studies.
In an attempt to reveal molecular mechanisms of the behavioral abnormalities observed in gestational-stress offspring, we focused on epigenetic regulation of BDNF expression, especially in the hippocampal region. Postmortem and animal studies found that insufficient BDNF leads to hippocampal shrinking, decreased LTP expression and downregulated learning and memory abilities and may be responsible for depressive phenotypes.36-41 Taken together, it suggests that the hippocampus is sensitive to gestational stress and is an important brain region in the pathophysiology of depression. BDNF has multiple important roles in brain development including supporting the survival and differentiation of selected neuronal populations, modulating dendritic growth, regulating synaptic transmission, and plasticity.36 Substantial evidence suggests that this neurotrophin also plays important roles in psychiatric disorders and is one of the primary targets of major depression.36 Since reduced brain BDNF is associated with the pathogenesis of depression,25,27-33,36 it is considered a candidate gene for this mental illness.
Our data show that gestational stress induces a significant decrease of BDNF protein expression in the hippocampus of offspring. Because total BDNF level is contributed from various individual transcripts, the decreased BDNF induced by the gestational-stress paradigm in the present study can be considered the result of downregulation of specific transcripts, such as BDNF-i, -iv, -vi, and -ix. This finding suggests that in the hippocampus, BDNF-i, -iv, -vi, and -ix transcripts are sensitive to environmental stress. In addition, the other BDNF variants measured, including -ii, -iii, and -v, failed to show changes in both stress and non-stress offspring, suggesting that these splice variants may be stable and do not respond to the stressors used in this study. It has been reported that unpredictable stress differentially regulates the expression of BDNF splice variants in hippocampal subfields and impacts their function.42 Repeated administration of antidepressant to rat induces differential expressions of BDNF in different hippocampal subregions.43 These findings suggest that the components of hippocampus function differently in response to stressors. In present study, we focused on BDNF expression in whole hippocampus. However, further studies are necessary to investigate the expressions of BDNF transcripts in different subregions of hippocampus under gestational-stress conditions in the future.
Recent studies show that different segregation of BDNF transcripts may provide a particular mechanism for the modulation of BDNF availability and function in specific hippocampal subfields during development or in response to environmental stimuli.37-39 In the present explorative study, we focused on the expression pattern of different BDNF transcripts in whole hippocampus under gestational-stress conditions. It is necessary to investigate expressions of BDNF transcripts in the different hippocampal regions to gain insight into their function in coping with environmental stressors.
To establish whether the downregulation of BDNF transcripts observed in gestational-stress offspring may be related to altered epigenetic mechanisms, we measured the expression of DNMT1 in the hippocampus of offspring at PND40. DNMT1 is a key epigenetic biomarker that affects DNA transcription by modification of CpG at promoter regions. Our results show that gestational stress induces overexpression of DNMT1 in the hippocampus of offspring. To establish in detail whether the altered expression of DNMT1 is expected to result in enrichment of CpG methylation at different regulatory regions of BDNF, we then measured levels of 5mC, a CpG methylation marker using MeDIP assay. As expected, there was significant methylation (high levels of 5mC) found on the promoters of BDNF-i, -iv, -vi, and -xi in gestational-stress offspring. Such increased promoter methylation is inversely correlated with the corresponding splice variants' expression (Fig. 4), suggesting a DNA hypermethylation mechanism involved in decreased BDNF expression induced by gestational stress. BDNF gene structures are species-dependent. However, some homology in BDNF structure between humans and rodents is identified. For example, exon-i and exon-iv in human BDNF are identical to those in mice.42 Regarding proximal promoter activity, the beginning exon of a gene is generally considered to be important for transcription.43,44 It has been shown that CpG methylation on exon-iv is involved in the regulation of the BDNF gene under pathological conditions.45–48 The findings from the present study may provide a clue and reference for human studies. Our data thus support the neurotrophic hypothesis of major depression.
Epigenetic regulation of gene transcription consists primarily of chromatin structure and function, including histone and DNA modifications such as cytosine methylation/demethylation. Histone tail modifications include acetylation, methylation, ubiquitination, phosphorylation, sumoylation, ribosylation, and citrullination. Among them, histone tail acetylation by histone acetyltransferases and deacetylation by histone deacetylases (HDACs) have received much attention because the former leads to opened chromatin facilitating gene transcription; the latter yields condensed chromatin causing transcription repression.49-51
Importantly, histone tail modification is associated with the pathophysiology of several psychiatric disorders including major depression.52 For instance, increased expression of HDAC2 was found in patients with major depression.53 Administered HDAC inhibitors such as valproate and MS-275 corrected depressive-like behaviors in rodent models.52 We measured the expression of HDAC1, HDAC2, and HDAC3 in the hippocampus to determine whether gestational stress induced post-translational modifications of chromatin remodeling and contributed to the epigenetic regulation of BDNF alteration detected in offspring. We found that the expression of HDAC1 and HDAC2 but not HDAC3 is elevated in the hippocampus of gestational-stress offspring, indicating that alteration of chromatin architecture at BDNF promoter regions may occur from gestational stress. Elevated HDAC1 and HDAC2 imply a decrease of histone H3 deacetylation. Histone H3 acetylation, especially at lyine-14 facilitates transcriptional activation 54 by loosening DNA-histone interactions and allowing transcriptional machinery to bind and facilitate gene expression. We observed that reduction in AcH3K14 mediates a lower binding on selective BDNF promoters in stress offspring. This provides evidence that reduced BDNF expression induced by gestational stress in the hippocampus is the result of comprehensive epigenetic dysfunction characterized by increased promoter methylation and altered histone H3 modification. Because DNMT1 associated with HDAC1 in a large molecular complex uses the deacetylase as a substrate for DNA methylation,55 the lower histone acetylation promotes higher DNA methylation, resulting in decreased gene expression. This notion is supported by an observed inverse correlation between levels of BDNF and HDAC1 in the present study.
It is not surprising that the level of BDNF positively and specifically correlates with sucrose preference behavior because as an essential molecular substrate in hippocampus, BDNF can regulate motivational behaviors.56 We also observed an inverse correlation between levels of HDAC1 and HDAC2 with sucrose preference. This effect of HDACs on behavior may be indirect, possibly through regulating the expression of BDNF or other related genes. Further experiments are needed to support this observation.
In conclusion, the mechanisms underlying the interplay between gene and environment are combinatorial with histone and DNA modifications presenting the diversity of epigenetic landscapes and contributing to complicated behavioral phenotypes induced by gestational stress. The results from the present study support the concept that young adult offspring born to gestational-stress dams has construct face validity and pharmacologic utility as an experimental model of depression. The gestational-stress model could be used both to predict the course of depressive-like behavioral pathology and also to investigate epigenetic mechanisms of depression and develop more effective treatments.
Materials and methods
Animals and gestation stress procedures
All animal experiments were performed in accordance with the Institutional Animal Care and Use Guidelines of Chongqing Medical University. Pregnant mice (Kunming species) were individually housed with food and water ad libitum. Control dams were left undisturbed throughout gestation with a 12-h light-dark cycle. The gestational-stress dams were housed in a separate room with fluorescent ceiling lights. The stress procedure consisted of restraining the pregnant dam in a transparent tube (12 cm × 3 cm) for 30 minutes three times per day from the fifth day of pregnancy until delivery,9 and 24-h constant light throughout gestation. After weaning (postnatal day 20), male mice were selected for the study and housed four-to-five per cage, separately by condition. At postnatal day (PND) 40, the following experiments were performed.
Forced swimming test
As described by Porsolt et al,57 in the pre-test session, mice were placed individually in a clear container 20 cm diameter, 50 cm height) that contained water (25°C ± 1°C) to a depth of 25 cm and forced to swim for 5 minutes. The water was replaced for each mouse. In the test-session, mice were placed back into the container for 5 minutes. Immobility was noted if the mouse remained floating without climbing. Struggling was defined as the mouse making vigorous movements. Swimming was defined as horizontal movement throughout the container with vigorous motion.
Tail suspension test
The experiment was carried out according to the method described by Steru et al.58 The mice were individually suspended by the tail taped on a stand above floor. The duration of the test was 6 minutes. Immobility was defined as the mouse remaining completely motionless.
Sucrose preference test
The test was carried out at PND40. As previously described,57,59 mice were given a free choice between two identical bottles, one with 1% sucrose solution and another with water for 24-h. The consumption of water and sucrose solution was estimated simultaneously in control and experimental groups by weighing the bottles. The sucrose preference was defined as a percentage of consumed sucrose solution of the total liquid intake.
Elevated plus-maze test
To examine the anxiety behavior of gestational-stress offspring, the elevated plus-maze was performed in similar way as described by Rodgers et al.60 Briefly, it consisted of two open and two closed arms (all arms: 30 cm × 5 cm) and was made of Plexiglas. The open arms were surrounded by 4 mm-high edges. The closed arms had transparent 14.5 cm high Plexiglas walls at the sides and end. The floor was made of black Plexiglas and elevated to a height of 50 cm above the floor. At the start of each test, mice were placed individually on the central platform and their behavior monitored by video camera for 10 min. The number of entries for each arm and the time spent in each arm were recorded and analyzed. The percentage of open arm entries (open arm entries x 100/total arm entries) and percentage of time spent in open arm (time spent in open arm x 100/time spent in open and closed arms) were used as indices of anxiety.
Quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction was performed using SYBR®Premix Ex TaqTMII (TaKaRa RR820A). Total RNA from the hippocampus was isolated using TRIzol reagent (Life Technologies, Grand Island, New York) and was further purified using the QIAGEN RNeasy kit (Qiagen, Valencia, California). cDNA synthesis was performed using PrimeScriptTM RT reagent Kit(TaKaRa RRO37A). The primer sequences for the genes analyzed are summarized in Table S1 in Supplement 1. Each sample was run in duplicate and repeated twice. For normalizing mRNA expression, two housekeeping genes (β-actin and GAPDH) were chosen as the internal control.
Western blot analysis
Total protein from hippocampus, extracted using RIPA lysis buffer and quantified by Enhanced BCA Protein Assay Kit (Beyotime P0010S), was separated by SDS-PAGE and transferred to PVDF membrane. After being blocked in TBS buffer containing 0.05% Tween-20 and 5% skim milk, the membranes were incubated overnight at 4°C with the following primary antibodies: anti-DNMT1 (Imagenex; 1:1000), anti-HDAC1, anti-HDAC2, anti-HDAC3 (Santa Cruz Biotechnology; 1:500), anti-BDNF (Santa Cruz Biotechnology; 1:500), anti-acetyl-histone H3 (lys14) (AcH3K14) antibody (Millipore, Billerica, MA 1: 2000). After incubation with the corresponding secondary antibody, the immunoreactive signals were visualized by ECL Plus Western Blotting Detection System and quantitated using Quantity One software. The levels of these proteins in the stress offspring vs. non-stress offspring were normalized by β-actin protein levels. In order to estimate background caused by non-specific binding of secondary antibody, a secondary control without the primary antibody was performed.
Methylated dna immunoprecipitation
BDNF promoter methylation was assessed using Methylated DNA Immunoprecipitation [MeDIP kit (Diagenode, Denville, New Jersey)], followed by quantitative real-time polymerase chain reaction. The procedures for sample treatment and immunoprecipitation are described in the kit instruction manuals. The percentage of methylated vs. unmethylated promoter was calculated using the following equation: % (meDNA IP/total input) = 2(Ct[10% input] − Ct [meDNA-IP] −3.32) ×100%.
Chromatin immunoprecipitation assay
ChIP assay was performed using commercially available kits (Millipore, Billerica, MA), as reported (9, 14). Approximately 10 mg of hippocampal tissue was used for this procedure. Briefly, tissue was treated with formaldehyde to crosslink acetylated histone 3 with the target genomic DNAs. After being washed with cold PBS containing protease inhibitors, slices were homogenized in SDS lysis buffer. To obtain consistent chromatin fragmentation, the lysates were sonicated by a Sonic Dismembrator, Model 500 (Fisher Scientific). An aliquot (1-2%) of the sonicated lysate without antibody (Input) was used to quantitate the total amount of DNA present in different sample extracts before immunoprecipitation. Immunoprecipitation was carried out using ChIP grade anti-acetyl-histone H3 (lys14) (AcH3K14) antibody (Millipore, Billerica, MA). The antibody concentration used was that suggested by the manufacturer. In preliminary experiments, it was empirically established that in a given amount of tissue extract, the amount of BDNF promoters precipitated by the antibody failed to increase when the antibody concentration was increased by 10-fold. At the end of the ChIP procedure, the protein/DNA cross-linked nucleosomal chromatin complex immunoprecipitated by AcH3 was reverse cross-linked. Samples were then treated with proteinase-K. Protein-free DNA was extracted in phenol/chloroform and precipitated and washed in ethanol. This extract was used for detection and quantification of BDNF.
Statistical analysis
Results are expressed as mean ± SEM. Experimental differences were assessed by Student t-test, one-way ANOVA followed by Bonferroni post-hoc comparisons, and Pearson correlation analysis using Predictive Analytics Software v.18 (SPSS, Inc., Chicago, Illinois). The criterion for significance was P < 0.05, 2-tailed.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments and Disclosures
This work is supported by National Natural Science Foundation of China. Grant Number: NSFC 81271264.
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Monk C, Spicer J, Champagne FA. Linking prenatal maternal adversity to developmental outcomes in infants: The role of epigenetic pathways. Dev Psychopathol 2012; 24:1361-76; PMID:23062303; http://dx.doi.org/ 10.1017/S0954579412000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markham JA, Koenig JI. Prenatal stress: Role in psychotic and depressive diseases. Psychopharmacology 2011; 214:89-106; PMID:20949351; http://dx.doi.org/ 10.1007/s00213-010-2035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillott A, Standen PJ. Levels of anxiety and sources of stress inadults with autism. J Intellect Disabil 2007; 11:359-70; PMID:18029412; http://dx.doi.org/ 10.1177/1744629507083585 [DOI] [PubMed] [Google Scholar]
- 4.Rice F, Jones I, Thapar A. The impact of gestational stress andprenatal growth on emotional problems in offspring: A review. Acta Psychiatr Scand 2007; 115:171-83; PMID:17302617; http://dx.doi.org/ 10.1111/j.1600-0447.2006.00895.x [DOI] [PubMed] [Google Scholar]
- 5.Walker FR, Knott B, Hodgson DM. Neonatal endotoxinexposure modifies the acoustic startle response and circulating levels of corticosterone in the adult rat but only following acute stress.J Psychiatr Res 2008; 42:1094-03; PMID:18406426; http://dx.doi.org/ 10.1016/j.jpsychires.2007.12.006 [DOI] [PubMed] [Google Scholar]
- 6.Charil A, Laplante DP, Vaillancourt C, King S. Prenatal stress and brain development. Brain Res Rev 2010; 65:56-79; PMID:20550950; http://dx.doi.org/ 10.1016/j.brainresrev.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 7.Matrisciano F, Tueting P, Dalal I, Kadriu B, Grayson DR, Davis JM, Nicoletti F, Guidotti A. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatalstress in mice. Neuropharmacology 2013; 68:184-94; PMID:22564440; http://dx.doi.org/ 10.1016/j.neuropharm.2012.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matrisciano F, Tueting P, Maccari S, Nicoletti F, Guidotti A. Pharmacological activation of group-II metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice. Neuropsychopharmacology 2012; 37:929-38; PMID:22089319; http://dx.doi.org/ 10.1038/npp.2011.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong E, Dzitoyeva SG, Matrisciano F, Tueting P, Grayson DR, Guidotti A. Brain-derived neurotrophic factor epigenetic modifications associated with schizophrenia-like phenotype induced by prenatal stress in mice. Biol Psychiatry 2015; 6:589-96; PMID:25444166; http://dx.doi.org/20053376 10.1016/j.biopsych.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGowan PO, Szyf M. The epigenetics of social adversity inearly life: Implications for mental health outcomes. Neurobiol Dis 2010; 39:66-72; PMID:20053376; http://dx.doi.org/ 10.1016/j.nbd.2009.12.026 [DOI] [PubMed] [Google Scholar]
- 11.Zhang TY, Labonté B, Wen XL, Turecki G, Meaney MJ. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology 2013; 38:111-23; PMID:22968814; http://dx.doi.org/ 10.1038/npp.2012.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boersma GJ, Lee RS, Cordner ZA, Ewald ER, Purcell RH, Moghadam AA, Tamashiro KL. Prenatal stress decreases Bdnf expression and increases methylation of Bdnf exon IV in rats. Epigenetics 2014; 9:1-11; PMID:24739672; http://dx.doi.org/ 10.4161/epi.27558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, Costa E. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophr Res 2007; 91:51-61; PMID:17270400; http://dx.doi.org/ 10.1016/j.schres.2006.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laserassisted microdissection. Mol Psychiatry 2007; 4:385-97; PMID:17264840; http://dx.doi.org/22948384 10.1038/sj.mp.4001954 [DOI] [PubMed] [Google Scholar]
- 15.Dong E, Gavin D, Chen Y, Davis J. Up-regulation of TET1 and down-regulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry 2012; 2:e159; PMID:22948384; http://dx.doi.org/ 10.1038/tp.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry 2014; 21:e349; PMID:24448211; http://dx.doi.org/22948975 10.1038/tp.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology 2013; 38:138-66; PMID:22948975; http://dx.doi.org/ 10.1038/npp.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009; 3:342-8; PMID:19234457; http://dx.doi.org/23020296 10.1038/nn.2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GenRED Consortium, Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, Murakami P, Lessard A, Yolken RH, Feinberg AP, et al.. Genome-Wide DNA Methylation Scan in Major Depressive Disorder. PLoS One. 2012; 4:e34451; PMID:22511943; http://dx.doi.org/23020296 10.1371/journal.pone.0034451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vialou J, Feng J, Robison AJ, Nestler EJ. Epigenetic mechanisms of depression and antidepressants action. Annu Rev Pharmacol Toxicol. 2013; 53:59-87; PMID:23020296; http://dx.doi.org/ 10.1523/jneurosci.1164-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menke A, Binder EB. Epigenetic alterations in depression and antidepressant treatment. Dialogues Clin Neurosci. 2014; 3:395-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature 2008; 455(7215):894-902; PMID:18923511; http://dx.doi.org/ 10.1038/nature07455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan V, Nestler EJ. Animal models of depression: molecular perspectives. Curr Top Behav Neurosci 2011; 7:121-47; PMID:21225412; http://dx.doi.org/ 10.1007/7854_2010_108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci 2006; 9(4):519-25; PMID:16501568; http://dx.doi.org/ 10.1038/nn1659 [DOI] [PubMed] [Google Scholar]
- 25.Autry AE, Monteggia LM. Brain-Derived Neurotrophic Factor and Neuropsychiatric Disorders Pharmacol Rev 2012; 2:238-58; PMID:24448211; http://dx.doi.org/22182689 10.1124/pr.111.005108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwivedi Y. Involvement of Brain-Derived Neurotrophic Factor in Late-Life Depression Am J Geriatr Psychiatry 2013; 5:433-49; PMID:23570887; http://dx.doi.org/22182689 10.1016/j.jagp.2012.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavin DP, Akbarian S. Epigenetic and post-transcriptional dysregulation of gene expression in schizophrenia and related disease. Neurobiol Dis 2012; 46:255-62; PMID:22182689; http://dx.doi.org/ 10.1016/j.nbd.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 28.Ikegame T, Bundo M, Murata Y, Kasai K, Kato T, Iwamoto K. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet 2013; 58:434-8; PMID:23739121; http://dx.doi.org/ 10.1038/jhg.2013.65 [DOI] [PubMed] [Google Scholar]
- 29.Wysokiński A. Serum levels of brain-derived neurotrophic factor (BDNF) and neurotrophin-3 (NT-3) in depressed patients with schizophrenia. Nord J Psychiatry 2015; 7:1-5; PMID:26548545; http://dx.doi.org/20194826 10.3109/08039488.2015.1087592 [DOI] [PubMed] [Google Scholar]
- 30.Keller S, Sarchiapone M, Zarrilli F, Ferraro A, Carli V, Sacchetti S, Lembo F, Angiolillo A, Jovanovic N, Pisanti F, et al.. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry 2010; 67:258-67; PMID:20194826; http://dx.doi.org/ 10.1001/archgenpsychiatry.2010.9 [DOI] [PubMed] [Google Scholar]
- 31.Karpova NN. Role of BDNF epigenetics in activity-dependent neuronal plasticity. Neuropharmacology 2014; 76(Pt C):709-18; PMID:23587647; http://dx.doi.org/ 10.1016/j.neuropharm.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 32.Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci 2007; 10:1089-93; PMID:17726474; http://dx.doi.org/ 10.1038/nn1971 [DOI] [PubMed] [Google Scholar]
- 33.Tadić A, Müller-Engling L, Schlicht KF, Kotsiari A, Dreimüller N, Kleimann A, Bleich S, Lieb K, Frieling H. Methylation of the promoter of brainderived neurotrophic factor exon IV and antidepressant response in major depression. Mol Psychiatry 2014; 19:281-3; PMID:23670489; http://dx.doi.org/ 10.1038/mp.2013.58 [DOI] [PubMed] [Google Scholar]
- 34.Shang H, Wei H, Yue B, Xu P, Huang H. Microsatellite analysis in two populations of Kunming mice. Lab Anim 2009; 43(1):34-40; PMID:19141464; http://dx.doi.org/ 10.1258/la.2008.008098 [DOI] [PubMed] [Google Scholar]
- 35.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res 2007; 85:525-35; PMID:17149751; http://dx.doi.org/ 10.1002/jnr.21139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulle F, van den Hove DL, Jakob SB, Rutten BP, Hamon M, van Os J, Lesch KP, Lanfumey L, Steinbusch HW, Kenis G. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry 20126:584-96; PMID:21894152; http://dx.doi.org/20890399 10.1038/mp.2011.107 [DOI] [PubMed] [Google Scholar]
- 37.Fonken LK, Finy MS, Walton JC, Weil ZM, Workman JL, Ross J, Nelson RJ. Influence of light at night on murine anxiety-and depressive-like responses. Behavioural Brain Res 2009; 2:349-54; PMID:19591880; http://dx.doi.org/20890399 10.1016/j.bbr.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 38.Mao LM, Fibuch EE, Wang JQ. Decoding BDNF-LTP coupling in cocaine addiction. Neuron 2010; 67:679-81; PMID:20890399; http://dx.doi.org/ 10.1016/j.neuron.2010.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bekinschtein P, Cammarota M, Katche C, Slipczuk L, Rossato JI, Goldin A, Izquierdo I, Medina JH. BDNF is essential to promote persistence of long-term memory storage. Proc Natl Acad Sci USA 2008; 105:2711-6; PMID:18263738; http://dx.doi.org/ 10.1073/pnas.0711863105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pencea V, Bingaman KD, Wiegand SJ, Luskin MB. Infusion of brain-derived neurotrophic factor into the lateral ventricle of the adult rat leads to new neurons in the parenchyma of the striatum, septum, thalamus, and hypothalamus. J Neurosci 2001; 21:6706-17; PMID:11517260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warner-Schmidt JL, Duman RS. Hippocampal neurogenesis: opposing effects of stress and antidepressant treatment. Hippocampus 2006; 3:239-49; PMID:16425236; http://dx.doi.org/17164818 10.1002/hipo.20156 [DOI] [PubMed] [Google Scholar]
- 42.Nair A, Vadodaria KC, Banerjee SB, Benekareddy M, Dias BG, Duman RS, Vaidya VA. Stressor-specific regulation of distinct brain-derived neurotrophic factor transcripts and cyclic AMP response element-binding protein expression in the postnatal and adult rat hippocampus. Neuropsychopharmacol 2007; 32(7):1504-19; PMID:17164818; http://dx.doi.org/ 10.1038/sj.npp.1301276 [DOI] [PubMed] [Google Scholar]
- 43.De Foubert G, Carney SL, Robinson CS, Destexhe EJ, Tomlinson R, Hicks CA, Murray TK, Gaillard JP, Deville C, Xhenseval V, et al.. Fluoxetine-induced change in rat brain expression of brain-derived neurotrophic factor varies depending on length of treatment. Neuroscience 2004; 128(3):597-604; PMID:15381288; http://dx.doi.org/10.10169838053 [DOI] [PubMed] [Google Scholar]
- 44.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 2007; 3:397-406; PMID:17629449; http://dx.doi.org/9838053 10.1016/j.ygeno.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sengupta PK, Smith BD. Methylation in the initiation region of the first exon suppresses collagen pro-alpha2 (I) gene transcription. Biochim Biophys Acta 1998; 1443:75-89; PMID:9838053; http://dx.doi.org/ 10.1016/S0167-4781(98)00188-2 [DOI] [PubMed] [Google Scholar]
- 46.Delgado MD, Leon J. Gene expression regulation and cancer. Clin Transl Oncol 2006; 8:780-7; PMID:17134965; http://dx.doi.org/ 10.1007/s12094-006-0132-7 [DOI] [PubMed] [Google Scholar]
- 47.Yasuda S, Liang MH, Marinova Z, Yahyavi A, Chuang DM. The mood stabilizers lithium and valproate selectively activate the promoter IV of brain-derived neurotrophic factor in neurons. Mol Psychiatry 2009; 14:51-9; PMID:17925795; http://dx.doi.org/ 10.1038/sj.mp.4002099 [DOI] [PubMed] [Google Scholar]
- 48.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 2003; 302:885-9; PMID:14593183; http://dx.doi.org/ 10.1126/science.1086446 [DOI] [PubMed] [Google Scholar]
- 49.Levenson J, Sweatt JD. Epigenetic mechanisms in memory formation. Nature 2005; 1:108-18; PMID:26915423; http://dx.doi.org/16596331 10.1038/nrn1604 [DOI] [PubMed] [Google Scholar]
- 50.Levenson J, Sweatt JD. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol Life Sci 2006; 63:1009-16; PMID:16596331; http://dx.doi.org/ 10.1007/s00018-006-6026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fortress AM, Frick KM. Epigenetic regulation of estrogen-dependent memory. Front Neuroendocrinol 2014; 35:530-49; PMID:24878494; http://dx.doi.org/ 10.1016/j.yfrne.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Covington HE, Maze I, LaPlant QC, Vialou VF, Yoshinori ON, Berton O, Fass DM, Renthal W, Rush AJ, Wu EY, et al.. antidepressant actions of HDAC inhibitors. J Neurosci. 2009; 37:11451-11460; PMID:19759294; http://dx.doi.org/10.152322692567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology Reviews 2013; 38:124-137; PMID:22692567; http://dx.doi.org/ 10.1038/npp.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crosio C, Heitz E, Allis C, Borelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J Cell Sci 2013; 116:4905-16; PMID:14625384; http://dx.doi.org/10325416 10.1242/jcs.00804 [DOI] [PubMed] [Google Scholar]
- 55.Robertson K, Uzvolgi E, Liang G, Talmadge C, Sumegi J, Gonzales F, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res 1999; 27:2291-8; PMID:10325416; http://dx.doi.org/ 10.1093/nar/27.11.2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gourley SL, Kiraly DD, Howell JL, Olausson P, Taylor JR. Acute hippocampal BDNF restores motivational and forced swim performance after corticosterone Biol Psychiatry. 2008; 10:884-890; PMID:18675955; http://dx.doi.org/204499 10.1016/j.biopsych.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porsolt RD, Anton G, Blavet N, Jalfre M. Behaviorual despair in rats: a new model sensitive to antidepressant treatments. Eur J Pharmacol 1978; 47:379-391; PMID:204499; http://dx.doi.org/ 10.1016/0014-2999(78)90118-8 [DOI] [PubMed] [Google Scholar]
- 58.Steru L, Chermat R, Thierry B, Simon P. The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology (Berl) 1985; 3:367-70; PMID:3923523; http://dx.doi.org/ 10.1007/BF00428203 [DOI] [PubMed] [Google Scholar]
- 59.Strekalova T, Spanagel R, Bartsch D, Henn FA, Gass P. Stress-induced anhedonia in mice is associated with deficits in forced swimming and exploration. Neuropsychopharmacology 2004; 11:2007-17; PMID:15266352; http://dx.doi.org/ 10.1038/sj.npp.1300532 [DOI] [PubMed] [Google Scholar]
- 60.Rodgers RJ, Cole JC, Aboualfa K, Stephenson LH. Ethopharmacological analysis of the effects of putative ‘anxiogenic’ agents in the mouse elevated plus-maze Pharmacol Biochem Behav 1995; 52, pp. 805-813; PMID:8587923; http://dx.doi.org/ 10.1016/0091-3057(95)00190-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.