Abstract
Background
Ischemic stroke is widely recognized as a major health problem and social burden worldwide, and it usually leads to dementia. In this study, we aimed to better understand the pathogenesis in the development of dementia following ischemic stroke.
Material/Methods
We exploited miRNA database to search for the target for miR-125a and validated the found target using luciferase assay. Further, we performed real-time PCR and Western blot analysis to examine the expression of miR-125a and its target in the tissue samples. In addition, a polymorphism was genotyped and its association with post-stroke dementia was analyzed.
Results
We identified enthothelin-1 (ET-1) as a target of miR-125a, and this relationship was validated using luciferase assay. Furthermore, transfection of miR-125a inhibitor substantially upregulated the expression of ET-1, while miR-125a and ET-1 siRNA caused downregulation of ET-1 in endothelial cells. In addition, we found that a polymorphism (rs12976445) interferes with the expression of miR-125a, which in turn caused an increase in the expression of ET-1 in human endothelial cells. Logistic regression analysis showed that rs12976445 is significantly associated with the risk of dementia after ischemic stroke.
Conclusions
Our study demonstrated the pathogenesis mechanism during the development of dementia after ischemic stroke by investigating the relationship between miR-125a and its target ET-1, promising a potential pathological solution for post-stroke dementia in the future.
MeSH Keywords: 14-alpha Demethylase Inhibitors, Endothelin-1, Stroke
Background
Stroke ranks among the leading causes of disability and death and is an important risk factor for dementia including Alzheimer’s disease (AD) and vascular dementia (VaD). Three months after stroke, about 30% patients had severe dementia [1]. Alzheimer’s disease (AD) and ischemic/reperfusion (I/R) injury are closely associated with dementia [2]. Cerebrovascular diseases related to subcortical infarction were found to significantly lower the threshold of AD [3]. Recently, increasing evidence indicates that amyloid pathology contributing to sporadic AD could be exacerbated by I/R [4]. Vascular stress has been shown to result in significantly more cognitive deficit in experimental animal models [5].
As a potent vasoconstrictor, Endothelin-1 (ET-1) is involved in the pathogenesis of stroke. Patients with acute ischemic stroke have 4-fold higher level of the plasma ET-1 [6] and the severity of neurologic outcomes corresponds well to the extent of its increase [7]. Substantial brain lesion surrounding the injection site could be induced by ET-1 peptide administrated by intrastriatal injection into the brain [8]. In the CSF of patients with multi-infarction dementia or stroke, there is an increased level of ET-1, indicating that ET-1 might play a role in the cognitive deficits related to stroke [9]
MicroRNAs (miRNAs) are noncoding, small RNAs (21 to 25 nucleotides in length) which are supposed to regulate gene activity via accelerating mRNA degradation and/or inducing translational repression, leading to downregulation of target mRNA or protein expression [10]. Blockade of miR-92a expression reduces endothelial inflammation in mice, whereas miR-181b regulates endothelial cell NF-κB signaling, suggesting that these miRs are important in the endothelial dysfunction observed in vascular inflammation [11]. Hergenreider et al. [12] reported that shear stress-stimulated high expression of the miR-143/145 cluster in human umbilical vein endothelial cells (HUVECs). Moreover, it was reported that HUVECs control gene expression of cocultured vascular smooth muscle cells through extracellular vesicles enriched in miR-143/145. Previous investigation have demonstrated that the ability of miRNAs targeting genes could be disrupted by the presence of polymorphism either in the pri-microRNA that may compromise the mature processing of the miRNAs and reduce the expression level of mature miRNA, or located within or near the binding site in the 3′-untranslated region (3′UTR) of the target gene, which may interfere with the interaction between the miRNA and its target mRNA [13,14]. MiRNA SNPs have been reported to be functionally involved in the pathogenesis of human diseases by directly downregulating the expression of mature miRNA or disrupting the binding of miRNA to the mRNA [15].
It has been reported that a polymorphism (rs12976445) in pri-microRNA-125a compromises the mature processing of the miRNA, causing a decrease in the expression of the mature miRNA [16]. In this study, we validated ET-1 as a target of miR-125a, and found that rs12976445 polymorphism is responsible for an increased risk of dementia after ischemic stroke by releasing the inhibited ET-1.
Material and Methods
Participants
The study enrolled untreated patients (N=1023 cases) who were identified as having ischemic stroke according to the WHO criteria [17]. All patients had computed tomography and/or MRI scan within 24 h after admission. The demographic and pathological data are shown in Table 1. Patients who met the following entry criteria were included: Han ethnicity, age >40, admission within 48 h after stroke onset, and the presence of a reliable informant. Patients with a history of psychiatric disorders (e.g., depression), concomitant neurodegenerative disorders (like Parkinson’s disease), or those transferred from another hospital, were excluded. Cognitive status was assessed within 48 h after stroke onset using the Informant Questionnaire on Cognitive Decline in the Elderly (IQCODE) [18]. Patients who scored more than 104 were classified as demented, and according to the study population were divided into 2 groups: dementia positive (N=534) and dementia negative (N=489). The study protocol was approved by the research ethics committee at Henan University of Science and Technology and written consent was obtained from each participant prior to the study.
Table 1.
The frequency of CC genotype was significantly lower in the control group than the case group.
| Ischemic stroke with dementia (N=534) | Ischemic stroke without dementia (N=489) | Adjusted OR (95%CI) | |
|---|---|---|---|
| CC | 363 (67.97%) | 361 (73.82%) | |
| CT | 160 (29.96%) | 122 (24.95%) | |
| TT | 11 (2.07%) | 6 (1.23%) | |
| CT/TT | 171 (32.03%) | 128 (26.18%) | 1.32 (1.01–1.74) P=0.04 |
Endothelial cells collection
Human umbilical cord vessels were obtained from the First Affiliated Hospital of Henan University of Science and Technology, and vessels were rinsed with lactated Ringer’s irrigation solution. We filled the vessel from the bottom-up with 0.1% collagenase solution, and use 2 syringes to perfuse the cord with a volume of Hank’s Balanced Salt Solution, followed by flushing from 1 syringe through the cord into the other syringe, which was repeated about 6 times. The effluent was transferred to a sterile tube and re-suspended in DMEM medium. The cells were passaged 2–3 times before the following functional analysis. The endothelial cells were collected from a total of 28 individuals and genotyped for rs12976445 polymorphism. The study protocol was approved by the research ethics committee at Henan University of Science and Technology and written consent was obtained from each participant prior to the study.
Cell culture
HUVEC were obtained from Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium-F12 (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (Gibco, Grandstand, NY) at 37°C in a 5% CO2 atmosphere. When cells reached 80% confluence at passages 2–7, miR-125a-5p mimic and inhibitor (mirVana™ miRNA Inhibitor, Ambion, Austin, TX) or ET1 siRNA (Ambion, Austin, TX) were transfected into HUVECs at a concentration of 20 nM using Oligofectamine transfection reagent (Life Technologies, Grand island, NY) according to the manufacturer’s instructions.
Genotyping genomic
DNA was extracted from the peripheral blood of the participants using QIAamp DNA extraction Kit (Qiagen, Dusseldorf, Hamburg, German) according to the manufacturer’s protocol. The concentration and quality of genomic DNA was measured by NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, DE, USA) and then stored at −80°C prior to use. The chromosome segment containing rs12976445 polymorphism was amplified though PCR (primer set: 5′-CCA TCG TGT GGG TCT CAA G-3′ and 5′-TCT TTC ACA GTG GAT CCT CTG AC-3′), and the polymorphism was genotyped using direct Sanger sequencing.
Quantitative real-time reverse transcription PCR (RT-PCR)
Total RNA was extracted and purified using the miRNeasy kit (Qiagen). NanoDrop 2000C spectrophotometer (ThermoScientific) was used for the quantification of miRNA levels according to the manufacturer’s protocol. The miScript Reverse Transcription Kit (Qiagen,) was used to reversely transcribe target miRNA into cDNA with a gene-specific RT primer. The PCR amplification for the quantification of the target RNA was performed using miScipt SYBR Green PCR kit (Qiagen) and miScript Primer Assays kit (miR-125a, and ET-1, all from Qiagen) on a StepOnePlus real-time thermal cycler from Applied Biosystems. Each sample was analyzed in duplicate and miR-125a expression analyzed using the 2−ΔΔCT method. SNORD61 and U6 snRNA was included as house-keeping reference RNA. The relative expression of miR-125a was shown as fold difference relative to U6.
Western blot analysis
We collected the cells and lysed them in lysis buffer (Biosharp, Hefei, Anhui, China) containing 5 g/l sodium deoxycholate, 10 ml/l NP-40, 1 μg/ml aprotinin, 100 μg/ml phenylmethylsulfonyl fluoride, 0.1 g/lSDS, 0.2 g/l NaN3, 150 mmol/l NaCl, and 50 mmol/lTris-HCl (pH 8.5). Cell lysates were clarified of cell debris by centrifugation at 20 800 × g for 15 min through a QIAshredder spin column assembly (Qiagen) at 4°C. The Bradford method was employed to determine the concentrations of protein. The proteins were loaded on a 12% polyacrylamide gel (SDS-PAGE) and electrophoresed. After blotting onto a polyvinylidene fluoride membranes (Millipore, Boston, Massachusetts), we blocked the proteins with 5% non-fat milk (Merck, Darmstadt, Darmstadt, Germany) in Tris-buffered saline/Tween-20 (0.1%, Bioeasy, Shanghai, China) at room temperature for 2 h. The membranes were then probed with primary antibodies against ET-1 (1:10,000; Z0334; Dako), and β-actin (1:10,000; Abcam, Cambridge, LON, UK). Immunoreactive bands were quantified individually by estimating the number of pixels using an image analysis system (Image J, NIH, MD). The band intensity of the target protein was first normalized with the band intensity of the β-actin and expressed as a ratio of the 2 bands.
Luciferase assay
The 3′-UTR sequence of ET-1 containing putative or mutated miR-125a-5p binding sites was amplified from normal human genomic DNA and subcloned into the pmir-RB-REPORTTM vectors (RibioBiotech, Guangzhou, Guangdong, China). HUVECs cells (3.5×104) were seeded in triplicate in 24-well plates and cotransfected with mir-125a-5p mimics and a negative control (pre-mir-125a-5p-NC) using Lipofectamine 2000 (RibioBiotech, Beijing, China). After 48 h of transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System Kit (Promega Biotech, Madison, WI). hR-luc activities were first normalized to the internal control (h-luc) to evaluate the transfection efficiency. All experiments were performed triplicate. miR-125a-5p mediated repression of hR-luc/h-luc activity was calculated as the ratio of hR-luc/h-luc in miR-125a-5p-transfected cells to hR-luc/h-luc in control of oligo-transfected cells.
Statistical analysis
Comparisons between groups were made by the χ2 test (nominal variables) or Mann-Whitney U test (continuous variables). A value of p <0.05 was considered statistically significant. Hardy-Weinberg equilibrium was tested by the χ2 method. All variables with p<0.05 in univariate analyses were subjected to 3 sets of multivariate analyses with a backward stepwise logistic regression to identify independent predictors of PSD. To control for age and sex, both variables were included in regression models. The odds ratios and 95% confidence intervals were estimated from the regression coefficients. The analyses were performed using SPSS for Windows version 10.0 (SPSS Inc, Chicago, IL).
Results
Reported to be actively involved in various cellular activities, miRNAs negatively regulate the expression of their specific target genes by binding to the “seed sequences” in the 3′ UTR of its target gene. Recently, ET-1 has been reported to be significantly upregulated among samples collected from patients with ischemic stroke with dementia.
To identify the regulatory miRNA that negatively regulates the expression of ET-1, we searched the miRNA database (www.mirdb.org) and identified ET-1 as its candidate target gene. As shown in Figure 1, the alignment of miR-125a with the putative binding site in ET-1 3′UTR was significantly conserved among species. To further verify ET-1 as a validated target gene of miR-125a, we set up a luciferase assay reporter system by transfecting wild-type and mutant ET-1 3′UTR constructs, respectively, into pGL3 that contained downstream firefly luciferase. As shown in Figure 2, relative luciferase activity of cells transfected with constructs carrying wild-type ET-1 3′UTR seed sequence was clearly lower than that of the scramble control group and the blank control group, while that of the cells transfected with constructs carrying mutant ET-1 3′UTR seed sequence showed little difference when compared with the scramble control groups, indicating ET-1 as the exact target of miR-125a.
Figure 1.

ET-1 was identified as a target of miR-125a, with its putative binding site on ET-1 3′UTR showing conserved consistency in different species.
Figure 2.
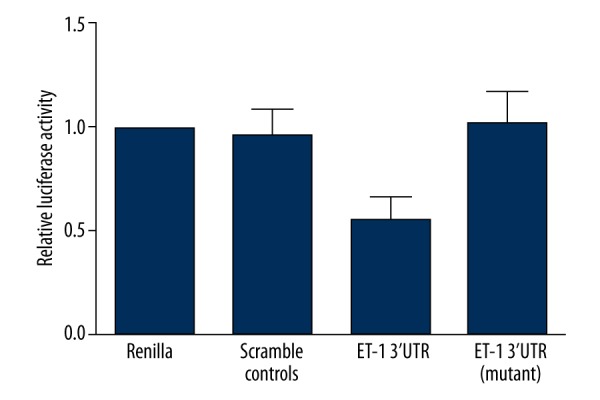
Relative luciferase activity of cells transfected with constructs carrying wild-type ET-1 3′UTR seed sequence was significantly lower compared with that of the scramble/blank controls and that of cells transfected with constructs carrying mutant ET-1 3′UTR seed sequence.
Furthermore, to validate ET-1 as a target of miR-125a as well as explore the interaction between miR-125a and ET-1, we also transfected miR-125a mimics, miR-125a inhibitors, and ET-1 siRNA into the cells carrying wild-type ET-1 3′UTR seed sequence. As shown in Figure 3, the protein (Figure 3A) and mRNA (Figure 3B) expression levels of ET-1 were similarly but significantly upregulated by miR-125a inhibitors as compared to the negative controls. On the contrary, presence of miR-125a mimics and ET-1 siRNA both downregulated the mRNA and protein expression levels when compared with the negative controls. The above findings all indicate a possible negatively associated relationship between miR-125a and its target ET-1.
Figure 3.
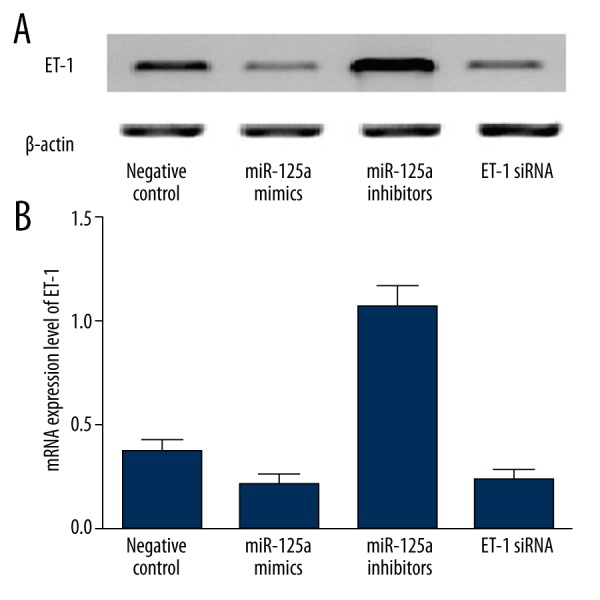
(A) ET-1 protein expression level of cells transfected with miR-125a mimics, miR-125a inhibitors, and ET-1 siRNA as compared with the negative control group. (B) ET-1 mRNA expression level of cells transfected with miR-125a mimics, miR-125a inhibitors, and ET-1 siRNA compared with the negative control group.
Also, in respect to the previous study of rs12976445 polymorphism in the pri-miR-125a, which was reported to cause a significant reduction in the amount of mature miR-125 by compromising the mature processing of the miRNA, we also explored the miRNA expression level of different rs12976445 polymorphism genotypes in human endothelial cells genotyped as CC (N=15), CT (N=9), and TT (N=4). As shown in Figure 4, expression levels of miR-125a were similar between TT and CT, both of which were significantly lower when compared with CC, indicating a dominant model of minor allele of rs12976445 polymorphism, which would lead to a less efficient inhibition of target gene ET-1. Consistent with the hypothesis that miR-125a negatively regulates the expression of ET-1, we found that the mRNA and protein expression levels were comparable between CT and TT, but were much lower in CC than in CT/TT groups.
Figure 4.
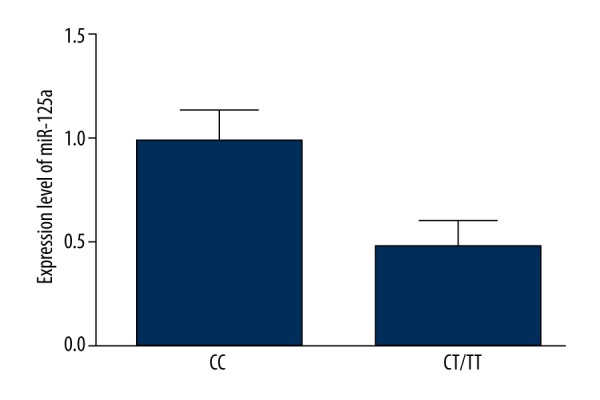
Expression level of miR-125a was significantly higher among CC samples compared with CT/TT samples.
To study the association between rs12976445 polymorphism and the risk of dementia after ischemic stroke, we enrolled 1023 ischemic stroke patients, 534 of who were also suffering from dementia (case group) while 489 were not (controls), as described in Table 1. Logistic regression analysis was used to investigate the association, and we found that the risk factors were comparable between the groups, and the frequency of CC genotype was significantly lower in the control group than the case group, indicating that the ability of miR-125a to suppress the expression of ET-1 helps to reduce to risk of dementia after ischemic stroke, and the presence of minor alleles of rs12976445 polymorphism caused a less efficient inhibition of ET-1, resulting in an increased risk of post-stroke dementia.
Discussion
In this study, we identified ET-1 as a target of miR-125a, and this relationship was validated using luciferase assay. Furthermore, transfection of miR-125a inhibitor substantially upregulated the expression of ET-1, while miR-125a and ET-1 siRNA caused downregulation of ET-1 in endothelial cells. In addition, we found that a polymorphism (rs12976445) interferes with the expression of miR-125a, which in turn caused an increase in the expression of ET-1 in human endothelial cells. The logistic regression analysis showed that rs12976445 is significantly associated with the risk of dementia after ischemic stroke.
Situated in chromosome 19q13.41, miR-125a is critical both in the adult tissues and organ development [19]. Increasing evidence has indicated that miR-125a is responsible for the pathogenesis of human diseases, especially malignancies such as lung cancers, gastric cancers, and breast cancers [20–22]. Two SNPs (rs12976445 and rs41275794) in the pre-miR-125a gene exist in the Han population of China and there is a complete linkage disequilibrium between them [23]. Rs12976445 is responsible for recurrent pregnancy loss [23] and the susceptibility to autoimmune thyroid diseases (24). Furthermore, poor prognosis in patients with esophageal squamous cell carcinoma (25) is associated with rs12976445. A recent investigation indicated that expression of mature miR-125a and processing of mature miRNA were impaired by the allele of rs12976445, consequently, target genes such as LIFR and ERBB2 [23,26] were induced upregulation [23,26]. In breast cancer [27,28], a poor prognosis and a more aggressive behavior are always associated with overexpression/Amplification of ERBB2, which makes ERBB2 a predictive biomarker of trastuzumab. Additionally, gefitinib might have better effect on nasopharyngeal carcinoma cells [29] when a higher level of miR-125a is achieved. Accordingly, the effectiveness of trastuzumab might be affected by miR-125a rs12976445. In this study, we found that relative luciferase activity of cells transfected with constructs carrying wild-type ET-1 3′UTR seed sequence was evidently lower than that of the scramble control group and the blank control group, while that of the cells transfected with constructs carrying mutant ET-1 3′UTR seed sequence showed mere difference when compared with the scramble control groups, indicating ET-1 as the exact target of miR-125a. In addition, we found that the protein (Figure 5A) and mRNA (Figure 5B) expression levels of ET-1 were similarly but significantly upregulated by miR-125a inhibitors as compared to the negative controls. On the contrary, presence of miR-125a mimics and ET-1 siRNA both downregulated the mRNA and protein expression levels when compared with the negative controls.
Figure 5.
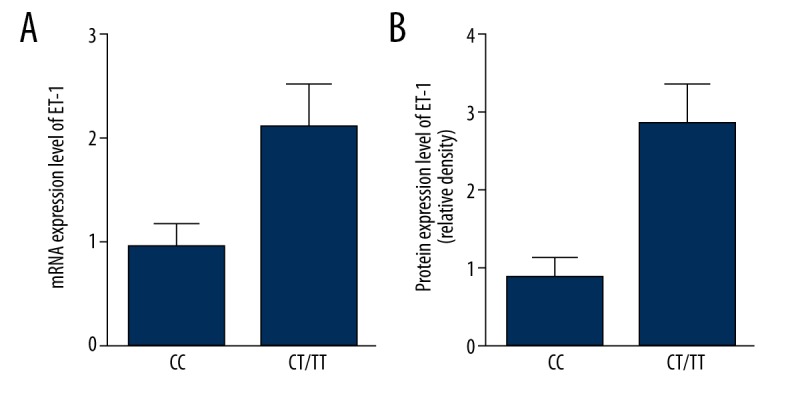
(A) MRNA expression level of ET-1 was significantly lower among CC samples compared with CT/TT samples. (B) Protein expression level of ET-1 was significantly lower among CC samples compared with CT/TT samples.
Initially separated from the supernatant of cultured aortic endothelial cells of pigs [29], endothelin-1 (ET-1) has been reported to be involved in the pathogenesis of many cerebral vascular and cardiovascular diseases, including stroke, because of its strong vasoconstriction effect. Previously, transgenic mice with an increased expression level of ET-1 in endothelial cells showed mild hypertension as well as altered vascular tone in the mesenteric artery and aorta, suggesting that overexpression of endothelial ET-1 had long-term detrimental effects on vascular and cardiac functions [30]. Compared to non-transgenic (NTg) mice, more severe brain damage and sensorimotor compromises occurred in the ET-1 transgenic mice after middle cerebral artery occlusion (MCAO) for 2 h followed by 22-h reperfusion [31], showing the effect of increased expression levels of endothelial ET-1 on cognitive function after stroke. Endothelial ET-1 increased vasoconstriction and permeability of the blood-brain barrier (BBB) when cerebral ischemia took place, by which endothelial ET-1 influenced neurological reactions [32–34]. In a recent study, ET-1 was injected to animals to induce cerebral ischemia. The Alzheimer disease (AD)-like cognitive deficit worsened when Aβ was injected to cerebral ventricle or the hippocampus of those mice. The level of amyloid precursor protein fragments such as Aβ was significantly elevated when ET-1 was injected to the striatal [35] indicating that overexpression of ET-1 during ischemic stroke may be involved in the accumulation and production of Aβ, which has neurotoxicity to the brain cells. Additionally, spatial reference memory and learning impairment and anxiety-like behavior was exacerbated by overexpression of ET-1 in the endothelial cells after a short-term ischemia followed by 7-day reperfusion [36], indicating that endothelial ET-1 plays a critical role in dementia and Aβ. It has been reported that a polymorphism (rs12976445) in pri-microRNA-125a compromises the mature processing of the miRNA, causing a decrease in the expression of the mature miRNA [16]. In the present study, we found that expression levels of miR-125a were similar between TT and CT, both of which were significantly lower when compared with CC, indicating a dominant model of minor allele of rs12976445 polymorphism, which would lead to a less efficient inhibition of target gene ET-1. Consistent with the hypothesis that miR-125a negatively regulates the expression of ET-1, we found the mRNA and protein expression levels were comparable between CT and TT, whereas they were much lower in CC than in CT/TT groups. Furthermore, we found that the risk factors were comparable between the groups, and the frequency of CC genotype was significantly lower in the control group than in the case group.
Conclusions
We elucidated the important role of ET-1 in the development of dementia after ischemic stroke, which was regulated by miR-125a, and found that the presence of minor allele of rs12976445 polymorphism reduces the expression of ET-1 in endothelial cells, causing less efficient inhibition of ET-1, and leading to an increased risk of cognitive dysfunction after stroke. Rs12976445 polymorphism could be a predictive biomarker for the risk of post-stroke dementia.
Footnotes
Source of support: Departmental sources
References
- 1.Pohjasvaara T, Erkinjuntti T, Ylikoski R, et al. Clinical determinants of poststroke dementia. Stroke. 1998;29:75–81. doi: 10.1161/01.str.29.1.75. [DOI] [PubMed] [Google Scholar]
- 2.Tatemichi TK, Desmond DW, Mayeux R, et al. Dementia after stroke: baseline frequency, risks, and clinical features in a hospitalized cohort. Neurology. 1992;42:1185–93. doi: 10.1212/wnl.42.6.1185. [DOI] [PubMed] [Google Scholar]
- 3.Snowdon DA, Greiner LH, Mortimer JA, et al. Brain infarction and the clinical expression of Alzheimer disease. The Nun Study. JAMA. 1997;277:813–17. [PubMed] [Google Scholar]
- 4.Song B, Ao Q, Niu Y, et al. Amyloid beta-peptide worsens cognitive impairment following cerebral ischemia-reperfusion injury. Neural Regen Res. 2013;8:2449–57. doi: 10.3969/j.issn.1673-5374.2013.26.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitehead SN, Cheng G, Hachinski VC, Cechetto DF. Progressive increase in infarct size, neuroinflammation, and cognitive deficits in the presence of high levels of amyloid. Stroke. 2007;38:3245–50. doi: 10.1161/STROKEAHA.107.492660. [DOI] [PubMed] [Google Scholar]
- 6.Ziv I, Fleminger G, Djaldetti R, et al. Increased plasma endothelin-1 in acute ischemic stroke. Stroke. 1992;23:1014–16. doi: 10.1161/01.str.23.7.1014. [DOI] [PubMed] [Google Scholar]
- 7.Moldes O, Sobrino T, Millan M, et al. High serum levels of endothelin-1 predict severe cerebral edema in patients with acute ischemic stroke treated with t-PA. Stroke. 2008;39:2006–10. doi: 10.1161/STROKEAHA.107.495044. [DOI] [PubMed] [Google Scholar]
- 8.Agnati LF, Zoli M, Kurosawa M, et al. A new model of focal brain ischemia based on the intracerebral injection of endothelin-1. Ital J Neurol Sci. 1991;12:49–53. [PubMed] [Google Scholar]
- 9.Nakajima M, Morimoto S, Takamoto S, et al. Endothelin-1 in cerebrospinal fluid in elderly patients with hypertension and dementia. Hypertension. 1994;24:97–100. doi: 10.1161/01.hyp.24.1.97. [DOI] [PubMed] [Google Scholar]
- 10.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–48. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 11.Loyer X, Potteaux S, Vion AC, et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ Res. 2014;114:434–43. doi: 10.1161/CIRCRESAHA.114.302213. [DOI] [PubMed] [Google Scholar]
- 12.Hergenreider E, Heydt S, Treguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nature Cell Biology. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 13.Lin JH, Huang Y, Zhang XL, et al. Association of miR-146a rs2910164 with childhood IgA nephropathy. Pediatr Nephrol. 2014;29:1979–86. doi: 10.1007/s00467-014-2818-3. [DOI] [PubMed] [Google Scholar]
- 14.Gao XB, Yang LP, Ma Y, et al. No association of functional variant in pri-miR-218 and risk of congenital heart disease in a Chinese population. Gene. 2013;523:173–77. doi: 10.1016/j.gene.2013.03.119. [DOI] [PubMed] [Google Scholar]
- 15.Yang B, Liu C, Diao L, et al. A polymorphism at the microRNA binding site in the 3′ untranslated region of C14orf101 is associated with non-Hodgkin lymphoma overall survival. Cancer Genet. 2014;207:141–46. doi: 10.1016/j.cancergen.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Jiao LH, Zhang JX, Dong YY, et al. Association between miR-125a rs12976445 and survival in breast cancer patients. Am J Transl Res. 2014;6:869–75. [PMC free article] [PubMed] [Google Scholar]
- 17.Hatano S. Experience from a multicentre stroke register: a preliminary report. Bull World Health Organ. 1976;54:541–53. [PMC free article] [PubMed] [Google Scholar]
- 18.Jorm AF, Korten AE. Assessment of cognitive decline in the elderly by informant interview. Br J Psychiatry. 1988;152:209–13. doi: 10.1192/bjp.152.2.209. [DOI] [PubMed] [Google Scholar]
- 19.Shaham L, Binder V, Gefen N, et al. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–18. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 20.Nishida N, Mimori K, Fabbri M, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–33. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 21.Zhu WY, Luo B, An JY, et al. Differential expression of miR-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Invest. 2014;32:394–401. doi: 10.3109/07357907.2014.922569. [DOI] [PubMed] [Google Scholar]
- 22.Jiang LL, Chang JH, Zhang QF, et al. MicroRNA Hsa-miR-125a-3p activates p53 and induces apoptosis in lung cancer cells. Cancer Invest. 2013;31:538–44. doi: 10.3109/07357907.2013.820314. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Liu CM, Qi L, et al. Two common SNPs in pri-miR-125a alter the mature miRNA expression and associate with recurrent pregnancy loss in a Han-Chinese population. Rna Biology. 2011;8:861–72. doi: 10.4161/rna.8.5.16034. [DOI] [PubMed] [Google Scholar]
- 24.Inoue Y, Watanabe M, Inoue N, et al. Associations of single nucleotide polymorphisms in precursor-microRNA (miR)-125a and the expression of mature miR-125a with the development and prognosis of autoimmune thyroid diseases. Clin Exp Immunol. 2014;178:229–35. doi: 10.1111/cei.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu CH, Li MJ, Hu C, Duan HB. Prognostic role of microRNA polymorphisms in patients with advanced esophageal squamous cell carcinoma receiving platinum-based chemotherapy. Cancer Chemother Pharmacol. 2014;73:335–41. doi: 10.1007/s00280-013-2364-x. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann TP, Korski K, Ibbs M, et al. rs12976445 variant in the pri-miR-125a correlates with a lower level of hsa-miR-125a and ERBB2 overexpression in breast cancer patients. Oncol Lett. 2013;5:569–73. doi: 10.3892/ol.2012.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16:413–28. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 28.Piccart MJ. Proposed treatment guidelines for HER2-positive metastatic breast cancer in Europe. Ann Oncol. 2001;12(Suppl 1):S89–94. doi: 10.1093/annonc/12.suppl_1.s89. [DOI] [PubMed] [Google Scholar]
- 29.Yanagisawa M, Kurihara H, Kimura S, et al. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411–15. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- 30.Leung JW, Wong WT, Koon HW, et al. Transgenic mice over-expressing ET-1 in the endothelial cells develop systemic hypertension with altered vascular reactivity. PLoS One. 2011;6:e26994. doi: 10.1371/journal.pone.0026994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung JW, Ho MC, Lo AC, et al. Endothelial cell-specific over-expression of endothelin-1 leads to more severe cerebral damage following transient middle cerebral artery occlusion. J Cardiovasc Pharmacol. 2004;44(Suppl 1):S293–300. doi: 10.1097/01.fjc.0000166277.70538.b0. [DOI] [PubMed] [Google Scholar]
- 32.Macrae IM, Robinson MJ, Graham DI, et al. Endothelin-1-induced reductions in cerebral blood flow: dose dependency, time course, and neuropathological consequences. J Cereb Blood Flow Metab. 1993;13:276–84. doi: 10.1038/jcbfm.1993.34. [DOI] [PubMed] [Google Scholar]
- 33.Matsuo Y, Mihara S, Ninomiya M, Fujimoto M. Protective effect of endothelin type A receptor antagonist on brain edema and injury after transient middle cerebral artery occlusion in rats. Stroke. 2001;32:2143–48. doi: 10.1161/hs0901.94259. [DOI] [PubMed] [Google Scholar]
- 34.Stanimirovic DB, Bertrand N, McCarron R, et al. Arachidonic acid release and permeability changes induced by endothelins in human cerebromicrovascular endothelium. Acta Neurochir Suppl (Wien) 1994;60:71–75. doi: 10.1007/978-3-7091-9334-1_18. [DOI] [PubMed] [Google Scholar]
- 35.Amtul Z, Whitehead SN, Keeley RJ, et al. Comorbid rat model of ischemia and beta-amyloid toxicity: striatal and cortical degeneration. Brain Pathol. 2015;25:24–32. doi: 10.1111/bpa.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Yeung PK, McAlonan GM, et al. Transgenic mice over-expressing endothelial endothelin-1 show cognitive deficit with blood-brain barrier breakdown after transient ischemia with long-term reperfusion. Neurobiol Learn Mem. 2013;101:46–54. doi: 10.1016/j.nlm.2013.01.002. [DOI] [PubMed] [Google Scholar]


