Abstract
Zygotes are totipotent cells that have the ability to differentiate into all cell types. It is believed that this ability is lost gradually and differentiation occurs along with the progression of preimplantation development. Here, we hypothesized that the loose chromatin structure is involved in the totipotency of one-cell stage embryos and that the change from loose to tight chromatin structure is associated with the loss of totipotency. To address this hypothesis, we investigated the mobility of eGFP-tagged histone H2B (eGFP-H2B), which is an index for the looseness of chromatin, during preimplantation development based on fluorescent recovery after photobleaching (FRAP) analysis. The highest mobility of eGFP-H2B was observed in pronuclei in 1-cell stage embryos and mobility gradually decreased during preimplantation development. The decrease in mobility between the 1- and 2-cell stages depended on DNA synthesis in 2-cell stage embryos. In nuclear transferred embryos, chromatin in the pseudopronuclei loosened to a level comparable to the pronuclei in 1-cell stage embryos. These results indicated that the mobility of eGFP-H2B is negatively correlated with the degree of differentiation of preimplantation embryos. Therefore, we suggest that highly loosened chromatin is involved in totipotency of 1-cell embryos and the loss of looseness is associated with differentiation during preimplantation development.
Keywords: Chromatin structure, FRAP, mouse embryo, preimplantation development, reprogramming
Introduction
Embryos at the 1-cell stage are totipotent and this totipotency is gradually lost during preimplantation development. When the embryos reach the blastocyst stage, blastomeres are differentiated into 2 types of cell lineages: inner cell mass (ICM) and trophectoderm (TE). The former is pluripotent, while the latter is differentiated and its developmental potential is restricted to form the placenta. One-cell stage embryos are also highly plastic. When the pronuclei from 1-cell stage embryos are transplanted into enucleated zygotes, 95% of these nuclear transferred oocytes develop to the blastocyst stage.1 However, when nuclei from embryos at the 2- and 4-cell stages were transplanted as the donor, only 19% and 5% of the zygotes, respectively, developed to the blastocyst stage. Thus, 1-cell stage embryos have totipotency and the highest plasticity. However, the mechanisms underlying these characteristics in 1-cell stage embryos and their loss during preimplantation development remain unclear.
It was suggested that chromatin structure is tightly associated with the differentiation state of cells. Numerous reports have shown that the chromatin structure of pluripotent embryonic stem (ES) cells is in a “loose” or “open” state, and changes to “tight” or “compact” when they differentiate. Heterochromatin regions appear as dispersed organizations in ES cells, but are distinct spots in neuron precursor cells.2 Sensitivity to DNase I and micrococcus nuclease (MNase) of ES cells decreased after the induction of differentiation by treatment with retinoic acid.3,4 Moreover, biochemical experiments revealed that the sensitivity of chromatin to high salt concentrations is higher in ES cells than neuron precursor cells,2 suggesting that chromatin structure becomes tighter or more compact when ES cells are differentiated. Thus, the loose chromatin structure characterizes the undifferentiated state in ES cells.3
It is thought that fluorescence recovery after photobleaching (FRAP) analysis with eGFP-tagged core histones reveals the extent of chromatin looseness.3 This analysis estimates the mobility of core histones, and it has been demonstrated that there is a positive correlation between the mobility of histones and the extent of chromatin looseness.2,3 Indeed, when cells were treated with trichostatin A (TSA), which is a potent inhibitor of histone deacetylases (HDACs) and induces a loosened chromatin state, the mobility of eGFP-H2B increased.5 In contrast, hypertonic treatment led to chromatin condensation and severely decreased the mobility of eGFP-H2B.6 These reports suggest that the mobility of eGFP-H2B can be used as an index of chromatin structure looseness.
We hypothesized that the chromatin structure was loosened in the 1-cell stage embryos and that this loosened state was involved in their totipotency and high plasticity. This loose chromatin structure is lost during preimplantation development. To examine these hypotheses, we investigated the mobility of eGFP-H2B in preimplantation embryos based on FRAP analysis.
Materials and methods
Collection and culture of oocytes and embryos
Unfertilized MII-stage oocytes were collected from the oviducts of 3-week-old ddY female mice (SLC, Inc., Shizuoka, Japan) that were superovulated by injection of 5 IU equine chorionic gonadotropin (eCG; ASKA Pharmaceutical Co., Ltd. Tokyo, Japan) and human chorionic gonadotropin (hCG; ASKA pharmaceutical) at 46–50 h intervals. Spermatozoa were collected from the cauda epididymis of male mature ICR mice (SLC) and cultured in HTF (human tubal fluid) medium7 supplemented with 10 mg/ml bovine serum albumin (BSA). Fourteen to 16 h after hCG injection, oocytes were collected and inseminated with capacitated spermatozoa that had been cultured for 2 h. Two hours after insemination, the fertilized oocytes were washed and cultured in KSOM medium8 supplemented with 3 mg/ml BSA in a humidified atmosphere of 5% CO2/95% air at 38°C. All procedures using animals were reviewed and approved by the University of Tokyo Institutional Animal Care and Use Committee and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals.
Vector construction
The coding sequence (CDS) of histone H2B (Hist1h2bc) was amplified using the pCRII topo vector (Life Technologies Corp., Carlsbad, CA, USA) encoding flag-tagged H2B that was used in our previous report9 as a template and PrimeSTAR Max DNA polymerase (Takara Bio Inc., Shiga, Japan). The sequences of the primer set were as follows: 5′-CCCAAGCTTATGCCTGAGCCTGCGAAGTC-3′ (forward), 5′-GGGGTACCTCACTTGGAGCTGGTGTACT-3′ (reverse). The forward and reverse primers contained HindIII and KpnI sites, respectively. PCR conditions were 37 cycles at 98°C for 10 s, 57°C for 15 s, and 72°C for 6 s. The amplified CDS of H2B was purified by phenol-chloroform extraction and digested with HindIII and KpnI. To construct the eGFP-H2B cassette, the digested fragment was purified from the agarose gel and cloned into the pEGFPC3 vector (Clontech, Shiga, Japan), which had been digested with the same restriction enzymes. Using this construct (pEGFP-H2B) as a template, the eGFP-H2B cassette was amplified using PrimeSTAR Max DNA polymerase and treated with Ex-taq (Takara) to add a deoxyadenosine at the 3′ terminus of the amplified fragment. The amplified fragment was cloned into the pCRII-TOPO vector (hereafter referred as pTOPO-eGFP-H2B).
In vitro transcription
pTOPO-eGFP-H2B was linearized with NotI and purified by phenol-chloroform extraction. Using the purified DNA fragment as a template, 5′-capped complementary RNA (cRNA) of eGFP-H2B was synthesized in vitro using the mMESSAGE mMACHINE sp6 kit (Life technologies: AM1340) according to the manufacturer's protocol. The synthesized cRNA was polyadenylated with a poly(A) tailing kit (Life technologies; AM1350). The cRNA with poly(A) tail was precipitated using lithium chloride precipitation solution. The cRNA was dissolved at 500 ng/μl in nuclease-free water and stored −80°C until use. The cRNA was diluted to 250 ng/μl prior to microinjection.
Microinjection
cRNA was microinjected into the cytoplasm of 1-cell stage embryos 2 h post-insemination. Microinjection was performed in KSOM-HEPES medium using borosilicate glass capillaries (GC 100 TF-10, Harvard Apparatus Ltd., Kent, UK) on an inverted microscope (Nikon Corp., Tokyo, Japan; Eclipse TE300) with a micromanipulator (Narishige, Tokyo, Japan) and microinjector (Narishige; IM300). After microinjection, the embryos were washed and cultured in KSOM medium.
Stable cell line preparation
NIH 3T3 cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Wako, Japan) supplemented with penicillin/streptomycin and 10% fetal bovine serum (Life technologies) at 37°C and a 5% CO2 atmosphere. Linearized pEGFPC3-H2B was prepared by digesting 5 µg of plasmid with FastDigest Eco31I (Thermo Scientific), as recommended by the manufacturer. Linear plasmid was purified by phenol/chloroform/isoamylalcohol and resuspended in H2O. The NIH 3T3 cells were transfected with 2.5 µg of linearized plasmid using Metafectene Easy according to the manufacturer's instructions (Biontex, Germany). Two days after transfection, G418 selection (600 µg/ml, Roche) was applied. GFP-positive colonies were individually picked up after 14 d of culture and expanded.
Nuclear transfer
ICR females (8 weeks old; SLC, Japan) were injected intraperitoneally with 7.5 IU eCG followed by 7.5 IU hCG 48 h later. Females were sacrificed by cervical dislocation 14–15 h post hCG and metaphase II oocytes were isolated in HTF-HEPES (Zenith Biotech, USA) supplemented with hyaluronidase (Sigma Aldrich) to remove the surrounding cumulus cells. Nuclear transfer was performed essentially as described previously10 with stable eGFP-H2B positive cells as donors. After the injection of donor nuclei, the reconstructed embryos were cultured for an additional 1 h in KSOM medium (Zenith Biotech, USA) before activation in Ca2+-free CZB supplemented with 10 mM SrCl2 and 5 µg/ml Cytochalsin B (Sigma Aldrich). After 6 h of activation, the reconstructed embryos were observed for the presence of the GFP signal and further cultured in KSOM medium until FRAP analysis.
Culture of CGR8 mouse ES cells
Mouse ES cells (CGR8) were cultured as described previously11 with minor modifications. After overnight culture, cells (2 × 105/well) were transiently transfected with pEGFP-H2B vector using Lipofentamine (Life technologies, #11668–027). After 6 h of culture, the medium was replaced with fresh medium. For culture of ES cells on glass-bottomed dishes (Greiner bio-one; #627860), 24 h after transfection 5 × 105 cells/well of ES cells were transferred onto mouse embryonic fibroblast (MEF) cells that had been treated with mitomycin C. Forty-eight hours after transfection, the cells were subjected to FRAP analysis.
Fluorescence recovery after photobleaching (FRAP)
The embryos were transferred into 20 μl of KSOM-HEPES medium covered with mineral oil on a glass-bottomed dish (Greiner bio-one). The chamber and lens of the confocal microscope LSM5 exciter (Carl Zeiss, Oberkochen, Germany) were warmed at 37°C with a microscope incubation system (Tokai Hit, Co., Shizuoka, Japan). The dishes in which the embryos were cultured were placed in the chamber and maintained for 15 min before FRAP analysis. The region of interest (ROI), reference region (REF), and background (BG) were set using LSM5 exciter software. Three pictures were taken at 5 s intervals, after which the ROI was photo-bleached with the laser: 100% excitation at 458, 488, and 514 nm for 10 s. A total of 27 pictures were taken at 5 s intervals. At the same time, the fluorescent intensity of ROI, REF, and BG in each picture was measured using LSM5 exciter software. The excitation of a 488 nm laser was set from 1 to 27%. Fluorescent intensity was set at around 2000. Recovery rate was calculated as described previously.12 For each picture, the relative intensity of ROI was determined as follows: the value of BG was subtracted from those of ROI and REF. The value of ROI was then divided by that of REF. The relative intensity at each time point was plotted in the recovery curve. The bleaching rate was determined by subtracting the relative intensity soon after bleaching from 1. The recovery rate at each time point was determined by dividing the relative intensity of ROI at each time point by the average of the relative intensity of ROI of 3 images taken before photo bleaching. Mobile fraction (MF) was calculated using the equation as follows13-15: MF = (Fend-Fpost)/(Fpre-Fpost), where Fend is the relative intensity of fluorescence at the endpoint, Fpost is immediately after photo-bleaching, and Fpre is before photo-bleaching.
Observation of the fluorescent signal of eGFP-H2B in preimplantation embryos
Embryos were fixed in 3.7% paraformaldehyde (PFA; Wako) in PBS for 15 min, after which the cells were washed in PBS containing 1% BSA (1% BSA/PBS). After washing, cells were permeabilized with 0.5% Triton X-100 in PBS for 15 min. The cells were washed with 1% BSA/PBS and mounted on a slide with Vectashield antibleaching solution (Vector laboratories) containing 3 μg/ml 4′, 6-diamino-2-phenylindole (DAPI; Dojindo). To wash away the unbound fraction of eGFP-H2B, embryos were permeabilized with Triton X before fixation. The embryos were treated with 0.2% Triton X-100 containing ice-cold permeabilization solution16 (50 mM NaCl, 3 mM MgCl2, 300 mM sucrose in 25 mM HEPES pH7.4) for 45 s. Cells were then washed with permeabilization solution without Triton X-100 3 times and fixed in 3.7% PFA in PBS at room temperature for 15 min. After being washed in 1% BSA/PBS, cells were mounted on a slide as described above. Fluorescent images were obtained using a Carl Zeiss LSM5 exciter laser scanning confocal microscope.
Results
Distribution of eGFP-H2B in the nucleus
The cRNA encoding eGFP-tagged histone H2B (eGFP-H2B) was injected into the cytoplasm of 1-cell stage embryos 2 h post-insemination. eGFP fluorescence was clearly observed in nuclei throughout preimplantation development (Supplemental Fig. 1A). eGFP-H2B appeared to be deposited in the nucleosomes, since the eGFP and DAPI (fluorescence of DNA stain) signals were well-merged; the intense eGFP signal was observed in heterochromatin regions where DNA is condensed and the signal intensity of DAPI was high. The exception for co-localization of eGFP-H2B and DNA signals was an intense eGFP-H2B signal in the nucleolar precursor body (NPB) where DNA is not present in 1-cell and 2-cell stage embryos.17,18 The deposition of eGFP-H2B in the nucleosome was assessed by treating the embryos with Triton-X100 before fixation to wash away free eGFP-H2B from the nucleus. Although this treatment cleared up the fluorescence signal from the nucleus, as well as the cytoplasm, of the embryos which had been injected with eGFP, the signal in the nucleus, but not NPB, remained in embryos injected with eGFP-H2B (Supplemental Fig. S2).
Figure 1.
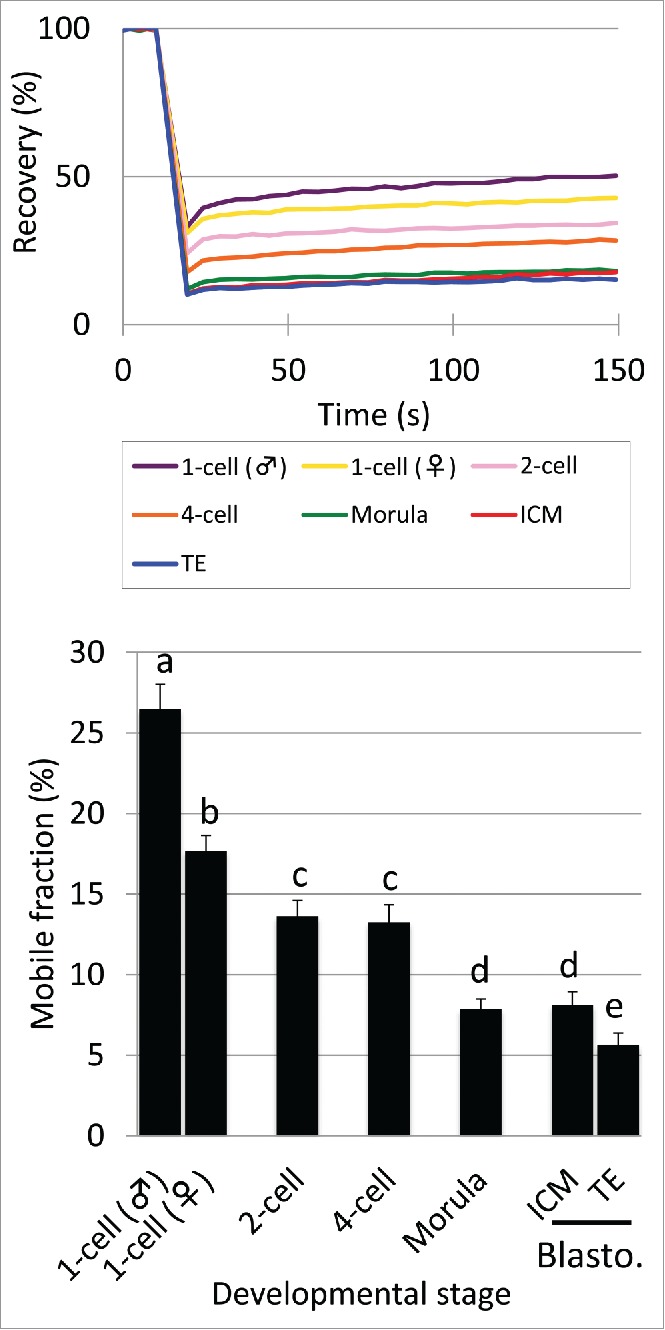
Mobility of eGFP-H2B during preimplantation development Complementary RNA (cRNA) encoding eGFP-tagged H2B (eGFP-H2B) was injected into the cytoplasm of 1-cell stage embryos 2 h post-insemination (hpi). The mobility of eGFP-H2B was analyzed using FRAP in embryos at the 1-, 2-, 4-, morula, and blastocyst stages during 10–13, 28–32, 45–48, 70–72, and 94–98 hpi, respectively. The recovery curve and mobile fraction are shown on the top and bottom, respectively. In the bar graph of the mobile fraction, different characters indicate statistical differences among developmental stages (P<0.05; by Student t-test) and the error bar indicates the standard error. More than 3 independent experiments were performed for each developmental stage and the data were accumulated. In total, at least 23 or more nuclei were analyzed at each developmental stage.
Changes in the mobility of eGFP-H2B during preimplantation development
In 1-cell stage embryos, the genomes of paternal and maternal origins form separate pronuclei (PNs) and differ in various aspects, e.g., pattern of DNA replication,19 epigenetic modifications,20,21 and regulation of gene expression.22,23 Therefore, chromatin structure may differ between paternal and maternal PNs and the mobility of eGFP-H2B would reflect this difference. The results of FRAP analysis showed that the recovery of fluorescence was always faster in the male pronucleus than the female throughout the measurement time: the recovery curve (RC) for male pronuclei was higher than that for females (Fig. 1). The mobile fraction (MF) was also significantly higher in male PN than female PN (Fig. 1), suggesting that chromatin in the male PN was looser than in the female PN.
As described above, an intense signal of free eGFP-H2B was observed in the NPB of 1-cell and 2-cell stage embryos (Supplemental Fig. S1). The mobility of eGFP-H2B was extremely high in the NPB compared to other regions in nuclei of 1-cell stage embryos: eGFP-H2B fluorescence recovered immediately after bleaching (Supplemental Fig. S3). Thus, it was confirmed that eGFP-H2B in the NPB is not associated with the nucleosome.
The mobility of eGFP-H2B was highest at the 1-cell stage and then gradually decreased during preimplantation development: both RC and MF decreased during development (Fig. 1). Although mobility was lower in female pronuclei than male pronuclei in 1-cell stage embryos, it was still higher in female pronuclei than nuclei of 2-cell stage embryos, suggesting that the chromatin structure is extremely loose in the 1-cell stage. After decreasing in the 2-cell stage, mobility was maintained in the 4-cell stage and decreased again at the morula stage. At the blastocyst stage, embryos consist of 2 types of cell lineages; namely, the inner cell mass (ICM) and trophectoderm (TE). Although the ICM is pluripotent, the TE is more differentiated because its cell fate has been determined to form the placenta. To investigate the correlation of looseness of chromatin with differentiation state, we compared the mobility of eGFP-H2B in the ICM and TE. The RC of the ICM was slightly faster than in the TE and the MF of the ICM was significantly higher than for TE (Fig. 1).
Previous studies demonstrated that chromatin structure was looser in pluripotent ES cells than differentiated somatic cells.2,3 Therefore, we compared the mobility of eGFP-H2B between pluripotent ES cells and totipotent 1-cell stage embryos. The results showed that the mobility was much higher in 1-cell stage embryos than ES cells (Fig. 2). These results, taken together, chromatin structure is extremely loose in 1-cell stage embryos and this looseness is gradually reduced during preimplantation development, which appears to be associated with the loss of totipotency and pluripotency.
Figure 2.
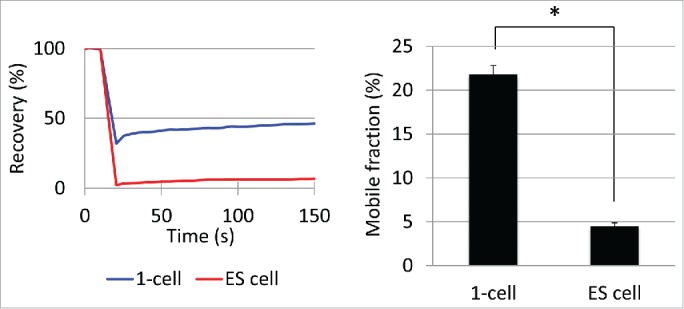
Mobility of eGFP-H2B in embryonic stem (ES) cells Mobility of eGFP-H2B in ES cells was analyzed 48 h after transfection with the eGFP-H2B expressing vector. The 1-cell stage embryos were analyzed between 10–13 hpi. The recovery curve and mobile fraction are shown on the left and right, respectively. Three independent experiments were performed and the data were accumulated. In total, 18 ES cells were examined. In the 1-cell stage embryos, 83 cells were analyzed and the averaged value of the male and female pronuclei is shown. Error bars indicate standard error. The asterisk indicates statistical difference (P<0.05; by Student t-test).
The loss of loose chromatin structure is associated with DNA replication at the 2-cell stage
Previous studies suggested that dynamic changes in chromatin structure occur during the 2-cell stage and that DNA replication is involved in this process.24,25 Therefore, we hypothesized that DNA replication is associated with loss of the extremely loose chromatin structure between the 1- and 2-cell stages. To explore this hypothesis, we examined the mobility of eGFP-H2B in late 2-cell stage embryos that were treated with aphidicolin, a DNA polymerase inhibitor. The RC and MF of aphidicolin-treated 2-cell stage embryos were higher than in the control and were at levels comparable to 1-cell stage embryos (Fig. 3). These results suggested that the decreases in looseness of chromatin between the 1- and 2-cell stages are associated with DNA replication at the 2-cell stage.
Figure 3.
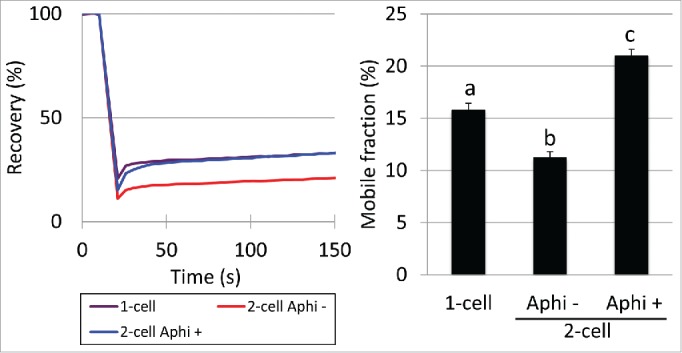
Mobility of eGFP-H2B in 2-cell stage embryos treated with aphidicolin Mobility of eGFP-H2B in 2-cell stage embryos treated with aphidicolin (Aphi +) was examined. Embryos treated with DMSO (Aphi -) served as the control. At 16 hpi, the embryos were transferred into medium containing aphidicolin or DMSO. Embryos at the 1- and 2-cell stages were analyzed between 10–13 and 28–32 hpi, respectively. The recovery curve and mobile fraction are shown on the left and right, respectively. In the bar graph of the mobile fraction, different characters indicate statistical differences among the experimental groups (P<0.05; by Student t-test). Error bars indicate the standard error. More than 3 independent experiments were performed and the data were accumulated. In total, more than 20 nuclei were examined in each experimental group.
Changes in the mobility of eGFP-H2B in euchromatin and heterochromatin regions during preimplantation development
As described above, the mobility of eGFP-H2B decreases during preimplantation development (Fig. 1). The amount of heterochromatin increases during preimplantation development26 (Supplemental Fig. S1). Since the region of interest (ROI), which is an objective area in FRAP analysis, was selected at random, it is possible that the ROI includes more heterochromatic regions at the later stages of implantation development. Therefore, if the mobility of eGFP-H2B differs between euchromatin and heterochromatin regions, the observed changes in mobility of eGFP-H2B during preimplantation development (Fig. 1) may reflect increases in heterochromatin during that period. To address this possibility, we conducted FRAP analysis using a small ROI, which allowed us to measure the fluorescence only in euchromatin or heterochromatin regions (Supplemental Fig. S4). In euchromatin regions, the mobility of eGFP-H2B was highest at the 1-cell stage and was higher in male PN than female PN (Fig. 4). It decreased at the 2-cell stage and did not change significantly until the morula stage. In heterochromatin regions, a minor but not significant decrease in mobility was observed between the 2- and 4-cell stages, after which it decreased significantly at the morula stage. Taken together, the results suggest that loss of the loose chromatin structure is due to the increase in heterochromatic regions between the 1-cell and morula stages and that condensation of both of euchromatin and heterochromatic regions contributes to the loss of loose chromatin structure at the 2-cell and morula stages.
Figure 4.
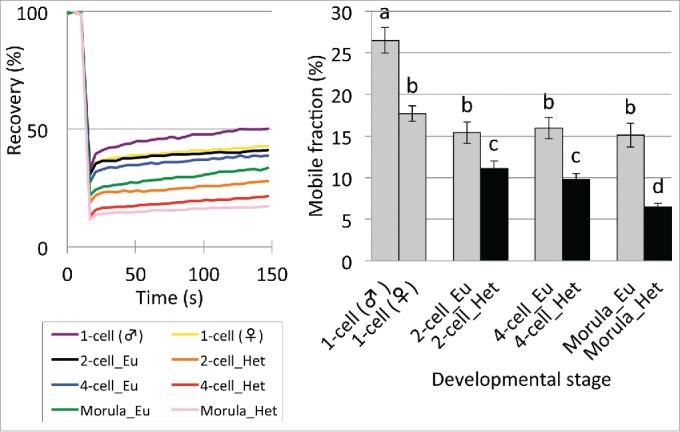
Mobility of eGFP-H2B in euchromatin (Eu) and heterochromatin (Het) regions in preimplantation embryos. Embryos at the 1-, 2-, 4-, and morula stage were analyzed at 10–13, 28–32, 45–48, and 70–72 hpi, respectively. Heterochromatin regions were not analyzed in the 1-cell stage embryos because they were present only around the nucleolar precursor body at this stage. At the blastocyst stage, since the blastocoel made it difficult to distinguish heterochromatin from euchromatin, these regions were not analyzed. In the bar graph of the mobile fraction, different characters indicate statistical differences among developmental stages (P<0.05; by Student t-test) and the error bar indicates the standard error. More than 3 independent experiments were performed for each developmental stage and the data were accumulated. In total, more than 23 nuclei were examined at each developmental stage.
The mobility of eGFP-H2B increased in nuclei of somatic cells after being transferred into enucleated unfertilized oocytes
Extremely high eGFP-H2B mobility was observed in 1-cell stage embryos and was lost during preimplantation development (Fig. 1), suggesting that the extremely loose chromatin structure is associated with totipotency in 1-cell stage embryos. Numerous studies have demonstrated that the nuclei of differentiated somatic cells acquired totipotency after they were transferred into enucleated unfertilized oocytes.27-29 Therefore, we hypothesized that the chromatin in a somatic nucleus that was transferred into an enucleated oocyte would acquire an extremely loose chromatin structure if such a structure is associated with totipotency. To address this hypothesis, we examined the mobility of eGFP-H2B in somatic cell nuclear transferred (SCNT) embryos. We prepared NIH3T3 cells stably expressing eGFP-H2B and transferred them into enucleated oocytes. The mobility of eGFP-H2B increased after the transfer: the RC and MF of eGFP-H2B in the pseudopronuclei of SCNT embryos were significantly higher than in the nuclei of the original 3T3 cells and at levels comparable to pronuclei in 1-cell stage embryos (Fig. 5). These results suggest that a highly loose chromatin structure is associated with totipotency in 1-cell stage embryos.
Figure 5.
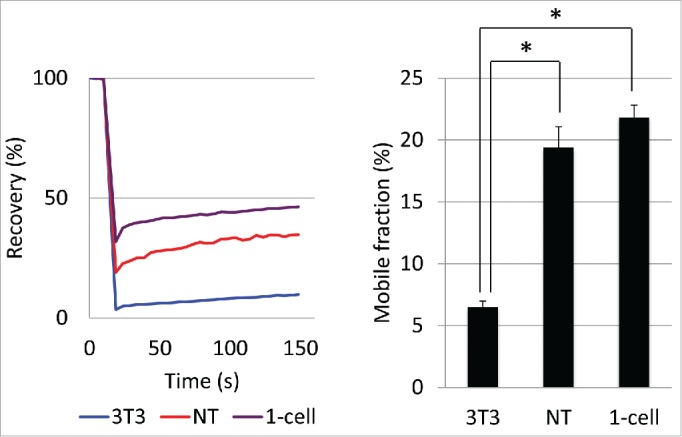
Mobility of eGFP-H2B in somatic cell nuclear transferred (SCNT) embryos Mobility of eGFP-H2B was examined in the pseudopronucleus of SCNT embryos. 3T3 cells stably expressing eGFP-H2B were trypsinized and then used as donors to reconstruct SCNT embryos. At the same time, the remaining cells were seeded on glass-bottomed dishes. The SCNT embryos, control 1-cell stage embryos, and 3T3 cells were analyzed 10–13 h after being transferred, inseminated, and seeded, respectively. More than 4 independent experiments were performed and the data were accumulated. In total, more than 19 cells were examined in each group. Error bars indicate standard errors. The asterisks indicate statistical differences (P<0.05; by Student t-test)
Discussion
In this study, we showed that chromatin is loosest at the 1-cell stage. Thereafter, the looseness of chromatin is gradually reduced during preimplantation development. The reduction in looseness between the 1- and 2-cell stages is due to DNA replication at the 2-cell stage. In SCNT embryos, chromatin from donor nuclei are loosened to a level comparable to that of 1-cell stage embryos. Thus, the looseness of chromatin is correlated with plasticity and the state of differentiation of embryos during preimplantation development.
The changes in chromatin structure between the 1- and 2-cell stages may be involved in the alteration of transcriptional regulation between these stages. Transcription that occurs at the 1- and 2-cell stages is called minor and major zygotic gene activation (ZGA), respectively.30 However, the regulatory mechanisms for minor and major ZGA differ. In minor ZGA, transcription occurs independently of enhancers. In contrast, enhancers are required for major ZGA. Here, we found that the chromatin structure is extremely loose at the 1-cell stage and becomes tighter at the 2-cell stage (Fig. 1). Generally, one important role of enhancers is to loosen the chromatin structure at promoter regions.31 Therefore, it is possible that enhancer-independent transcription occurs in 1-cell stage embryos because the chromatin structure is extremely loose, whereas transcription depends on enhancers at the 2-cell stage because the chromatin structure is tighter. In support of this hypothesis, both changes in the looseness of chromatin structure and the alteration of enhancers from independent to dependent transcription are associated with DNA replication at the 2-cell stage (Fig. 3).32-34 In addition, we previously reported that intergenic regions were pervasively transcribed in 1-cell stage embryos, but suppressed in 2-cell embryos. This pervasive transcription was still observed at the late 2-cell stage when DNA replication was inhibited.35 These results suggest that the extensively loose chromatin structure, which was specifically observed in 1-cell stage embryos, allows for pervasive transcription, but the tighter chromatin structure, which is associated with DNA replication, represses this type of transcription in 2-cell stage embryos.
A previous study used FRAP analysis to compare the chromatin structure between 2- and 8-cell stage embryos without discriminating between euchromatin and heterochromatin regions and found that it was tighter at the 8-cell stage.36 However, our study revealed that the heterochromatin regions indeed become tighter, but not the euchromatin regions between the 2-cell and morula stages. Therefore, the results of the earlier study appear to be due to the increase in heterochromatin regions, which become tighter during preimplantation development. If the ROI is randomly selected in FRAP analysis, it should include more heterochromatin regions in 8-cell embryos; the amount of heterochromatin regions increases during preimplantation development.26
In this study, we found that the chromatin structure became tighter after cleavage into 2-cell embryos, which may be part of the difference between 1- and 2-cell embryos. Although both the 1- and 2-cell embryos are totipotent, their characteristics differ in many ways. Embryos are still totipotent at the 2-cell stage, since a single blastomere can develop to term; however, the gene expression patterns differ from those of 1-cell embryos in which the pervasive transcription occurs from intergenic regions, as well as from genes.35 In addition, the nuclear architecture differs between them: the homogeneous euchromatin, which is devoid of obvious heterochromatin structure, is observed in 1-cell stage embryos, while heterochromatin appears at the late 2-cell stage.26 When the nuclei from 2-cell stage embryos are transferred into enucleated zygotes, the percentages of embryos developing to the blastocyst stage are lower than that of embryos transplanted with pronuclei from other zygotes,1 suggesting that plasticity decreases after cleavage into 2-cell embryos. Various changes occur between the 1- and 2-cell stages during progression of the developmental program. These changes are reminiscent of those that occur during the differentiation of ES cells. ES cells have a hyperactive transcriptional state in which repeat elements are transcribed and have highly dispersed heterochromatin structure, but these distinct characteristics are lost after they differentiate.2,37 It is thought that these changes are associated with the loss of chromatin looseness.3 Therefore, the difference in various characteristics between 1- and 2-cell embryos may also be associated with the change in the looseness of the chromatin structure.
The loss of the extremely loosened chromatin structure between the 1- and 2-cell stages may be mediated by CAF1. In the colony of ES cells, some cells acquire the loosened chromatin structure stochastically.36 Although this acquisition of loosened chromatin structure occurs very infrequently, it increased when CAF1 was downregulated.38 Consequently, CAF1 is likely to be involved in chromatin compaction, and the loss of CAF1 may cause the formation of a loosened chromatin structure. Considering that CAF1 is a chaperon of histone variants H3.1/H3.226 and histones H3.1 and H3.2 are not incorporated into the nucleus at the 1-cell stage, but start being incorporated at the 2-cell stage (unpublished data, F.A.), CAF1 does not seem to function at the 1-cell stage. It becomes functional later, and could be involved in the dramatic change in the looseness of the chromatin structure between the 1- and 2-cell stages. In addition, the nuclear deposition of H3.1 and H3.2 is responsible for the tight chromatin structure after cleavage into the 2-cell stage, since these variants are enriched in some histone modifications associated with the tight chromatin structure, i.e., H3K9me2, H3K9me3, and H3K27me3. Consistent with this hypothesis, the incorporation of H3.1 and H3.2 into nucleosomes is DNA replication dependent26,39 and the inhibition of DNA replication with aphidicolin kept the chromatin structure in the extremely loosened state in the 2-cell embryos (Fig. 3).
In SCNT embryos, the chromatin of donor cells acquired a highly loosened structure (Fig. 5). The rapid incorporation of histones H1oo and/or H3.3 into chromatin might be involved in this change in chromatin structure. Both of these proteins are associated with the loosened chromatin structure. H1oo is a linker histone H1, which is specifically expressed in oocytes and early preimplantation embryos,40 and its ectopic expression caused the formation of open chromatin in ES cells.41 In SCNT embryos, the somatic linker histone H1 was rapidly replaced with H1oo.42 H3.3 is abundant in euchromatin regions.11 All histone H3 variants forming the chromatin of somatic nuclei were rapidly removed and replaced with H3.3 derived from the oocyte cytoplasm after SCNT.43 Therefore, rapid replacement of the linker and core histones is involved in the acquisition of highly loosened chromatin in SCNT embryos, although the mechanism regulating this remodeling of chromatin structure remains to be elucidated.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported in part by Grants-in-Aid (to F. A.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (#26112507, #25252054). H.F. was supported by a grant from the Czech Science Foundation (#13–03269S).
References
- 1.Mcgraph J, Solter D. inability of mouse blastomere nuclei transerred ot enucleate zygotes to support development in vitro. Science 1984; 226:1317-9; PMID:6542249; http://dx.doi.org/ 10.1126/science.6542249 [DOI] [PubMed] [Google Scholar]
- 2.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. H plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006; 10:105-16; PMID:16399082; http://dx.doi.org/ 10.1016/j.devcel.2005.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 2011; 12:36-47; PMID:21179060; http://dx.doi.org/ 10.1038/nrm3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaniel C, Ang Y-S, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in Mouse embryonic stem cells. Stem Cells 2009; 27:2979-91; PMID:19785031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalut KJ, Höpfler M, Lautenschläger F, Boyde L, Chan CJ, Ekpenyong A, Martinez-Arias A, Guck J. Chromatin decondensation and nuclear softening accompany nanog downregulation in embryonic stem cells. Biophys J 2012; 103:2060-70; PMID:23200040; http://dx.doi.org/ 10.1016/j.bpj.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin RM, Cardoso MC. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J 2010; 24:1066-72; PMID:19897663; http://dx.doi.org/ 10.1096/fj.08-128959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinn P, Begley AJ. Effect of human seminal plasma and mouse accessory gland extracts on Mouse fertilization in vitro. Aust J Biol Sci 1984; 37:147-52; PMID:6517760 [DOI] [PubMed] [Google Scholar]
- 8.Lawitts JA, Biggers JD. Culture of preimplantation embryos. Meth Enzymol 1993; 225:153-64; PMID:8231853; http://dx.doi.org/ 10.1016/0076-6879(93)25012-Q [DOI] [PubMed] [Google Scholar]
- 9.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 2010; 137:3785-94; PMID:20943707; http://dx.doi.org/ 10.1242/dev.051805 [DOI] [PubMed] [Google Scholar]
- 10.Kishigami S, Wakayama S, Thuan NV, Ohta H, Mizutani E, Hikichi T, Bui H-T, Balbach S, Ogura A, Boiani M, et al.. Production of cloned mice by somatic cell nuclear transfer. Nat Protoc 2006; 1:125-38; PMID:17406224; http://dx.doi.org/ 10.1038/nprot.2006.21 [DOI] [PubMed] [Google Scholar]
- 11.Yukawa M, Akiyama T, Franke V, Mise N, Isagawa T, Suzuki Y, Suzuki MG, Vlahovicek K, Abe K, Aburatani H, et al.. Genome-wide analysis of the chromatin composition of histone H2A and H3 variants in mouse embryonic stem cells. PLoS ONE 2014; 9:e92689; PMID:24658136; http://dx.doi.org/ 10.1371/journal.pone.0092689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura H, Hieda M, Cook PR. Measuring histone and polymerase dynamics in living cells. Meth Enzymol 2004; 375:381-93; PMID:14870679; http://dx.doi.org/ 10.1016/S0076-6879(03)75024-1 [DOI] [PubMed] [Google Scholar]
- 13.Subramanian V, Mazumder A, Surface LE, Butty VL, Fields PA, Alwan A, Torrey L, Thai KK, Levine SS, Bathe M, et al.. H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS Genet 2013; 9:e1003725-21; PMID:23990805; http://dx.doi.org/ 10.1371/journal.pgen.1003725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae J, Sung BH, Cho IH, Song WK. F-Actin-dependent regulation of NESH dynamics in Rat hippocampal neurons. PLoS One 2012; 7:e34514-12; PMID:22496823; http://dx.doi.org/ 10.1371/journal.pone.0034514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieteren CEJ, Willems PHGM, Swarts HG, Fransen J, Smeitink JAM, Koopman WJH, Nijtmans LGJ. Defective mitochondrial translation differently affects the live cell dynamics of complex I subunits. Biochim Biophys Acta 2011; 1807:1624-33; PMID:21978538; http://dx.doi.org/ 10.1016/j.bbabio.2011.09.013 [DOI] [PubMed] [Google Scholar]
- 16.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 2010; 329:78-82; PMID:20595612; http://dx.doi.org/ 10.1126/science.1187945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biggiogera M, Martin TE, Gordon J, Amalric F, Fakan S. Physiologically inactive nucleoli contain nucleoplasmic ribonucleoproteins: immunoelectron microscopy of mouse spermatids and early embryos. Exp Cell Res 1994; 213:55-63; PMID:8020606; http://dx.doi.org/ 10.1006/excr.1994.1172 [DOI] [PubMed] [Google Scholar]
- 18.Fléchon JE, Kopecný V. The nature of the “nucleolus precursor body” in early preimplantation embryos: a review of fine-structure cytochemical, immunocytochemical and autoradiographic data related to nucleolar function. Zygote 1998; 6:183-91; PMID:9770784; http://dx.doi.org/ 10.1017/S0967199498000112 [DOI] [PubMed] [Google Scholar]
- 19.Aoki E, Schultz RM. DNA replication in the 1-cell mouse embryo: stimulatory effect of histone acetylation. Zygote 1999; 7:165-72; PMID:10418111; http://dx.doi.org/ 10.1017/S0967199499000532 [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Kim J-M, Aoki F. Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development 2004; 131:2269-80; PMID:15102709; http://dx.doi.org/ 10.1242/dev.01116 [DOI] [PubMed] [Google Scholar]
- 21.Burton A, Torres-Padilla ME. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief in Funct Genomics 2011; 9:444-54; PMID:21186177; http://dx.doi.org/ 10.1093/bfgp/elq027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol 1993; 159:366-78; PMID:8365573; http://dx.doi.org/ 10.1006/dbio.1993.1248 [DOI] [PubMed] [Google Scholar]
- 23.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181:296-307; PMID:9013938; http://dx.doi.org/ 10.1006/dbio.1996.8466 [DOI] [PubMed] [Google Scholar]
- 24.Majumder S, Miranda M, DePamphilis ML. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J 1993; 12:1131-40; PMID:8458327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho T, Sakai S, Nagata M, Aoki F. Involvement of chromatin structure in the regulation of mouse zygotic gene activation. Anim Sci J 2002; 73:113-22; http://dx.doi.org/ 10.1046/j.1344-3941.2002.00017.x [DOI] [Google Scholar]
- 26.Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet 2011; 7:e1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 1998; 394:369-74; PMID:9690471; http://dx.doi.org/ 10.1038/28615 [DOI] [PubMed] [Google Scholar]
- 28.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature 1997; 385:810-3; PMID:9039911; http://dx.doi.org/ 10.1038/385810a0 [DOI] [PubMed] [Google Scholar]
- 29.Meissner A, Jaenisch R. Mammalian nuclear transfer. Dev Dyn 2006; 235:2460-9; PMID:16881069; http://dx.doi.org/ 10.1002/dvdy.20915 [DOI] [PubMed] [Google Scholar]
- 30.Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays 1993; 15:531-8; PMID:8135766; http://dx.doi.org/ 10.1002/bies.950150806 [DOI] [PubMed] [Google Scholar]
- 31.Stees J, Varn F, Huang S, Strouboulis J, Bungert J. Recruitment of transcription complexes to enhancers and the role of enhancer transcription. Biology 2012; 1:778-93; PMID:23919179; http://dx.doi.org/ 10.3390/biology1030778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conover JC, Temeles GL, Zimmermann JW, Burke B, Schultz RM. Stage-specific expression of a family of proteins that are major products of zygotic gene activation in the mouse embryo. Dev Biol 1991; 144:392-404; PMID:2010038; http://dx.doi.org/ 10.1016/0012-1606(91)90431-2 [DOI] [PubMed] [Google Scholar]
- 33.Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol 1991; 147:403-14; PMID:1916016; http://dx.doi.org/ 10.1016/0012-1606(91)90298-H [DOI] [PubMed] [Google Scholar]
- 34.Majumder S, DePamphilis ML. A unique role for enhancers is revealed during early mouse development. Bioessays 1995; 17:879-89; PMID:7487969; http://dx.doi.org/ 10.1002/bies.950171010 [DOI] [PubMed] [Google Scholar]
- 35.Abe K-I, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J 2015; 34:1523-37; PMID:25896510; http://dx.doi.org/ 10.15252/embj.201490648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bošković A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EVS, Meshorer E, Torres-Padilla M-E. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev 2014; 28:1042-7; PMID:24831699; http://dx.doi.org/ 10.1101/gad.238881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RDG, Buetow KH, et al.. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008; 2:437-47; PMID:18462694; http://dx.doi.org/ 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishiuchi T, Enriquez-Gasca R, Mizutani E, cacute ABSK, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla M-E. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 2015; 22:662-671; PMID:26237512; http://dx.doi.org/ 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- 39.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc Natl Acad Sci USA 2006; 103:6428-35; PMID:16571659; http://dx.doi.org/ 10.1073/pnas.0600803103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 2001; 128:655-64; PMID:11171391 [DOI] [PubMed] [Google Scholar]
- 41.Hayakawa K, Ohgane J, Tanaka S, Yagi S, Shiota K. Oocyte-specific linker histone H1foo is an epigenomic modulator that decondenses chromatin and impairs pluripotency. Epigenetics 2012; 7:1029-36; PMID:22868987; http://dx.doi.org/ 10.4161/epi.21492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teranishi T, Tanaka M, Kimoto S, Ono Y, Miyakoshi K, Kono T, Yoshimura Y. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev Biol 2004; 266:76-86; PMID:14729479; http://dx.doi.org/ 10.1016/j.ydbio.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 43.Nashun B, Akiyama T, Suzuki MG, Aoki F. Dramatic replacement of histone variants during genome remodeling in nuclear-transferred embryos. Epigenetics 2011; 6:1489-97; PMID:22139579; http://dx.doi.org/ 10.4161/epi.6.12.18206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


