Abstract
White adipose tissue (WAT) is the major site of energy storage in bony vertebrates, and also serves central roles in the endocrine regulation of energy balance. The cellular and molecular mechanisms underlying WAT development and physiology are not well understood. This is due in part to difficulties associated with imaging adipose tissues in mammalian model systems, especially during early life stages. The zebrafish (Danio rerio) has recently emerged as a new model system for adipose tissue research, in which WAT can be imaged in a transparent living vertebrate at all life stages. Here we present detailed methods for labeling adipocytes in live zebrafish using fluorescent lipophilic dyes, and for in vivo microscopy of zebrafish WAT.
I. Introduction
A. Biomedical Relevance of Adipose Tissue
The prevalence of obesity has reached epidemic proportions in developed as well as developing countries (Yach et al., 2006). Furthermore, adult obesity is recognized as a major risk factor for the development of metabolic disease including insulin resistance, type II diabetes, and nonalcoholic fatty liver disease (Morse et al., 2010). New methods for primary prevention and treatment of obesity are therefore urgently required to address this global public health crisis. Obesity develops when energy intake exceeds energy expenditure, resulting in increased storage of excess energy in adipose tissues (Rosen and Spiegelman, 2006). Adipose tissues also serve active roles in endocrine control of energy balance and thermogenic energy expenditure (Cypess and Kahn, 2010; Poulos et al., 2010). Therefore, manipulation of adipose tissue structure and function is an attractive strategy to alleviate obesity and its associated morbidities. In this chapter, we discuss zebrafish as a novel model system in adipose tissue research, with emphasis on methods for fluorescent labeling and in vivo imaging of zebrafish adipose tissues.
B. Cellularity of White Adipose Tissue
The major constituent of adipose tissue, adipocytes, can be of two distinct lineages: white and brown. Brown adipocytes are thermogenic and help regulate body temperature by converting energy to heat through the action of the mitochondrial anion transporter uncoupling protein 1 (UCP1) (Cypess and Kahn, 2010). Although UCP1 homologs have been identified in zebrafish and other poikilothermic vertebrates (Hughes and Criscuolo, 2008; Jastroch et al., 2005), zebrafish are not considered to possess brown adipose tissue; therefore, brown adipocytes will not be discussed further in this chapter. White adipocytes, within white adipose tissue (WAT), store surplus energy in the form of triacylglyceride (TG), and hydrolyze accumulated TG for use as a fuel source during times of nutrient deprivation (Redinger, 2009). They are present in bony vertebrates (e.g., fish, amphibians, reptiles, birds, and mammals), and WAT is considered the primary site of energy storage in these vertebrate taxa (Gesta et al., 2007). Stored TG within mature white adipocytes (hereafter referred to as “adipocytes”) is contained in a single, large (up to rv100 mm in diameter) lipid droplet (LD) (Farese and Walther, 2009). Manipulation of WAT metabolism can induce whole-animal insulin resistance (Abel et al., 2001), whereas removal of WAT can improve insulin sensitivity (Barzilai et al., 1999; Carvalho et al., 2005; Gabriely et al., 2002). Therefore, modification of WAT form and function may represent viable strategies for primary prevention of obesity and mitigation of associated metabolic disease.
Mammalian animal and cell culture systems have contributed the majority of our current information on WAT cell biology. However, the importance of WAT to commercial aquaculture and the increasing use of zebrafish as a biomedical model system have provided a rapidly expanding knowledge base on WAT in teleost fish (the largest subclass of Actinopterygii or ray-finned fish). All teleost species analyzed accumulate lipid within WAT (Figs. 1 and 2) (Flynn et al., 2009; Imrie and Sadler, 2010; Sheridan, 1988; Song and Cone, 2007). Deposition and mobilization of lipid within teleost WAT is altered in response to nutritional manipulation (Fig. 2), suggesting the energy storage functions of WAT have been conserved between teleosts and mammals (Albalat et al., 2007; Bellardi et al., 1995; Company et al., 1999; Flynn et al., 2009; Imrie and Sadler, 2010; Om et al., 2001). Furthermore, histological analysis reveals evolutionarily conserved morphological features of teleost adipocytes, including large cytoplasmic LDs, caveolae, and close association with capillaries (Flynn et al., 2009; Imrie and Sadler, 2010). In addition, teleost adipocytes express genes associated with adipocyte differentiation (fatty acid–binding protein 11a, fabp11a; peroxisome proliferator–activated receptor gamma, pparg; and CCAAT/enhancer-binding protein alpha, cebpa) (Fig. 3) (Flynn et al., 2009; Ibabe et al., 2005; Imrie and Sadler, 2010; Oku and Umino, 2008; Vegusdal et al., 2003), adipocyte lipolysis (lipoprotein lipase, lpl) (Oku et al., 2006), and adipocyte endocrine function (leptin, lep; adiponectin, acrp30; and adipsin, cfd) (Imrie and Sadler, 2010; Vegusdal et al., 2003). Like mammals, fish WAT also possesses a stromal–vascular fraction (SVF, defined as a heterogeneous population of stromal cells isolated by enzymatic digestion of WAT), which contains adipocyte progenitors (Rodeheffer et al., 2008; Tang et al., 2008; Todorcevic et al., 2010; Vegusdal et al., 2003). Together, the considerable functional, morphological, and molecular homology between teleost and mammalian WAT suggests new insights into WAT biology gained in the zebrafish system will be directly translatable to humans and other vertebrates.
Fig. 1.
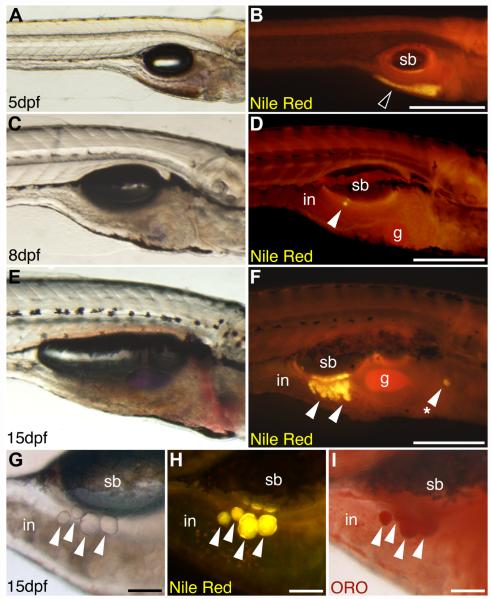
Fluorescent lipophilic dyes reveal lipid droplet accumulation in live zebrafish. Live zebrafish at 5 dpf (3.6 ± 0.1 mm SL), 8 dpf (4.3 ± 0.1 mm SL), or 15 dpf (5.3 ± 0.1 mm SL) were stained with the fluorescent lipophilic dye Nile Red, and imaged on a fluorescence stereomicroscope using a GFP longpass emission filter set. Bright field (A, C, E, G) and corresponding fluorescence images (B, D, F, H) are shown. Nile Red fluorescence emission maxima are shifted to shorter wavelengths when incorporated into neutral lipid, so neutral lipid depots in yolk (black arrowhead in B) and adipocytes (white arrowheads in D, F, H) appear yellow-orange. (A and B) The yolk is the major neutral lipid depot in 5 dpf larvae. (C and D) After yolk resorption, the first adipocyte neutral lipid droplets form in the right viscera by 8 dpf in association with the pancreas. (E and F) By 15 dpf, adipocyte lipid droplets have increased in number within the viscera, and also appear in other locations (asterisk in F). An individual 15 dpf zebrafish stained with Nile Red (G and H) then stained with Oil Red O (ORO; I) reveals colocalization of Nile Red and ORO staining in neutral lipid droplets of visceral adipocytes (white arrowheads). Swim bladder (sb), gall bladder (gb), and intestine (in) are indicated. Anterior is to the right, and dorsal to the top in all images. Scale bars: 400 mm (A and B), 300 mm (C–F), and 100 mm (G–I). This research was originally published in Flynn et al. (2009). © The American Society for Biochemistry and Molecular Biology.
Fig. 2.
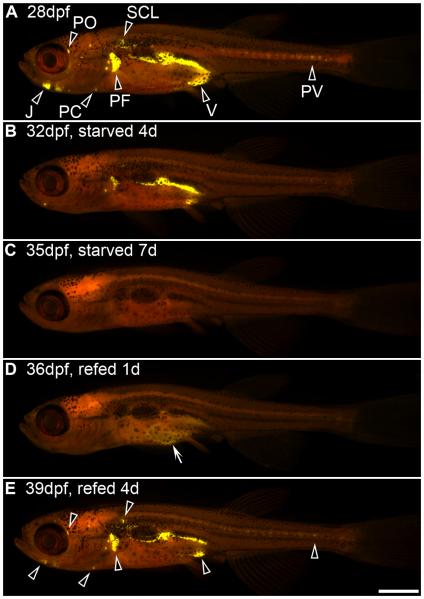
Longitudinal analysis of zebrafish adipose depots can be used to visualize fat mobilization in response to starvation, and fat deposition in response to refeeding. Zebrafish were starved for 7 days beginning at 28 dpf (9.6 ± 0.4 mm SL) and then refed for 4 days. Individual zebrafish were stained with Nile Red and imaged daily using a fluorescence stereomicroscope with a GFP longpass emission filter set to monitor neutral lipid deposits (black arrowheads in A and E). Panels A–E depict an individual representative animal. (A) Zebrafish fed normally through 28 dpf store neutral lipid in multiple salient adipose depots, including visceral (V), pectoral fin plate (PF), pericardial (PC), jaw (J), perivertebral (PV), periorbital (PO), and subcutaneous lateral (SCL) depots. Note that zebrafish begin depositing fat in additional adipose depots [e.g., subcutaneous dorsal (SCD), subcutaneous ventral (SCV), and at the base of fins] at subsequent developmental stages (Imrie and Sadler, in press; J. Minchin, unpublished). (B) When starved for 4 days, neutral lipid depots were reduced in all locations, although the larger visceral and pectoral fin plate depots were the last to be exhausted. (C) After 7 days of starvation, all neutral lipid depots were depleted. (D) Refeeding for 1 day was sufficient to form transient neutral lipid deposits in the intestine (white arrow in D), and refeeding for 4 days was sufficient to reestablish neutral lipid depots in the same locations as before starvation (black arrowheads in E and A). Anterior is to the left, and dorsal to the top in all images. Scale bar: 1 mm. This research was originally published in Flynn et al. (2009). © The American Society for Biochemistry and Molecular Biology.
Fig. 3.
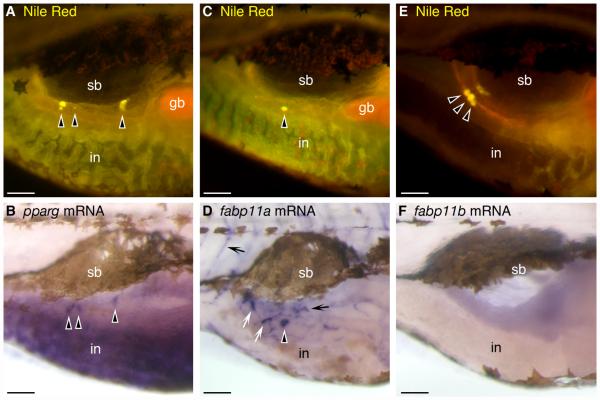
Coupling of fluorescent lipophilic dyes with in situ hybridization reveals expression of pparg and fabp11a mRNA in zebrafish visceral adipocytes. Individual zebrafish were stained with Nile Red at 15 dpf (5.3 ± 0.1 mm SL), imaged using a fluorescence stereomicroscope with a GFP longpass emission filter set, and then processed for whole-mount in situ hybridization (WISH) using riboprobes directed against zebrafish pparg, fabp11a, or fabp11b mRNA. Cells labeled by WISH stain purple, in contrast to the brown melanin pigment contained within melanophores. Nile Red and WISH staining patterns were compared to detect colocalization. Whole-mount images from the trunk of the same individuals are shown. Both fabp11a and pparg mRNA colocalize with visceral neutral lipid droplets associated with the pancreas (black arrowheads). fabp11a mRNA is also observed in nearby cells lacking neutral lipid droplets (white arrows in D) and blood vessels (black arrows in D), while pparg mRNA is also found in the intestinal epithelium (B). In contrast, visceral adipocytes did not express fabp11b (F). Swim bladder (sb), gall bladder (gb), and intestine (in) are indicated. Dorsal is to the top and anterior is to the right in all images. Scale bars: 100 μm. This research was originally published in Flynn et al. (2009). © The American Society for Biochemistry and Molecular Biology.
Adipose tissue is not a single homogeneous tissue in mammals and fishes, but rather is distributed at specific anatomical locations throughout the body. In humans, the largest sites of WAT deposition are either subcutaneous (defined as between muscle and skin) or intra-abdominal (within the abdominal cavity) (Gesta et al., 2007; Shen and Chen, 2008). Anatomically distinct depots display different molecular and physiological characteristics (Gesta et al., 2006; Peinado et al., 2010; Vidal, 2001; Vohl et al., 2004), and have different risk associations for obesity-related disorders (Kissebah and Krakower, 1994). In particular, visceral adipose tissue (VAT, intra-abdominal WAT surrounding internal organs) is associated with more adverse risk factors than subcutaneous WAT (Despres, 1998; Fox et al., 2007). Teleost WAT is also deposited in both subcutaneous and intra-abdominal positions, including VAT locations (Figs. 1 and 2A), raising the possibility that developmental programs responsible for WAT anatomy have been maintained during vertebrate evolution (Flynn et al., 2009; Imrie and Sadler, 2010; Umino et al., 1996; Weil et al., 2009). Recent work from our lab and others has provided a provisional nomenclature for zebrafish adipose depots (Fig. 2A) (Flynn et al., 2009; Imrie and Sadler, 2010), but an expanded evaluation of adipose depot anatomy and heterogeneity is needed to comprehensively enumerate and categorize zebrafish adipose depots. Both terminally differentiated adipocytes and the SVF within distinct WAT locations contribute to the diverse physiologies of WAT (Peinado et al., 2010; Wajchenberg, 2000), although the molecular and cellular basis for disease risk associations based on WAT anatomic location is unclear. Therefore, it is apparent that investigation of multiple sites of WAT deposition within whole animals is desperately needed.
II. Rationale
Current analysis of WAT is predominantly conducted after fixation and histological sectioning of adipose tissues. This often results in incomplete preservation of WAT architecture and limited information on cellular interactions and dynamics (Xue et al., 2010). In addition, imaging of whole-animal WAT deposition in mammals is technically challenging, is typically restricted to low-resolution views, and has only been undertaken on a limited scale (Shen and Chen, 2008). Moreover, most of our knowledge of mammalian adipose tissues is derived from adult stages, due in part to the difficulty of accessing adipose tissues during the gestational stages when they initially develop (Ailhaud et al., 1992). As a consequence, outstanding questions regarding the spatial and temporal dynamics of in vivo adipose tissue formation and growth remain understudied. Innovative approaches have been developed to address these gaps in our knowledge, such as high-resolution imaging of resected adipose tissue cultured in vitro (Nishimura et al., 2007), and in vivo imaging of adipocyte precursors introduced into mice fitted with an implanted coverslip (Nishimura et al., 2008). However, these approaches do not permit imaging of adipose tissues within the intact physiological context of a living organism. Mathematical modeling has also been used to predict in vivo mechanisms of adipose tissue growth, but these models remain largely untested due to a paucity of suitable in vivo model systems (Jo et al., 2009). There is, therefore, a pressing need for new experimental platforms for image analysis of WAT formation and function in live animals.
The features of the zebrafish system are especially well suited to meet these needs. Zebrafish develop externally and are optically transparent from fertilization to the onset of adulthood, permitting in vivo imaging of dynamic cellular events during adipose tissue formation and growth. This provides new opportunities to investigate the earliest stages of WAT morphogenesis (Fig. 1), a process poorly understood in mammals with potentially high relevance for obesity and metabolic disease. The small size of the zebrafish also facilitates wholeanimal imaging of multiple adipose depots (Fig. 2), unlike mammalian systems in which specific adipose depots are difficult to access. Real-time imaging of living adipose tissues is also possible in the zebrafish, enabling observation of molecular and cellular events over short time scales. Furthermore, the amenability of the zebrafish to in vivo imaging permits longitudinal imaging of WAT in individual animals, which can be used to mitigate complications from interindividual variation in adiposity (Fig. 2). As described above, the identification of extensive conserved homologies between teleost and mammalian adipose tissue suggests that insights gained in the zebrafish system could be applicable to humans and other vertebrates.
These diverse imaging strategies require robust methods for labeling the cellular constituents of WAT in live animals. In this chapter, we present methods for labeling adipocytes in zebrafish using fluorescent lipophilic dyes that specifically incorporate into adipocyte LDs, and for imaging adipose tissues in live zebrafish using stereomicroscopy and confocal microscopy. These methods can be used in combination with transgenic zebrafish expressing fluorescent proteins (FP) in specific WAT cell types to reveal the dynamic cellular interactions underlying WAT formation and function.
III. Materials
Adult zebrafish. Any strain of adult zebrafish can be used for this protocol. Zebrafish lines may be obtained from the Zebrafish International Resource Center (ZIRC, http://zebrafish.org/zirc). All experiments should be performed in accordance with protocols approved by the user’s Institutional Animal Care and Use Committee.
Large nets (Aquatic Eco-Systems, cat. no. AN8).
Zebrafish aquarium (system) water.
Breeding tanks (Laboratory Product Sales, cat. no. T233792).
Plastic tea strainer, 7 cm (Comet Plastics, cat. no. strainer 0).
Scienceware pipette pump (Fisher Scientific, cat. no. 13-683C).
Wide-bore Pasteur pipettes (Kimble Chase, cat. no. 63A53WT).
100 × 15 mm Petri dishes (Fisher Scientific, cat. no. 0875712).
Methylene blue stock solution (0.01%) (Sigma, cat. no. M9140). Dissolve 50 mg methylene blue in 500 mL distilled water (dH2O). Dilute this stock solution 1:200 in fresh zebrafish aquarium system water to prevent growth of bacteria and mold during embryonic development.
dH2O.
Fluorescence stereomicroscope (e.g., Leica MZ 16F or M205 FA) equipped with an eyepiece graticule and the following Leica emission filter sets: GFP2 (510LP) for the green fluorescent lipophilic dyes (i.e., BODIPY 505/515, 500/510, NBT-Cholesterol, BODIPY FL C5, and the yellow-orange dye, Nile Red); YFP (535-630BP) for the yellow, orange, and orange-red dyes (i.e., BODIPY 530/550, 558/ 568, and Cholesteryl BODIPY 576/589); and Texas Red (610LP) for HCS LipidTOX Red/Deep Red. See Table I for a full description of fluorescent lipophilic dyes. Equivalent fluorescence stereomicroscopes and filter sets can be used from alternative manufacturers.
Air incubator set at 28.5 °C (Powers Scientific Inc., cat. no. IS33SD).
Two-liter fish tanks (Marine Biotech, cat. no. 10198-00A).
Assorted mesh drainage plugs for 2-L fish tanks (Marine Biotech, 425 mm, cat. no. 10222-01A; 600 mm, cat. no. 10222-02A; 1600 mm, cat. no. 10222-03A; 4000 mm, cat. no. 10222-04A).
Brine shrimp (Artemia franciscana) cysts (Utah strain; Aquafauna Bio-Marine Inc., cat. no. ABMGSL-TIN90). Detailed brine shrimp hatchery methods are included in The Zebrafish Book (Westerfield, 1995). Briefly, 80 mL of brine shrimp cysts is momentarily immersed in bleach before rinsing with system water. After rinsing, the cysts are added to 12 L of system water supplemented with 10 g sodium bicarbonate and 155 g sodium chloride. The cysts are aerated vigorously for 24 h, under continuous light. The hatched brine shrimp are filtered through a 105-mm mesh sieve and diluted in 2 L of system water. Although brine shrimp hatching rates can vary, we typically find this procedure generates rv4 × 107 brine shrimp per 24 h.
Sodium bicarbonate (Aquatic Eco-Systems, cat. no. SC12).
Sodium chloride (Fisher Scientific, cat. no. S96860).
Brine shrimp net (Aquatic Eco-Systems, cat. no. BSN1).
Fifteen-milliliter conical tubes (polystyrene or polypropylene) (Becton Dickinson, cat. no. 35-2099).
Plastic transfer pipettes (Samco Scientific, cat. no. 225).
Fluorescent lipophilic dyes (see Table I for details). Chloroform:methanol (MeOH) (2:1) is typically used as a solvent when making stock solutions of lipophilic dyes. However, chloroform:MeOH cannot be added directly to system water containing zebrafish. Therefore, the desired quantity of chloroform:MeOH stock solution containing lipophilic dye is air dried in a 1.6-mL microcentrifuge tube for rv10 min before being resuspended in 10 mL of 100% ethanol (EtOH). The EtOH/dye solution can be added directly to system water containing zebrafish. Alternatively, dimethyl sulfoxide (DMSO) or acetone can be used as solvents when making stock solutions, and can be added directly to system water containing zebrafish. However, use of DMSO and acetone as solvents is not advised as the resulting stock solution is less stable over long periods of storage. Stock solutions of fluorescent lipophilic dyes are kept in the dark at −20 °C.
Chloroform (Fisher Scientific, cat. no. BP1145-1).
MeOH (VWR, cat. no. BDH1135-4LP).
EtOH (Decon Labs, Inc., cat. no. 2716).
DMSO (Fisher Scientific, cat. no. D128-1).
Acetone (Mallinckrodt Chemicals, cat. no. 2440-02).
Ethyl 3-aminobenzoate methanesulfonate salt (tricaine or MS222) stock solution (24x) (Sigma, cat. no. A5040-110G). Combine 0.8 g of tricaine, 4.2 mL of 1 M Tris (pH 9.0), and 195.8 mL of dH2O. Adjust pH to between 7.0 and 7.5, and store at 4 °C. Anesthetizing concentration is 1x, and euthanizing concentration is 5x.
Methyl cellulose (4%) (Sigma, cat. no. MO387-100G). Dissolve 4 g methyl cellulose in 100 mL dH2O. Make 1-mL aliquots in 1.6-mL microcentrifuge tubes and freeze at −20 °C.
Low-melting-point (LMP) agarose (1%) (Fisher Scientific, cat. no. BP165-25). Dissolve 1 g LMP agarose in 100 mL of 1× phosphate buffered saline (PBS). Make 1-mL aliquots in 1.6-mL microcentrifuge tubes and freeze at −20 °C.
1.6-mL microcentrifuge tubes (Genesee Scientific, cat. no. 22-282A).
Heat block set at 65 and 42 °C (Denville Scientific, Inc., D1100).
Twenty-four-well plastic culture plates (Greiner Bio-One, cat. no. 662160).
Metal dissection probe (Fine Science Tools, cat. no. 10140-01).
Thirty-five-millimeter Petri dish with glass coverslip as base (MatTek Corp., cat. no. P35G-1.5-10-C).
Thirty-five-millimeter Petri dish (Becton Dickinson, cat. no. 351008).
Epinephrine stock solution (100 mg/mL) (Sigma, cat. no. E4375-5G). Dissolve 1 g epinephrine powder in 10 mL of dH2O. Store at 4 °C in the dark. Add 3 mL of stock solution to 30 mL system water (10 mg/mL final concentration) containing fish for 5 min to contract melanosomes (Rawls and Johnson, 2003).
Paraformaldehyde (PFA) stock solution (4%) (Acros, cat. no. 30525-89-4). Dissolve 4 g PFA powder in 99 mL prewarmed 1× PBS. Once cooled, add 1 mL of DMSO.
PBS stock solution (25x). Dissolve 200 g sodium chloride, 5 g potassium chloride, 36 g sodium dihydrogen phosphate, and 6 g monopotassium phosphate in 1 L dH2O and autoclave.
Tween-20 (Fisher Scientific, cat. no. BP337-500).
Laser scanning confocal microscope (e.g., Zeiss LSM 510), equipped with argon 488 nm, HeNe1 543 nm, and HeNe2 633 nm excitation lasers. Other platforms, such as spinning disk confocal and multiphoton fluorescence microscopes, can also be used.
Table I.
Lipophilic fluorescent dyes for staining lipid droplets in zebrafish
| Color | Name | IUPAC name | Cat. no.a | Solvent | Stock concentration |
Working concentrationb |
Absorption/emission maxima (nm)c |
Additional notesd |
|---|---|---|---|---|---|---|---|---|
| Green | BODIPY 500/510 |
4,4-Difluoro-5-methyl-4-bora- 3a,4a-diaza-s-indacene-3- dodecanoic acid |
D3823 | Chloroform: MeOH |
1 mg/mL | 0.5 μg/mL (2,000×) |
500/510 (use filter sets appropriate for Alexa Fluor 488) |
Very bright LD stain, weak gall bladder stain |
| Green | BODIPY 505/515 |
4,4-Difluoro-1,3,5,7- tetramethyl-4-bora-3a,4a- diaza-s-indacene |
D3921 | DMSO | 1 mg/mL | 1 μg/mL (1,000×) |
505/515 (use filter sets appropriate for Alexa Fluor 488) |
Very bright LD stain, bright gall bladder stain, bright intestine stain, stains blood plasma, significant fluorescence emission in red channel after 488 nm laser excitation |
| Green | BODIPY FL C5 |
4,4-Difluoro-5,7-dimethyl-4- bora-3a,4a-diaza-s- indacene-3-pentanoic acid |
D3834 | Chloroform: MeOH |
0.5 μg/mL | 0.25 ng/mL (2,000×) |
503/512 (use filter sets appropriate for Alexa Fluor 488) |
Weak LD stain, bright gall gladder stain, bright intestine stain |
| Green | NBT- Cholesterol |
22-(N-(7-Nitrobenz-2-oxa-1,3- diazol-4-yl)amino)-23,24- bisnor-5-cholen-3β-ol |
N1148 | Chloroform: MeOH |
5 μg/mL | 0.5 ng/mL (10,000×) |
Absorption/emission maxima are dependent on solvent and environment. However, we use filter sets appropriate for Alexa Fluor 488 |
Bright LD stain, weak gall bladder stain |
| Yellow | BODIPY 530/550 |
4,4-Difluoro-5,7-diphenyl-4- bora-3a,4a-diaza-s- indacene-3-dodecanoic acid |
D3832 | Chloroform: MeOH |
1 mg/mL | 10 μg/mL (100×) |
530/550 (use filter sets appropriate for Alexa Fluor 532) |
Very weak LD stain, high non-LD background |
| Yellow- orange |
Nile Red | 9-Diethylamino-5H-benzo [alpha]phenoxazine-5-one |
N1142 | Acetone | 1.25 mg/mL | 0.5 μg/mL | 510/580 (excite with argon 514 nm laser, collect emission with a longpass 530 filter)e |
Very bright LD stain, red phospholipid background stain |
| Orange | BODIPY 558/568 |
4,4-Difluoro-5-(2-thienyl)-4- bora-3a,4a-diaza-s- indacene-3-dodecanoic acid |
D3835 | Chloroform: MeOH |
1 mg/mL | 2 μg/mL (500×) |
558/568 (use filter sets appropriate for Alexa Fluor 546) |
Very bright LD stain, very bright non-LD background |
| Red-orange | Cholesteryl BODIPY 576/ 589 |
Cholesteryl 4,4-difluoro-5- (2-pyrrolyl)-4-bora-3a, 4a-diaza-s-indacene-3- undecanoate |
C12681 | Chloroform: MeOH |
1 mg/mL | 10 μg/mL (100×) |
576/589 (use filter sets appropriate for Alexa Fluor 568) |
Weak LD stain |
| Red | HCS LipidTOX RED |
– | H34476 | DMSO | – | (5000×) | 577/609 (use filter sets appropriate for Alexa Fluor 594 or Texas Red) |
Very bright LD stain, bright gall bladder stain, stains blood plasma |
| Far-red | HCS LipidTOX Deep Red |
– | H34477 | DMSO | – | (5,000×) | 637/655 (use filter sets appropriate for Alexa Fluor 647 or Cy5 dye) |
Very bright LD stain, bright gall bladder stain, stains blood plasma |
All catalog numbers (cat. no.) are from Invitrogen.
Dilution of stock solution to achieve working concentration is in parentheses.
Alexa Fluor dyes are from Invitrogen.
Staining patterns correspond to wild-type fish raised under the husbandry protocols described here. These staining patterns could potentially be altered as a function of fish genotype, dietary status, and other exposures.
Nile Red bound to phospholipid bilayer has absorption/emission maxima of ~550/640 nm.
IV. Methods
A. Obtaining Zebrafish Embryos
-
Day 1, duration ~60 min. The day before embryos are required, use a large net to place suitable breeding pairs of adult zebrafish in specialized breeding tanks filled with fresh system water. Adults remain in breeding tanks overnight and typically spawn once the aquarium lights turn on the following morning. Be sure to label each breeding tank with the genotype and stock number(s) of the respective breeding pair.
Optional: Specific fluorescent lipid probes can be used in conjunction with transgenic zebrafish lines expressing FP to facilitate cell localization studies (see Table I for excitation/emission properties of lipid probes). If required, obtain transgenic zebrafish from ZIRC (see Section III).
-
Day 2, duration ~60 min. To collect fertilized embryos, remove adult zebrafish to a different tank. Maintain labeling system of adult fish to track parentage of embryos. Collect embryos by pouring through a tea strainer. Clean embryos by rinsing multiple times in fresh system water, and dispense groups of 20–40 embryos into each 100-mm Petri dish filled with 30 mL of fresh system water. View embryos on a light stereomicroscope and remove unfertilized embryos using a wide-bore Pasteur pipette and pipette pump. Place fertilized embryos within an air incubator at 28.5 °C.
Optional: If natural breeding is unsuccessful, embryos can alternatively be generated by in vitro fertilization (or “squeezing”) using established protocols (Westerfield, 1995).
Optional: Methylene blue stock solution can be added to system water (0.01% final concentration) to inhibit fungal and bacterial growth during zebrafish development.
Optional: If using FP-expressing transgenic zebrafish, use a fluorescence stereomicroscope to screen for fluorescent embryos at a suitable developmental stage when FP expression is known to be observed. This procedure is preferentially done during embryonic stages to minimize rearing and feeding of unnecessary nontransgenic fish.
B. Rearing Zebrafish to Postembryonic Stages in Preparation for Fluorescent Lipid Staining
Days 2–6; duration 15–30 min/day. Continue to raise embryos/larvae at 28.5 °C in 100-mm Petri dishes until 5 days postfertilization (dpf). During these first 5 days of development, check daily for, and remove, dead embryos using a widebore Pasteur pipette and pipette pump (dead embryos are typically white in appearance). In addition, replace ~50% of water with fresh system water once every 2–3 days using a plastic transfer pipette. Zebrafish do not need exogenous nutrition until 5 dpf; therefore, do not feed during this period.
Day 6; static tank stage; duration ~15 min. At 5 dpf, transfer ~40 larvae to 1 L of fresh system water contained within a clean 2-L tank and fitted with μ25-mm mesh drainage plugs.
Day 6 onwards; duration ~15 min/day. From 5 dpf, and once larvae are transferred to 2-L tanks, feeding can commence. It is routine procedure for laboratory zebrafish facilities to feed young larvae (typically 5–10 dpf) either a Paramecium multimicronucleatum diet (http://zfin.org/zf_info/zfbook/chapt3/3.3.html) or commercial powdered food (reviewed in Lawrence, 2007). Although these diets can support adipogenesis in developing zebrafish, we observe increased larval survival rate and improved water quality with a diet consisting exclusively of live brine shrimp from 5 dpf (see Section IV). We feed each 2-L tank containing 20–40 fish with 0.5 mL of ~1000 brine shrimp/mL concentration once per day. Dead brine shrimp and debris collecting at the bottom of the tank should be removed every few days with a plastic transfer pipette.
Day 9; duration ~15 min. At 8 dpf, add 1 L of fresh system water to the existing 1 L containing larvae in each 2-L tank.
Day 12; low-flow tank stage; duration ~15 min/day. At 12 dpf, the 2-L tanks are placed on slow running system water with fine mesh (600 μm) drainage plugs. It is important to clean the drainage plugs every day to ensure unobstructed water flow. Continue feeding each tank with 0.5 mL of ~1000 brine shrimp/mL concentration once per day.
Day 16; duration 15 min/day. Increase strength of water flow from 15 dpf. It is important to continue swapping mesh drainage plugs for ones with a larger mesh (typically 1600 and 4000 mm can be used) concomitant with growth of larvae/ juveniles. Continue feeding each tank with 0.5 mL of ~1000 brine shrimp/mL concentration once per day.
Zebrafish that are fed using this protocol begin storing neutral lipid in adipocytes from ~10 to 15 dpf (Fig. 1). However, once independent feeding initiates (5 dpf), subsequent larval and juvenile growth rates vary considerably. Physical measurements such as standard length (SL; defined as distance from snout tip to caudal peduncle) provide more accurate metrics of postembryonic zebrafish development and growth (Parichy et al., 2009). Adipose tissue development in wild-type zebrafish is robustly correlated with SL (Imrie and Sadler, 2010; J. Minchin, unpublished). SL can be measured directly on live larvae/juveniles using a stereomicroscope equipped with an eyepiece graticule. Alternatively, the specimen can be imaged on a stereomicroscope using known magnification, and subsequently measured using suitable image analysis software (e.g., ImageJ or Adobe Photoshop).
Day 16 onwards; duration 5 min. To undertake a lipid stain at a selected time point, larvae/juveniles must be transferred to a smaller vessel such as a 15-mL conical tube. Larval and juvenile zebrafish are very delicate; therefore, during this step, it is essential to handle them with care. To transfer larvae/juveniles from 2-L tanks to a suitable vessel, fill a clean 2-L tank with system water. Then place a brine shrimp net into the freshly prepared tank of system water, and gently pour your larvae/ juvenile sample into the net partially submerged within the freshly prepared tank. Pour carefully so larvae are not buffeted by water, as vigorous pouring will damage the fish. Individually remove each larvae/juvenile from the net while partially submerged in the tank of system water using a plastic transfer pipette, and place in a 15-mL conical tube filled with an appropriate volume of fresh system water.
C. Staining Live Zebrafish Larvae/Juveniles with Fluorescent Lipophilic Dyes
A distinguishing feature of white adipocytes in fish as well as mammals is the presence of large cytoplasmic neutral LDs that consist largely of TG (Tobias and Farese, 2009). As described below, these characteristic adipocyte organelles can be unambiguously labeled in live zebrafish using any of several commercially available fluorescent lipophilic probes (see Table I).
Duration 5 min. Wash unanesthetized larvae/juveniles in fresh system water three times at room temperature. This is accomplished by removing 80% of the system water from the 15-mL conical tube with a transfer pipette before adding fresh system water. This step removes debris included with larvae. After the final wash, adjust volume of each 15-mL conical tube containing zebrafish to 5 mL with fresh system water.
Duration 5 min. Add lipid probe, at appropriate concentration (see Table I), to 5 mL fresh system water containing larvae/juveniles. Lipid probe dissolved in DMSO can be added directly to system water. However, lipid probe dissolved in chloroform:MeOH must first be dried by chloroform:MeOH evaporation, and then resuspended in 100% EtOH (see Section IV). It is important to minimize the volume of 100% EtOH added to system water containing specimen; therefore, we typically add 10 mL to 5 mL system water containing fish.
-
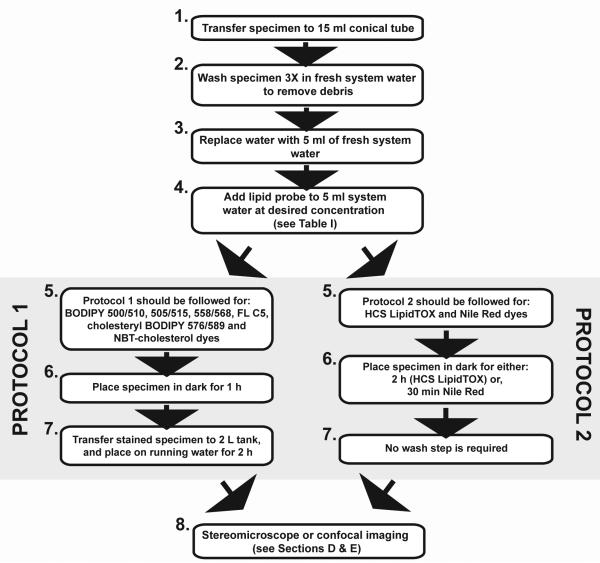
The experimental procedure for zebrafish LD staining varies depending on the lipid probe used. Based on fluorescent lipophilic dyes that we have used for staining LDs in zebrafish (see Table I), we have devised two main protocols (see Fig. 4 for details). Protocol 1 should be followed for BODIPY dyes (both nonpolar and fatty acid/cholesteryl conjugates) as these require extensive wash steps to reduce background staining. Protocol 2 should be followed for Nile Red and HCS LipidTOX stains as these dyes do not require a wash step. Subsequent to the lipid staining protocol, all specimens are imaged using standard techniques regardless of protocol followed (see Sections D and E).
Optional: It is common for ingested food and bile within the intestine to autofluoresce. In order to reduce these unwanted fluorescence signals, fish can be starved overnight to “clear” the intestine of food before beginning the lipid stain.
Fig. 4.
Flow diagram depicting protocols for staining zebrafish with fluorescent lipophilic dyes. Protocol 1 includes wash steps and should be used for BODIPY dyes. Protocol 2 does not contain wash steps and should be used for HCS LipidTOX and Nile Red dyes. See Table I for information on lipophilic dyes.
D. In vivo Imaging of Neutral Lipid on a Fluorescence Stereomicroscope
Imaging fluorescently labeled neutral lipid on a stereomicroscope allows for relatively high-throughput analysis of whole-animal fat deposition (Figs. 1 and 2). Using the following imaging procedure, it is possible to acquire multiple images of distinct adipose depots in an individual fish within 5 min. Therefore, stereoscopic imaging of fluorescently stained zebrafish adipose tissues is suitably quick for use as a viable phenotyping assay during chemical or genetic screens for factors that influence lipid storage in adipose tissues.
To image and measure stained zebrafish, 1× tricaine is used as a standard anesthetic in zebrafish research. Embryonic and larval zebrafish are particularly amenable to dosing with 1× tricaine, and recovery after 72 h of anesthesia is commonplace. However, older larval (larger than ~7 mm SL), juvenile, and adult zebrafish have more difficulty recovering after tricaine anesthesia. Presumably this is due to an increasing requirement for gill respiration as the zebrafish grows due to increased body size and epidermal thickness, combined with the absence of active gill respiration while under tricaine anesthesia. The consequence of increased sensitivity to tricaine anesthesia is higher rates of death. We try to minimize exposure to tricaine by imaging as quickly as possible, and we increase animal number to compensate for any death that might occur.
Fill a 100-mm Petri dish with 30 mL of fresh system water and add 1.25 mL of 24× tricaine.
Place a 3 × 3 mm drop of 4% methyl cellulose at the center of a 100-mm Petri dish lid. Cover the 4% methylcellulose drop with system water containing 1× tricaine.
Transfer stained unanesthetized fish from 15-mL conical tube to fish water containing tricaine. Allow fish to sit in tricaine-containing system water for ~5 min, or until fish has symptoms of being under anesthesia (i.e., belly up swimming, reduced gill ventilation). Do not keep fish in tricaine for longer than required as this may impede recovery from anesthesia.
Immediately transfer anesthetized fish to Petri dish lid containing 4% methyl cellulose droplet. Gently position tail of larvae/juvenile in the methyl cellulose and orientate appropriately using a metal dissection probe. Do not completely embed specimen in 4% methyl cellulose as this increases likelihood of damaging fish when subsequently releasing it. In zebrafish, neutral lipid within adipocytes is first deposited in association with the pancreas at ~5 mm SL, which is asymmetrically located on the right-hand side of the visceral cavity of larvae/juveniles (Fig. 1) (Flynn et al., 2009). Therefore, it is usually important to orient the specimen so the right-hand side is observable by the microscope objective.
-
Once the fish is positioned correctly, measure SL using an eyepiece graticule.
Optional: The zebrafish pigment pattern can obscure imaging of adipose depots. Before imaging, the confounding effects of the zebrafish pigment pattern can by reduced by treating animals with 10 mg/mL epinephrine for 5 min to contract the melanosomes into the center of the cell (Rawls and Johnson, 2003). This step is not optimal as epinephrine is known to stimulate lipolysis in adipocytes (Fain and Garcija-Sainz, 1983); however, our imaging procedure is sufficiently brief to prevent any salient effect on fat storage. Furthermore, for imaging visceral/pancreatic adipose tissue, melanin is usually not a problem during larval and early juvenile stages (i.e., through ~8 mm SL). Alternative solutions include using zebrafish mutants that fail to develop pigment cells, but the potential effects of these mutations on adipose development and function have not been explored. Although inhibition of melanin synthesis during zebrafish development can be achieved by treatment with phenylthiourea (PTU), this is not practical for fish kept in tanks maintained on flowing recirculating water.
E. In vivo Imaging of Neutral Lipid on a Confocal Microscope
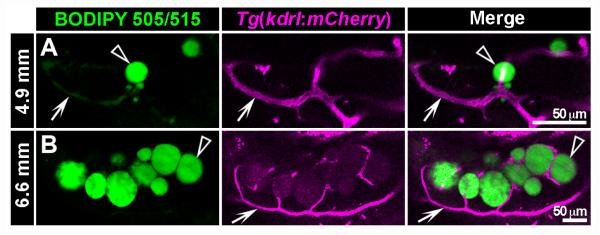
Although considerably more time consuming than stereomicroscopic analysis, imaging fluorescently stained neutral lipid by confocal microscopy provides much greater resolution. Furthermore, if FP transgenic lines are used, exact colocalization of adipocyte LDs with fluorescent cell types of interest is achievable (Fig. 5). The following protocol takes ~35 min per zebrafish imaged (based on a 15-min Z-stack); however, exact timing will vary depending on length of confocal scan taken:
Fig. 5.
Confocal imaging of live zebrafish stained with BODIPY 505/515 fluorescent lipophilic dye reveals close interplay between lipid droplets and the vascular system in visceral adipose tissue. Tg(kdrl: mCherry) transgenic zebrafish express mCherry fluorescent protein specifically in endothelial cells of the vasculature (Jin et al., 2005). Live Tg(kdrl:mCherry) transgenic zebrafish were stained with BODIPY 505/515 according to protocol 1 (see Fig. 4) at 4.9 mm SL (rv14 dpf) and 6.6 mm SL (rv18 dpf). Stained zebrafish were imaged on a Zeiss LSM510 confocal microscope using filter settings for mCherry and BODIPY 505/515 (see Table I). Individual optical sections from a Z-stack are presented in this figure. At 4.9 mm, LDs (arrowheads) are small and loosely associated with vasculature (arrows, A). At 6.6 mm, LDs (arrowheads) are larger and interspersed with mCherry+ vasculature (arrows, B). Note: BODIPY 505/515 has weak fluorescence in the red channel (B), and also labels lipid in vasculature (A). Scale bars: 50 μm.
Anesthetize fish in tricaine (see Section D).
Measure SL of each fish to be imaged (see Section B7).
-
Thaw 1% LMP agarose aliquots by placing 1-mL aliquot at 65 °C until completely melted. Cool LMP agarose to 42 °C using a heat block or water bath for 1 h until needed.
Upright confocal microscopes. It is preferential to use a water dipping objective. First, deposit a 3 × 3 mm droplet of 4% methyl cellulose in the center of a 35-mm Petri dish. Anesthetize larvae/juveniles in 1× tricaine (see Section D), transfer to methyl cellulose droplet, and roughly orientate. Remove excess system water with plastic transfer pipette and replace with 1% LMP agarose cooled to 42 °C. Using a stereoscope, quickly orientate sample into correct position with a metal dissecting probe. Continuously observe specimen as LMP solidifies to ensure correct positioning. It is important for specimen to be only lightly covered by LMP agarose. If too much LMP is added, it will prevent focusing the objective to regions deep within zebrafish adipose tissue. Once the LMP agarose has solidified, add ~2 mL of 1× tricaine diluted in system water to the Petri dish in order to keep fish anesthetized.
Inverted confocal microscopes. It is common to use either air or immersion objectives. We mount the specimen in a 35-mm Petri dish with a glass coverslip as base. When using this method, it is important to orientate specimen as close as possible to the coverslip. Anesthetize zebrafish in 1× tricaine (see Section D) and place the specimen on the glass coverslip within the 35-mm Petri dish. Remove excess system water carried over from transfer of specimen. Quickly add 1% LMP agarose cooled to 42 °C to cover the fish, and orient into correct position using a metal dissection probe. Continuously observe orientation of the fish as LMP solidifies to ensure correct positioning. Once LMP agarose is solidified, add ~2 mL of 1% tricaine diluted in system water to the Petri dish.
Table I contains excitation and emission information for each fluorescent lipid stain used to visualize neutral lipid. Due to the size of late larvae/juveniles, best results are obtained using 10× or 20× objectives that have large working distances but a numerical aperture (NA) of ~1.
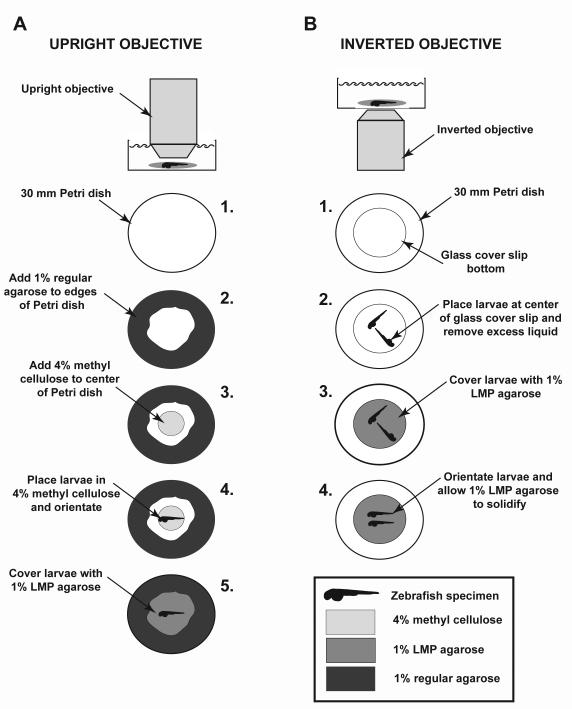
Note: For confocal imaging, there are multiple methods for stably mounting zebrafish in 1% LMP agarose during imaging. Depending on microscope design (inverted or upright) and the type of objective to be used (air, immersion, or dipping), we have found the following methods for mounting to be most successful (see Fig. 6):
Fig. 6.
Schematic illustrating stable mounting procedures for regular and inverted microscopes. (A) When imaging using an upright objective, a 60-mm Petri dish is used (step 1), and 1% regular agarose is used to fill in around the edges of the Petri dish (step 2). Four percent methyl cellulose is then placed in center of Petri dish (step 3), before the anesthetized animal is placed in 4% methyl cellulose and oriented (step 4). Once specimen is correctly oriented, it is covered with 1% LMP agarose and allowed to solidify (step 5). (B) For imaging using an inverted objective, a 60-mm Petri dish with a fitted glass coverslip as a base is used (step 1). Anesthetized zebrafish are place onto the glass coverslip base (step 2) and excess liquid is removed. The specimen is then covered with 1% LMP agarose (step 3) and quickly oriented while agarose is solidifying (step 4). For both methods, 2 mL of system water, containing 1× tricaine, is added to specimen. When undertaking imaging using the upright objective method, be sure to use a water dipping objective.
F. Recovery of Sample After Imaging of Fluorescent Neutral Lipid
Zebrafish are amenable to longitudinal analyses of fat storage within individual fish (Fig. 2) (Flynn et al., 2009). Therefore, once imaging has been completed, it is often necessary to recover larvae and allow development to proceed. Under a dissecting microscope, gently cut LMP agarose away from tail of larvae with a metal dissection probe. We find it is easier to recover fish from 1% LMP agarose if it is first immersed in fresh system water. Once tail is free, it may be possible to release larvae by gently squeezing clean system water over the specimen with a plastic transfer pipette. If necessary, carefully remove more of LMP agarose from anterior regions until larvae are released.
Once larvae are free and recovered from anesthesia, house each fish individually in a well of a 24-well plate filled with 1 mL system water to keep record of subsequent larvae/juvenile growth during longitudinal analysis. It is necessary to change 80% of system water within each well daily and to feed ~30 live brine shrimp per well per day.
G. Methods for Analyzing Fixed Zebrafish Adipose Tissues
Zebrafish adipose tissues can be analyzed in fixed specimens using the HCS LipidTOX series of fluorescent stains (J. Minchin, unpublished). Furthermore, HCS LipidTOX can be utilized after histological sectioning of zebrafish (Stoletov et al., 2009), which indicates that equivalent analytical techniques undertaken in mammalian adipose tissue research can be attempted in zebrafish. In vivo imaging of zebrafish adipocyte LDs can also be coupled with subsequent in situ hybridization analysis to assess gene expression in adipocytes and other WAT cell types (Fig. 3) (Flynn et al., 2009). Moreover, analysis of zebrafish adipose tissues within fixed specimens permits testing of the extensive range of antibodies used in mammalian adipose tissue research (Ibabe et al., 2005). Thus, when used in conjunction with in vivo observation of cell dynamics, analysis of fixed zebrafish adipose tissues offers several useful opportunities:
Zebrafish larvae/juveniles of an appropriate SL are transferred to a suitable vessel, such as a 15-mL conical tube. Specimen is washed 3× in fresh system water to remove any debris. System water is removed and freshly prepared 4% PFA/1% DMSO in PBS is added to the conical tube. Larvae/juveniles are fixed, while gently shaking, overnight at 4 °C.
Prior to staining, fixed specimen is washed 5× in 1× PBS/0.1% Tween-20 at room temperature.
A 200× dilution of HCS LipidTOX (in 1× PBS/0.1% Tween-20) is added to the specimen and incubated, in the dark, at room temperature for 2 h.
Specimen can be imaged immediately using techniques described in Sections D and E. No wash step is required.
In addition to the fluorescent HCS LipidTOX staining of fixed zebrafish LDs described above, numerous additional methods of analyzing fixed zebrafish WAT can be employed. These methods include, but are not limited to, in situ hybridization on whole (Flynn et al., 2009) and sectioned zebrafish (Imrie and Sadler, 2010), histological analysis of adipose tissues by hemotoxylin and eosin staining of paraffin sections (Imrie and Sadler, 2010; Song and Cone, 2007), immunohistochemical analysis (Ibabe et al., 2005), extraction and analysis of lipid from zebrafish tissues (Flynn et al., 2009; Song and Cone, 2007), and oil red O (ORO) staining of lipid within adipose tissues in whole (Flynn et al., 2009; Li et al., 2010) and sectioned zebrafish (Imrie and Sadler, 2010). Although ORO staining of LDs within adipocytes is usually unambiguous and can be confidently identified (Flynn et al., 2009; Imrie and Sadler, 2010), it should be noted that ORO readily stains nonadipogenic cells (Kinkel et al., 2004) and should thus be used with caution. A detailed description of these methods is beyond the scope of this chapter. However, coupled with fluorescent analysis of live and fixed zebrafish, these methods complement available tools for investigating zebrafish adipose tissues.
V. Summary
Investigation of WATs has only recently been initiated in the zebrafish, and consequently we have a relatively limited knowledge of adipose tissue development and physiology in this important vertebrate model system. However, the amenability of the zebrafish to high-resolution in vivo imaging presents exciting opportunities to address longstanding questions about WAT formation and function that have been difficult to resolve using available mammalian models. Moreover, systematic genetic and chemical tests in the zebrafish model could be used to identify novel factors regulating distinct aspects of adipose tissue biology.
Here we have provided detailed methods for in vivo imaging of zebrafish WATs in order to support the use of the zebrafish as a model for adipose tissue research. We anticipate that the fluorescence stereomicroscopy methods presented here will be useful for rapid phenotypic assessments required for genetic and chemical screens, while the confocal microscopy methods will facilitate high-resolution analysis of cellular and molecular events within adipose tissues. Furthermore, we expect that these methods will also be generally applicable to in vivo imaging of adipose tissues in other fish species. Deployment of these methods in the zebrafish system will be enriched by the use of existing and forthcoming zebrafish lines expressing FP and other transgenes in adipocyte lineages and other cellular constituents of adipose tissues. Use of zebrafish as a model for adipose tissue biology will also be enhanced by identification of genetic alterations, dietary and environmental manipulations, and chemicals that modify zebrafish adipose tissue formation and function.
Acknowledgments
We are grateful to Dru Imrie and Kirsten Sadler for sharing data prior to publication, and to Edward Flynn for valuable technical support. This work was supported by NIH grant DK056350 to the University of North Carolina at Chapel Hill’s Nutrition Obesity Research Center, as well as NIH grants DK081426 and DK073695, a Pilot Research Project Award from the UNC University Cancer Research Fund, and a Pew Scholars Program in the Biomedical Sciences Award to J.F.R.
References
- Abel ED, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- Ailhaud G, et al. Cellular and molecular aspects of adipose tissue development. Annu. Rev. Nutr. 1992;12:207–233. doi: 10.1146/annurev.nu.12.070192.001231. [DOI] [PubMed] [Google Scholar]
- Albalat A, et al. Insulin regulation of lipoprotein lipase (LPL) activity and expression in gilthead sea bream (Sparus aurata) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007;148:151–159. doi: 10.1016/j.cbpb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Barzilai N, et al. Surgical removal of visceral fat reverses hepatic insulin resistance. Diabetes. 1999;48:94–98. doi: 10.2337/diabetes.48.1.94. [DOI] [PubMed] [Google Scholar]
- Bellardi S, et al. Effect of feeding schedule and feeding rate on size and number of adipocytes in rainbow trout Oncorhynchus mykiss. J. World Aquacult. Soc. 1995;26:80–83. [Google Scholar]
- Carvalho E, et al. Adipose-specific overexpression of GLUT4 reverses insulin resistance and diabetes in mice lacking GLUT4 selectively in muscle. Am. J. Physiol. Endocrinol. Metab. 2005;289:E551–E561. doi: 10.1152/ajpendo.00116.2005. [DOI] [PubMed] [Google Scholar]
- Company R, et al. Growth performance and adiposity in gilthead seabream (Sparus aurata): risks and benefits of the high energy diets. Aquacult. Res. 1999;171:279–292. [Google Scholar]
- Cypess AM, Kahn CR. Brown fat as a therapy for obesity and diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2010;17:143–149. doi: 10.1097/MED.0b013e328337a81f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres JP. The insulin resistance-dyslipidemic syndrome of visceral obesity: effect on patients’ risk. Obes. Res. 1998;6(suppl. 1):8S–17S. doi: 10.1002/j.1550-8528.1998.tb00683.x. [DOI] [PubMed] [Google Scholar]
- Fain JN, Garcija-Sainz JA. Adrenergic regulation of adipocyte metabolism. J. Lipid Res. 1983;24:945–966. [PubMed] [Google Scholar]
- Farese RV, Jr., Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn EJ, 3rd, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) J. Lipid Res. 2009;50(8):1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CS, et al. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation. 2007;116:39–48. doi: 10.1161/CIRCULATIONAHA.106.675355. [DOI] [PubMed] [Google Scholar]
- Gabriely I, et al. Removal of visceral fat prevents insulin resistance and glucose intolerance of aging: an adipokine-mediated process? Diabetes. 2002;51:2951–2958. doi: 10.2337/diabetes.51.10.2951. [DOI] [PubMed] [Google Scholar]
- Gesta S, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, et al. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Hughes J, Criscuolo F. Evolutionary history of the UCP gene family: gene duplication and selection. BMC Evol. Biol. 2008;8:306. doi: 10.1186/1471-2148-8-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibabe A, et al. Expression of peroxisome proliferator-activated receptors in zebrafish (Danio rerio) depending on gender and developmental stage. Histochem. Cell Biol. 2005;123:75–87. doi: 10.1007/s00418-004-0737-2. [DOI] [PubMed] [Google Scholar]
- Imrie D, Sadler K. White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev. Dyn. 2010;239:3013–3023. doi: 10.1002/dvdy.22443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastroch M, et al. Uncoupling protein 1 in fish uncovers an ancient evolutionary history of mammalian nonshivering thermogenesis. Physiol. Genomics. 2005;22:150–156. doi: 10.1152/physiolgenomics.00070.2005. [DOI] [PubMed] [Google Scholar]
- Jin SW, et al. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Jo J, et al. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput. Biol. 2009;5:e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel AD, et al. Oil red-O stains non-adipogenic cells: a precautionary note. Cytotechnology. 2004;46:49–56. doi: 10.1007/s10616-004-3903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissebah AH, Krakower GR. Regional adiposity and morbidity. Physiol Rev. 1994;74:761–811. doi: 10.1152/physrev.1994.74.4.761. [DOI] [PubMed] [Google Scholar]
- Lawrence C. The husbandry of zebrafish (Danio rerio): a review. Aquaculture. 2007;269:1–20. [Google Scholar]
- Li N, et al. Regulation of neural crest cell fate by the retinoic acid and Pparg signalling pathways. Development. 2010;137:389–394. doi: 10.1242/dev.044164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse SA, et al. The obesity paradox and cardiovascular disease. Curr. Hypertens. Rep. 2010;12:120–126. doi: 10.1007/s11906-010-0099-1. [DOI] [PubMed] [Google Scholar]
- Nishimura S, et al. Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels. Diabetes. 2007;56:1517–1526. doi: 10.2337/db06-1749. [DOI] [PubMed] [Google Scholar]
- Nishimura S, et al. In vivo imaging in mice reveals local cell dynamics and inflammation in obese adipose tissue. J. Clin. Invest. 2008;118:710–721. doi: 10.1172/JCI33328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku H, et al. Effects of insulin, triiodothyronine and fat soluble vitamins on adipocyte differentiation and LPL gene expression in the stromal–vascular cells of red sea bream, Pagrus major. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006;144:326–333. doi: 10.1016/j.cbpb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Oku H, Umino T. Molecular characterization of peroxisome proliferator-activated receptors (PPARs) and their gene expression in the differentiating adipocytes of red sea bream Pagrus major. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;151:268–277. doi: 10.1016/j.cbpb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Om AD, et al. The effects of dietary EPA and DHA fortification on lipolysis activity and physiological function in juvenile black sea bream Acanthopagrus schlegeli (Bleeker) Aquacult. Res. 2001;32:255–262. [Google Scholar]
- Parichy DM, et al. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dyn. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado JR, et al. The stromal–vascular fraction of adipose tissue contributes to major differences between subcutaneous and visceral fat depots. Proteomics. 2010;10:3356–3366. doi: 10.1002/pmic.201000350. [DOI] [PubMed] [Google Scholar]
- Poulos SP, et al. The development and endocrine functions of adipose tissue. Mol. Cell. Endocrinol. 2010;323:20–34. doi: 10.1016/j.mce.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Johnson SL. Temporal and molecular separation of the kit receptor tyrosine kinase’s roles in zebrafish melanocyte migration and survival. Dev. Biol. 2003;262:152–161. doi: 10.1016/s0012-1606(03)00386-5. [DOI] [PubMed] [Google Scholar]
- Redinger RN. Fat storage and the biology of energy expenditure. Transl. Res. 2009;154:52–60. doi: 10.1016/j.trsl.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, et al. Identification of white adipocyte progenitor cells In vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Chen J. Application of imaging and other noninvasive techniques in determining adipose tissue mass. Methods Mol. Biol. 2008;456:39–54. doi: 10.1007/978-1-59745-245-8_3. [DOI] [PubMed] [Google Scholar]
- Sheridan MA. Lipid dynamics in fish: aspects of absorption, transportation, deposition and mobilization. Comp. Biochem. Physiol. B. 1988;90:679–690. doi: 10.1016/0305-0491(88)90322-7. [DOI] [PubMed] [Google Scholar]
- Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J. 2007;21:2042–2049. doi: 10.1096/fj.06-7503com. [DOI] [PubMed] [Google Scholar]
- Stoletov K, et al. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ. Res. 2009;104:952–960. doi: 10.1161/CIRCRESAHA.108.189803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias CW, Farese RV., Jr The life of lipid droplets. Biochim Biophys. Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorcevic M, et al. Gene expression profiles in Atlantic salmon adipose-derived stromo-vascular fraction during differentiation into adipocytes. BMC Genomics. 2010;11:39. doi: 10.1186/1471-2164-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umino T, et al. Development of adipose tissue in juvenile red sea bream. Fisheries Sci. 1996;62:520–523. [Google Scholar]
- Vegusdal A, et al. An In vivo method for studying the proliferation and differentiation of Atlantic salmon preadipocytes. Lipids. 2003;38:289–296. doi: 10.1007/s11745-003-1063-3. [DOI] [PubMed] [Google Scholar]
- Vidal H. Gene expression in visceral and subcutaneous adipose tissues. Ann. Med. 2001;33:547–555. doi: 10.3109/07853890108995965. [DOI] [PubMed] [Google Scholar]
- Vohl MC, et al. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes. Res. 2004;12:1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- Wajchenberg BL. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr. Rev. 2000;21:697–738. doi: 10.1210/edrv.21.6.0415. [DOI] [PubMed] [Google Scholar]
- Weil C, et al. Differentially expressed proteins in rainbow trout adipocytes isolated from visceral and subcutaneous tissues. Comp. Biochem. Physiol. D Genomics Proteomics. 2009 doi: 10.1016/j.cbd.2009.05.002. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book – A Guide for the Laboratory Use of Zebrafish (Danio rerio) University of Oregon Press; Eugene, OR: 1995. [Google Scholar]
- Xue Y, et al. Adipose angiogenesis: quantitative methods to study microvessel growth, regression and remodeling In vivo. Nat. Protoc. 2010;5:912–920. doi: 10.1038/nprot.2010.46. [DOI] [PubMed] [Google Scholar]
- Yach D, et al. Epidemiologic and economic consequences of the global epidemics of obesity and diabetes. Nat. Med. 2006;12:62–66. doi: 10.1038/nm0106-62. [DOI] [PubMed] [Google Scholar]