Abstract
The digestive tracts of vertebrates are colonized by complex assemblages of micro-organisms, collectively called the gut microbiota. Recent studies have revealed important contributions of gut microbiota to vertebrate health and disease, stimulating intense interest in understanding how gut microbial communities are assembled and how they impact host fitness (Sekirov et al. 2010). Although all vertebrates harbour a gut microbiota, current information on microbiota composition and function has been derived primarily from mammals. Comparisons of different mammalian species have revealed intriguing associations between gut microbiota composition and host diet, anatomy and phylogeny (Ley et al. 2008b). However, mammals constitute <10% of all vertebrate species, and it remains unclear whether similar associations exist in more diverse and ancient vertebrate lineages such as fish. In this issue, Sullam et al. (2012) make an important contribution toward identifying factors determining gut microbiota composition in fishes. The authors conducted a detailed meta-analysis of 25 bacterial 16S rRNA gene sequence libraries derived from the intestines of different fish species. To provide a broader context for their analysis, they compared these data sets to a large collection of 16S rRNA gene sequence data sets from diverse free-living and host-associated bacterial communities. Their results suggest that variation in gut microbiota composition in fishes is strongly correlated with species habitat salinity, trophic level and possibly taxonomy. Comparison of data sets from fish intestines and other environments revealed that fish gut microbiota compositions are often similar to those of other animals and contain relatively few free-living environmental bacteria. These results suggest that the gut microbiota composition of fishes is not a simple reflection of the micro-organisms in their local habitat but may result from host-specific selective pressures within the gut (Bevins & Salzman 2011).
Keywords: aquaculture, bacteria, coevolution, comparative biology, fish, microbial biology
Approximately 28 000 fish species comprise nearly half of all vertebrate diversity and represent a broad range of physiologies, ecologies and natural histories (Nelson 2006). Fishes therefore represent an important vertebrate group for understanding the evolution and ecology of host–microbiota interactions (Nayak 2010). Our current information on the intestinal microbiota of fishes has been largely derived from culture-based approaches, which often reveal only a limited range of microbial diversity. Although culture-independent DNA sequence-based approaches have been deployed to define the intestinal bacterial diversity of some fishes, few studies have compared multiple species and only one (Roeselers et al. 2011) compared the intestinal microbiotas of a large number of fish species. However, the diversity of fish species compared and statistical methods used in these studies were limited, and we still understand little about how ecological and environmental factors impact fish gut microbiota composition. This article by Sullam et al. (2012) is significant because it presents phylogenetic and statistical meta-analyses of intestinal microbiotas from the largest number of fish species to date. Their analysis included 16S rRNA gene sequences from 24 published culture-dependent and culture-independent libraries plus a new library from Trinidadian guppy, Poecilia reticulata. The 18 fish species represented in these libraries inhabit a wide range of water salinities and trophic levels, permitting unprecedented insight into the ecological and environmental factors shaping gut microbiota composition in fishes.
The authors’ comparison of freshwater, saltwater or estuarine fish revealed that intestinal microbiota composition is strongly correlated with salinity. Principal coordinate analysis (PCoA) revealed that intestinal bacterial communities from freshwater and saltwater fish formed significantly different clusters. Consistent with the findings of Roeselers et al. (2011), they found that these PCoA differences were associated with increased representation of operational taxonomic units (OTUs) from the bacterial order Aeromonadales in freshwater fishes and Vibrionales in saltwater fishes. As environmental water microbiota composition has previously been shown to correlate strongly with salinity (Lozupone & Knight 2007), additional research is needed to determine whether the differences between intestinal microbiotas of freshwater and saltwater fish are attributed to the diversity of environmental micro-organisms available for intestinal colonization and/or to host physiologic differences caused by different water salinities.
The authors also report significant associations between fish intestinal microbiota composition and host trophic level. Carnivores, omnivores and herbivores formed significant overlapping clusters in PCoA space. Enrichments of OTUs from specific taxa accompanied these differences between bacterial communities in fish from distinct trophic levels. Intriguingly, Sullam et al. (2012) also identified a significant association between intestinal microbiota composition and fish taxonomy. This association was significant only when considering all fish intestinal data sets, but not when only considering culture-based data sets. This suggests potential co-evolution of fishes and their gut microbiotas. This is consistent with a previous report showing that colonization of germ-free zebrafish with a Firmicutes phylum-dominant mouse gut microbiota results in enrichment of phylum Proteobacteria, which normally dominates zebrafish intestines (Rawls et al. 2006). Additionally, in accordance with previous studies noting few differences between animals raised in artificial and natural environments (Ley et al. 2008a; Roeselers et al. 2011), the analysis of Sullam et al. (2012) detected no significant effects of rearing environment on gut microbiota composition. Together, these data support several emerging themes in fish gut microbial ecology: microbiota composition is strongly associated with host trophic level, habitat salinity and perhaps taxonomy, and but with relatively little impact of host provenance.
To broaden the context for understanding diversity within and between the intestinal microbiotas of different fishes, Sullam et al. (2012) compared the fish libraries to a large collection of 16S rRNA gene sequences from bacteria associated with a diverse range of other animals and environments (Ley et al. 2008b). PCoA revealed that, instead of clustering together, the gut communities of fish were distributed across the range of other samples (Fig. 1). While omnivorous fish all clustered near the free-living and invertebrate-associated communities, carnivorous fish generally clustered with carnivorous mammals, and three of the four herbivorous fish communities were similar to mammalian communities. Moreover, the fact that these three herbivorous fish differed in the presence and location of specialized fermentation chambers suggests that these communities can assemble in a diversity of digestive tract anatomies. Additionally, top BLASTN hits of representative sequences from each OTU in the fish libraries were included in a phylogenetic analysis with the fish sequences, and the grouping of each fish bacterial sequence with each other and with those from other hosts and habitats was used to categorize the closest relatives of fish gut microbes to a projected source environment or ‘lifestyle’. The two most frequently observed bacterial taxa were enriched in, and associated almost exclusively with, freshwater fishes. Supporting the PCoA results, fish intestinal bacteria were often closely related to bacteria from land animals and not to free-living environmental bacteria. Surprisingly, more than half the OTUs from herbivorous fishes were more closely related to bacteria in mammalian and bird intestines than to bacteria from fish intestines. The authors also observed that only two of the 25 fish libraries in their study consisted primarily of sequences from bacteria most closely related to free-living bacteria, demonstrating the scarcity of bacteria related to free-living species in some fish guts. While this nearest neighbour approach provides a useful perspective on gut microbiota membership, the utility of this information is limited by availability of closely related 16S rRNA gene sequences in the nr/nt database and the accuracy of lifestyle information associated with those sequences. Nevertheless, these results suggest that intestinal microbiotas from different fishes usually share greater similarity with animal microbiotas than with environmental microbiotas.
Fig. 1.
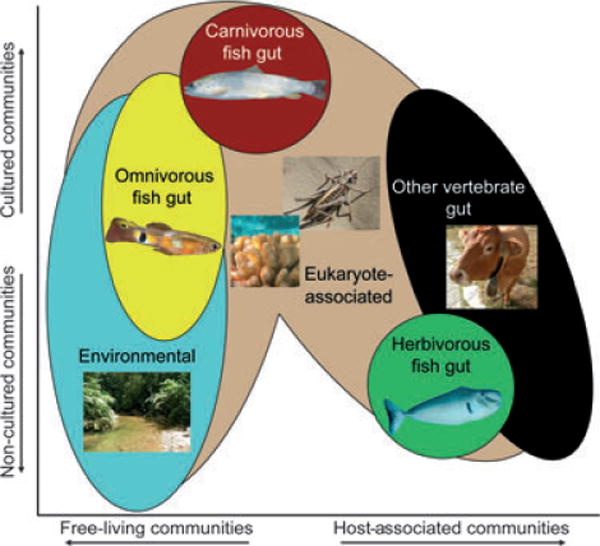
Similarity among gut bacterial communities in fishes from different trophic levels compared to non-fish-associated bacterial communities from various sources. This figure is a representation of a PCoA from fig. 3 in Sullam et al. (2012). Bacterial communities vary along the X-axis (PCoA1) according to their associations with organisms. Environmental bacteria, from both aquatic and terrestrial habitats, fall on the left-hand side of the X-axis (represented by blue area). Bacterial communities derived from eukaryotic animal hosts tend to fall further right along the X-axis (represented by beige area), with gut communities from mammals and other vertebrates found furthest right (represented by black area). Herbivorous fish gut bacterial communities (represented by green area), which were all derived from marine species, are more similar to mammalian gut bacterial communities. In contrast, omnivorous fish gut bacterial communities (represented by yellow area) are more similar to environmental samples, and carnivorous fish bacterial communities (represented by red area) are more similar to those found in insects and other eukaryotic habitats. Bacterial communities also differentiate along the ϒ-axis (PCoA2) depending in part on whether they are derived from culture-based or culture-independent approaches. Photo credits: Omnivorous fish (Poecilia reticulata), Paul Bentzen; Carnivorous fish (Salmo trutta), Emilie Person; Herbivorous fish (Naso tonganus), Kendall Clements. Other pictures from left to right: Trinidadian stream, Karen Sullam; Coral from French Polynesia (Pocillopora verrucosa), Adrian Stier; Orthopteran, Karen Sullam; Cow, Karen Sullam.
Overall, the authors’ results indicate that ecological and environmental features associated with fish host biology might facilitate selection for specific microbial memberships in their intestinal tracts, as has been suggested by studies in mammals (Bevins & Salzman 2011). It will be important in the future to include additional freshwater herbivores and fish from taxa, trophic levels and water salinities not included in this study. Moreover, the potential impact of other environmental parameters (e.g. water depth and temperature, diet composition, food chain dynamics, geographic location), host physiological parameters (e.g. developmental stage, immunity, digestive anatomy and physiology) and the interaction between these factors on gut microbiota composition needs to be evaluated. Furthermore, the degree to which individuals of a fish species possess bacterial communities with both taxonomic and functional similarity needs to be defined (Roeselers et al. 2011). As with any meta-analysis, the work of Sullam et al. (2012) relied largely on published libraries generated using inconsistent methods that could introduce apparent variation between samples. It will therefore be important to conduct future comparisons of intestinal microbiotas from different fishes using consistent methods for sample collection and analysis. Improved understanding of the determinants of fish gut microbiota composition and function could lead to new approaches for preventing and treating pathogenesis relevant to fish culture and conservation. Moreover, principles governing microbial ecology in the fish intestine might be active in mammals, permitting translation from fish models to relevant human disease states. Also, observed differences between the gut microbial ecologies of fish and mammals would yield important insights into how the underlying principles vary among different vertebrate lineages. This information would provide an essential foundation for exploring the impact of gut microbiota composition and function on the ecology, fitness and evolution of their respective hosts.
References
- Bevins CL, Salzman NH. The potter’s wheel: the host’s role in sculpting its microbiota. Cellular and Molecular Life Sciences. 2011;68:3675–3685. doi: 10.1007/s00018-011-0830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and the gut microbes. Science. 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Reviews Microbiology. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak SK. Role of gastrointestinal microbiota in fish. Aquaculture Research. 2010;41:1553–1573. [Google Scholar]
- Nelson JS. Fishes of the World. 4th. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2006. [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, et al. Evidence for a core gut microbiota in the zebrafish. The ISME Journal. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Atunes LCM, Finlay BB. Gut microbiota in health and disease. Physiological Reviews. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Sullam KE, Essinger SD, Lozupone CA, et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Molecular Ecology. 2012;21:3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


