Abstract
Epithelial to mesenchymal transition (EMT) is a key process associated with tumor progression and metastasis. To define molecular features associated with EMT states, we undertook an integrative approach combining mRNA, microRNA, DNA methylation and proteomic profiles of 38 cell populations representative of the genomic heterogeneity in lung adenocarcinoma were integrated with functional profiles consisting of cell invasiveness, adhesion and motility. A subset of cell lines that were readily defined as epithelial or mesenchymal based on their morphology and E-cadherin and vimentin expression elicited distinctive molecular signatures. However, most cell populations displayed intermediate/hybrid states of EMT, with mixed epithelial and mesenchymal characteristics. A dominant proteomic feature of aggressive hybrid cell lines was upregulation of cytoskeletal and actin binding proteins, a signature shared with mesenchymal cell lines. Cytoskeletal reorganization preceded loss of E-cadherin in epithelial cells in which EMT was induced by TGFβ. A set of transcripts corresponding to the mesenchymal protein signature enriched in cytoskeletal proteins was found to be predictive of survival in independent datasets of lung adenocarcinomas. Our findings point to an association between cytoskeletal and actin-binding proteins, a mesenchymal or hybrid EMT phenotype and invasive properties of lung adenocarcinomas.
Keywords: epithelial to mesenchymal transition, lung adenocarcinoma, proteomics, genomics, cancer survival
INTRODUCTION
Epithelial-to-mesenchymal transition (EMT) is a process in embryonic development that allows polarized epithelial cells to convert to loosely organized mesenchymal cells (1). The transition from an epithelial to a mesenchymal phenotype fosters cell movement during gastrulation and later during morphogenetic events such as neural crest ontogeny. Mesenchymal cells reaching the target site engage in a new differentiation program allowing development of diverse tissue types (2–4). The same features of EMT, namely loss of cell adhesion, increased migration and invasion that aid metazoan development provide a likely mechanism for tumor progression with loss of an epithelial phenotype in more aggressive tumors (5–8).
Regulation of EMT is complex and multi-layered, with diverse growth factors, microRNAs, genetic mutations and epigenetic alterations all having been shown to play a role. Transforming growth factor beta (TGFβ), hepatocyte growth factor, Notch, or Wnt can serve as initiating factors of EMT (9–13). Several microRNAs regulate EMT through inhibition of either effector genes or the signaling axis. The miR-200 family and mir-34 inhibit EMT while miR-21 has an opposite effect (14–16). Several studies demonstrated a role for DNA methylation in regulating miR-200 and an altered DNA methylation profile associated with EMT (17–20). Other EMT regulatory mechanisms include zinc-finger transcription factors Snail1, Snail2, Zeb1, Zeb2 and the basic helix-loop-helix family members Twist1 and Twist2 which control expression of downstream genes and cellular features associated with EMT, such as cell adhesion and polarity (21–23). The transient nature of EMT has been considered as aiding distant site metastasis through a reverse mesenchymal-epithelial transition after invading cells have colonized distant sites (8, 24). Recent work by Lu et al. proposed occurrence of a hybrid epithelial-mesenchymal state with a determination between the three phenotypes being regulated by a circuit composed of two interconnected chimeric modules-the miR-34/SNAIL and the miR-200/ZEB mutual-inhibition feedback circuits (25).
We have undertaken a study of lung adenocarcinoma to determine molecular and phenotypic features associated with EMT states and their relevance to survival in early stage disease. Extensive characterization of lung adenocarcinoma cell lines, including protein, mRNA, microRNA, DNA methylation, cell invasiveness, adhesion and motility analysis, revealed the occurrence of intermediate/hybrid phenotypes between epithelial and mesenchymal states. Gene and protein signatures associated with functional characteristics helped to define these hybrid states. Up-regulation of cytoskeletal-related proteins was a common feature between mesenchymal and aggressive hybrid types. A set of transcripts enriched for cytoskeletal and actin binding proteins was found to be predictive of survival in independent lung adenocarcinoma datasets.
MATERIALS AND METHODS
Cell culture
A panel of 38 lung adenocarcinoma cell lines selected to encompass known drivers in lung adenocarcinoma were grown in RPMI 1640 with 10% FBS and 1% penicillin/streptomycin unless otherwise noted. The identity of each cell line was confirmed by DNA fingerprinting via short tandem repeats at the time of mRNA and total protein lysate preparation using the PowerPlex 1.2 kit (Promega). Fingerprinting results were compared with reference fingerprints maintained by the primary source of the cell line. For SILAC labeling of cell lines, cells were grown for 7 passages in RPMI 1640 supplemented with 13C-lysine and 10% dialyzed FBS according to the standard SILAC protocol (26).
Mass spectrometric analysis
Proteomic analysis was performed as previously described (27). Detailed methods for mass spectrometric analysis can be found in the Supplementary Information section.
Protein datasets are available as Supplementary Table S1.
Determination of EMT status for cell lines
A protein ratio of surface localized CDH1 (CDH1_S) and VIM from total cell extracts was calculated from (CDH1n+1)/ (VIMn+1) where n is the number of spectral counts. High CDH1_S/VIM was considered to be a log2-transformed ratio > 0 and low to be log2-transformed ratio < 0. Cell morphology was assessed by plating cells at 25–50% confluence and acquiring phase contrast images on day 1, 2, 3 and 4 after plating. Cells were assessed for individual cell shape (spindle for mesenchymal or cuboid for epithelial) as well cell-cell interaction. Cells were classified as having epithelial morphology if the individual cells were cuboid and cells grouped to form discrete clusters with smooth edges indicative of tight junctions. Epithelial-like morphology lacked complete cuboid morphology or failed to form discrete clusters. Mesenchymal morphology required primarily spindle shape and no cell-cell adhesion. Cells with mesenchymal-like morphology were primarily spindle shaped but demonstrated cell-cell adhesion by forming clusters. Cells with log2-transformed CDH1_S/VIM ratios > 0 and an epithelial morphology were classified as epithelial, while mesenchymal cells had log2-transformed CDH1_S/VIM ratios < 0 and a mesenchymal morphology.
Profiling of mRNA, microRNA and DNA methylation
Gene expression data were obtained using Illumina Human WG-6 v3.0 Expression BeadChips (Illumina) and expression values log2 normalized. MicroRNA profiling was performed using a real-time PCR-based approach using miRCURY LNA Universal RT miRNA PCR (panel I+II) (Exiqon, Inc.). MicroRNA profiling was not available for cell lines H1299 and H1703. Illumina Infinium HumanMethylation27 BeadChips were used for DNA methylation analysis. DNA methylation profiling was not available for cell lines H1385 and H1703. mRNA, DNA methylation and microRNA datasets were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo).
Data analysis
Detailed methods for data analysis can be found in the Supplementary Information section.
Invasion, migration and aggregation assays
Detailed methods for Invasion, migration and aggregation assays can be found in the Supplementary Information section.
Western Blot Analysis
Western blot analysis was performed according to standard procedures using polyvinylidene difluoride membranes and an Enhanced Chemiluminescence system (GE Healthcare). Following antibodies were used for Western blot analysis: ISYNA1 (Sigma Aldrich), FBXO2 (Novus), TCEAL4 (Novus), FKBP65 (BD Biosciences), Vimentin (BD Biosciences), CDH1 (BD Biosciences), and AKAP12 (Abcam). α-tubulin (Sigma) was used as a loading control.
Immunofluorescence analysis
Detailed methods for immunofluorescence analysis and immunohistochemial analysis can be found in the Supplementary Information section.
RESULTS
Characterization of cell lines based on their morphology and CDH1/VIM ratios
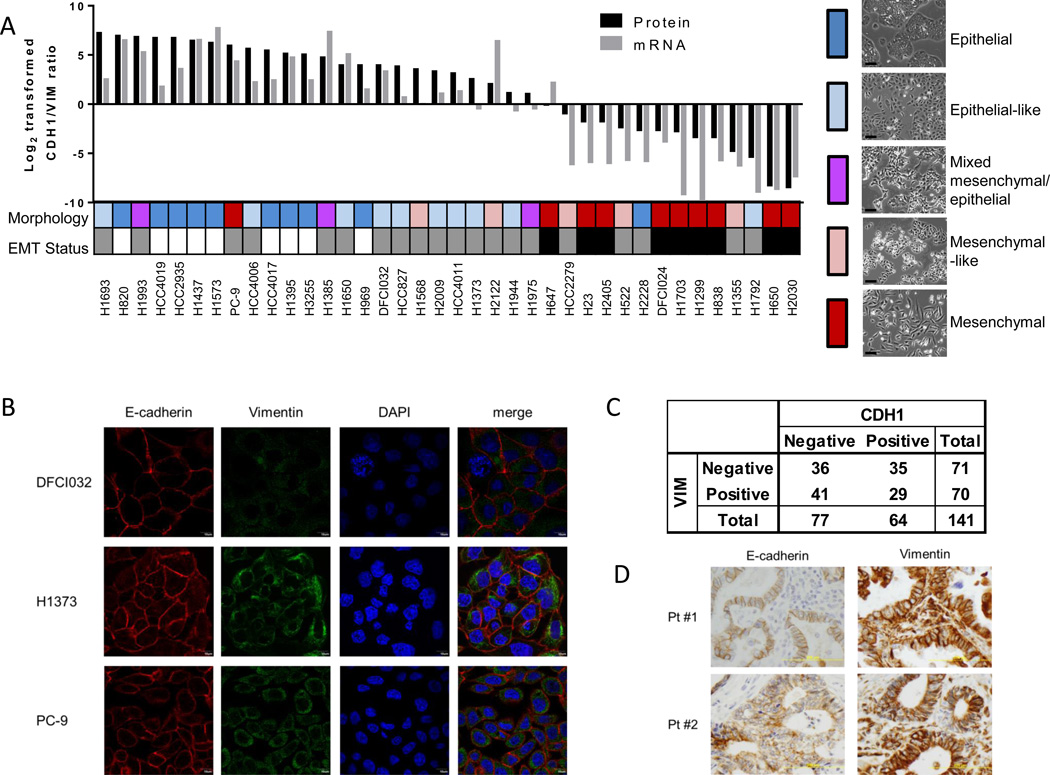
To define molecular features that distinguish epithelial from mesenchymal cells, a panel of 38 lung adenocarcinoma cell lines representative of the genomic diversity of this disease was subjected to proteomic, gene expression, microRNA, and DNA methylation profiling (Supplementary Figure S1A). Changes in CDH1 and Vimentin (VIM) have been considered hallmarks of EMT. Expression of CDH1 on the cell surface and VIM in whole cell lysates was determined based on normalized spectral counts from mass spectrometry data (27). We assessed ratios of cell surface-localized CDH1 (CDH1_S) and VIM from whole cell lysates along with cell morphology, and identified a subset of cell lines with a distinct mesenchymal or epithelial phenotype (28). Nine cell lines with a log2-transformed CDH1_S/VIM ratio > 0 and an epithelial morphology were classified as epithelial, while nine cell lines with a log2-transformed CDH1_S/VIM ratio < 0 and a mesenchymal morphology were classified as mesenchymal (Figure 1A and Supplementary Figure S1B). Log2-transfomed CDH1_S/VIM protein ratio were significantly correlated with CDH1/VIM ratios of mRNA expression (r = 0.8650, P < 0.0001; Spearman correlation). Common somatic gene mutations that occur in lung adenocarcinoma (Kras, TP53, EGFR) were not associated with a distinct EMT phenotype, with the exception of a negative correlation between EGFR mutation and a mesenchymal type as previously reported (29). The remaining cell lines could not be readily classified as epithelial or mesenchymal due to discordance between CDH1_S/VIM ratios and morphology and were investigated further for their hybrid properties. Immunofluorescence analysis of CDH1 and VIM revealed that both CDH1 and VIM were stained in the same cells in hybrid cell lines (Figure 1B). We further investigated CDH1 and VIM protein expression in lung adenocarcinoma tissues. Among 141 lung adenocarcinoma tissues in the tissue microarray, 29 (20.6%) tumors were both CDH1 and VIM positive (Figure1C and 1D), indicative of a hybrid transcriptional program.
Figure 1. Experimental design and classification of NSCLC cell lines.
A. Log2-transformed ratios of spectral counts of CDH1 on cell surface and VIM from whole cell lysates, log2-transformed ratios of CDH1/VIM mRNA, and morphology, with representative images. Cell lines are color-coded to indicate morphology: red = mesenchymal, pink = mesenchymal-like, purple = mixed mesenchymal/epithelial morphology, light blue = epithelial-like, blue = epithelial; and EMT status: white = epithelial, gray = hybrid, black = mesenchymal. Scale = 40 µm. B. Immunofluorescence analysis of E-cadherin and Vimentin in hybrid cell lines. C. CDH1 and VIM expression in lung adenocarcinoma tissue microarray. P values were calculated by chi-square test. D. Representative images of immunohistochemical analysis of E-cadherin and Vimentin.
Identification of distinctive gene and protein signatures for mesenchymal and for epithelial cell lines
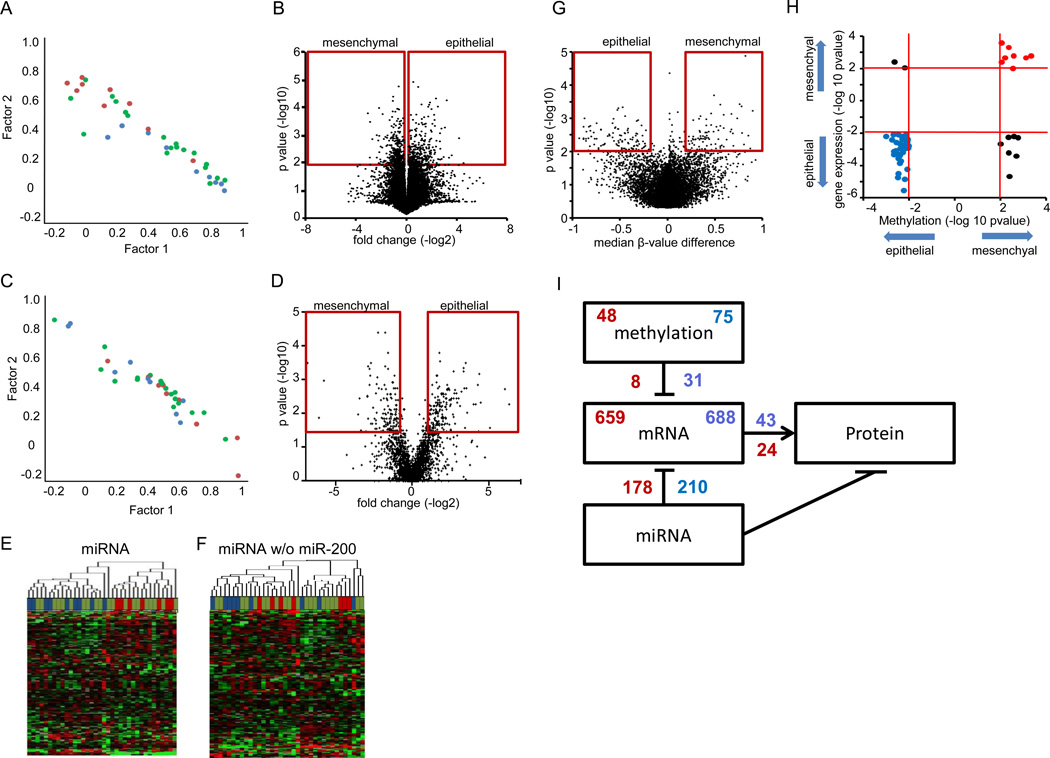
We performed a factor analysis to determine modality of gene expression patterns among the cell lines, which revealed a continuous rather than a modal distribution (Figure 2A). Genes comprised in the discriminating factors included known markers of epithelial or mesenchymal cells, including CDH1, VIM, and EpCAM (data not shown). Comparison of mRNA expression between the nine epithelial and nine mesenchymal cell lines yielded 1,347 genes with a P value < 0.01 (t test), consisting of 659 with higher expression in mesenchymal and 688 in epithelial cell lines (Figure 2B and Supplementary Table S2A). Gene ontology enrichment using DAVID (28) was applied to differentially expressed genes. The epithelial genes were enriched for genes encoding proteins localized to the cell surface, many of which play a role in cell adhesion while mesenchymal genes were enriched for nuclear localized proteins and regulators of transcription (Supplementary Table S2B). Concordant findings were observed when gene ontology for 1,347 epithelial and mesenchymal genes was analyzed with using Gene Set Enrichment Analysis (GSEA) (30) (Supplementary Table S2C).
Figure 2. Molecular characterization of mesenchymal and epithelial cell lines.
A. Factor analysis of gene expression showing distribution of cell lines. B. Volcano plot of the differential gene expression analysis. Red box indicates significance cutoffs. C. Factor analysis of protein expression showing distribution of cell lines. D. Volcano plot of the differential protein expression analysis. Red box indicates significance cutoffs. E. Unsupervised hierarchical clustering of microRNA expression from 36 lung adenocarcinoma cell lines separates the epithelial and mesenchymal cell lines into separate clusters. F. Unsupervised hierarchical clustering of microRNA expression without miR-200 family members. G. Volcano plot of the differential DNA methylation analysis. Red box indicates significance cutoffs. H. Starburst plot integrating differential DNA methylation and gene expression analyses. I. Overlaping regulation of gene and protein expression. For sub figures A, C, E and F, mesenchymal cell lines are indicated by red, epithelial by blue, and hybrid cell lines by green.
Extensive proteomic analysis of cell lysates by LC-MS/MS for all 38 cell lines identified a total of 12,808 distinct proteins with an average of 3,690 proteins identified in each cell line, pointing to substantial heterogeneity between cell lines (Supplementary Table S1 and Supplementary Figure S2A). Factor analysis performed with proteomic data similarly produced a continuum rather than a modal distribution (Figure 2C). Proteins comprised in the discriminating factors included VIM but not CDH1 nor EpCAM (data not shown). Proteomic analysis resulted in 232 proteins expressed more highly in epithelial cell lines and 166 proteins expressed more highly in mesenchymal cell lines with a P value < 0.05 (Mann-Whitney U test) and fold change > 1.5 (Figure 2D and Supplementary Table S2D). The epithelial and mesenchymal mRNA signatures significantly overlapped with corresponding protein signatures, respectively (P = 3.29×10−20, and P = 6.61×10−10, Fisher’s exact test) (Supplementary Figure S2B). Significantly enriched GO categories in the proteomic mesenchymal signature consisted predominantly of cytoskeleton and actin organization (GO: 0005856~cytoskeleton, GO: 0008092~cytoskeletal protein binding, GO: 0003779~actin binding) (Supplementary Table S2E). The enrichment in cytoskeletal and actin related proteins was also prominent by Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com/) (Supplementary Figures S2C and S2D). The GO terms enriched in the protein signature for epithelial cells included those associated with translation and metabolism (Supplementary Table S2E). To confirm functional relevance of cytoskeletal proteins in mesenchymal signature, we performed knockdown experiments of AKAP12, which is associated with actin-cytoskeleton reorganization (31) and identified in both mRNA and protein mesenchymal signatures (Supplementary Tables S2A and S2C). While no obvious change was observed in EMT status, cell invasion was inhibited in H1299 cells by treatment with AKAP12 shRNA (Supplementary Table S2E), indicating the functional relevance of cytoskeletal proteins in mesenchymal signature.
To explore potential regulatory factors for gene and protein expression, microRNA and DNA methylation profiling was performed. Unsupervised hierarchical clustering of microRNA data resulted in two clusters that separated the epithelial and mesenchymal cell lines and significantly differed in CDH1_S/VIM expression (P = 5.6×10−6, t test) (Figure 2E). Separation of the cell lines into two clusters was principally due to expression of the miR-200 family, as removal of miR-200 family members from the dataset resulted in lack of clustering into two clusters (Figure 2F). Restricting comparison of the microRNA data to the epithelial and mesenchymal cell lines resulted in 31 differentially expressed microRNAs at P < 0.01 (t test), with 10 expressed at lower levels in mesenchymal cell lines and 21 expressed at lower levels in epithelial cell lines (Supplementary Table S2F). Of the 10 miRNAs with lower expression in the mesenchymal cell lines, six had predicted binding sites in the mesenchymal gene signature (mir-200a, 200b, 200c, 429, 135b and148a) using the prediction algorithms miRanda and Targetscan (data not shown). Of the 21 miRNAs with lower expression in epithelial cell lines, four had predicted binding sites in the epithelial gene signature (miR-30a, 330-3p, 425, 455-3p). The miRNAs identified by both differential expression and algorithmic analysis of signatures are predicted to regulate 178 and 210 genes in the epithelial and mesenchymal signatures respectively (Supplementary Table S2F).
Comparison of the DNA methylation status of the mesenchymal and epithelial cell lines using Infinium HumanMethylation27 BeadChips identified 75 hypo-methylated genes in the epithelial cell lines and 48 hypo-methylated genes in the mesenchymal cell lines (Figure 2G and Supplementary Table S2G). Thirty one of the 75 hypo-methylated genes in the epithelial signature were also represented in the epithelial mRNA signature (P = 2.41×10−26, Fisher’s exact test). Conversely, eight of the 48 hypo-methylated genes in the mesenchymal cell lines were represented in the mesenchymal mRNA signature (P = 1.281×10−4, Fisher’s exact test) (Figure 2H). Assessment of the relative contribution of DNA methylation, microRNA and gene expression to protein levels suggested greater concordance between gene and protein expression for epithelial than mesenchymal gene sets (Figure 2I). Comparison of transcript and protein variance between mesenchymal and epithelial cell lines further revealed increased variance of protein expression in mesenchymal versus epithelial cell lines, compared to gene expression (Supplementary Table S2G).
Heterogeneity among cell lines with a hybrid phenotype
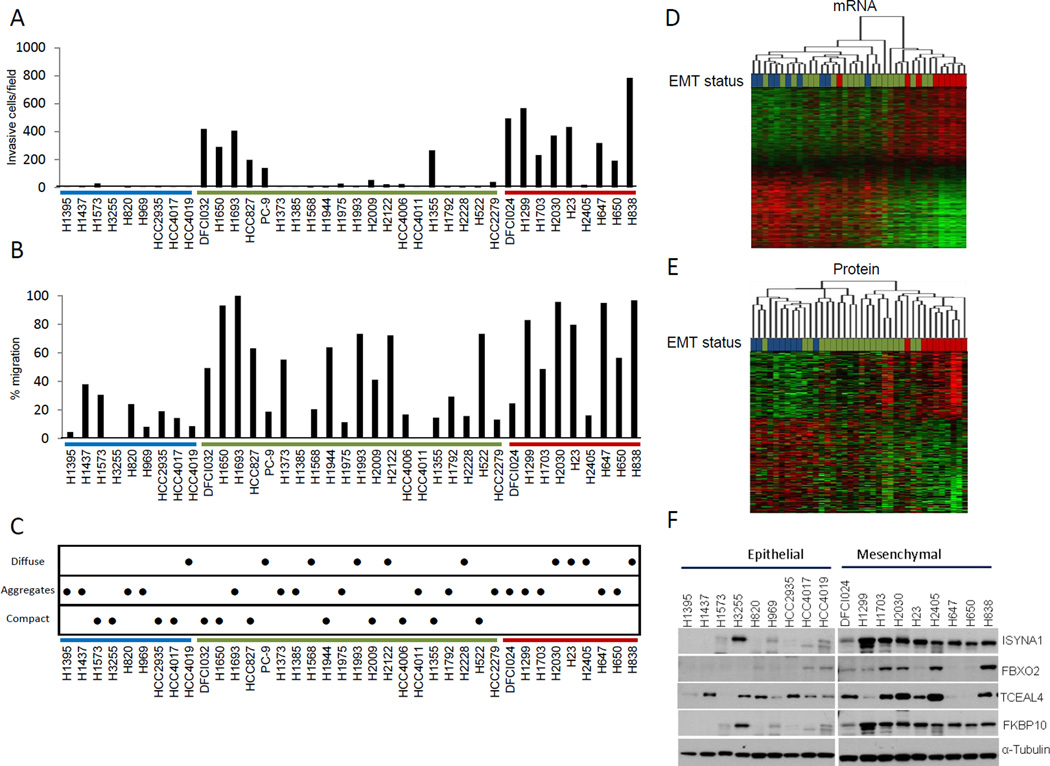
We next explored phenotypic differences and similarities between epithelial, hybrid and mesenchymal cell lines. We observed a significant difference in invasiveness between the epithelial and mesenchymal cell lines based on a Matrigel invasion assay (P = 0.0001, Mann-Whitney U test; Figure 3A). Hybrid cell lines were heterogeneous in their invasive properties. Expression of the mesenchymal proteins (r = 0.736, Spearman correlation) and genes (r = 0.715, Spearman correlation) had significantly higher correlation with invasiveness than CDH1_S/VIM expression alone (r = −0.483, Spearman correlation) (P = 0.041, Fisher r-to-z transformation). We observed a significant difference in cell migration between the epithelial and mesenchymal cell lines based on scratch wound assays (P = 0.0012, Mann-Whitney U test; Figure 3B). Expression of mesenchymal signature proteins and genes but not CDH1_S/VIM expression significantly correlated with migration (r = 0.546, Spearman correlation; P = 7.0 ×10−4, Mann-Whitney U test and r = 0.599, Spearman correlation; P = 2.0 ×10−4, Mann-Whitney U test, respectively). Cell-cell adhesion is a hallmark of epithelial cells and loss of cell-cell adhesion is considered to be a critical step in metastasis (4). Analysis of cell aggregation in a liquid culture allows assessment of the strength of cell-cell cadherin/catenin complex binding, as cells do not have a solid surface on which to bind. We observed significantly more cell aggregation in epithelial cell lines compared to mesenchymal cell lines (P = 0.0251, Mann-Whitney U test), with the hybrid cell lines distributed across the spectrum (Figure 3C). Cell line growth rates were also assessed. We did not observe a significant difference in cell growth rates between epithelial, hybrid, and mesenchymal cell lines as the variance in growth rates within each class was greater than the difference between them (data not shown).
Figure 3. Phenotypic characterization of cell lines.
A. Cell invasion through Matrigel. In at least 4 fields from 6 replicate wells the number of cells was counted for each cell line. B. Cell migration measured by a scratch wound assay, with 6 scratches measured per cell line. C. Cell aggregation after 24 hours in liquid culture over agarose as a measure of cell-cell adhesion. Six replicates, with assay performed in three individual wells on different days. D. Hierarchical clustering of differentially expressed genes in the epithelial and mesenchymal cell lines. E. Hierarchical clustering of differentially expressed proteins in the epithelial and mesenchymal cell lines. For all sub figures, mesenchymal cell lines are indicated by red, epithelial by blue, and hybrid cell lines by green. F. Confirmation of protein expression of novel EMT-related proteins. Western blotting of several EMT-related proteins showed increased expression of novel markers in mesenchymal cell lines compared to epithelial cell lines.
Hierarchical clustering was performed based on the gene and protein signatures to determine their ability to discriminate between epithelial, mesenchymal and hybrid cell lines. Clustering based on the mRNA signature separated the mesenchymal cell lines into one group and eight of the nine epithelial cell lines into the other (Figure 3D). The two clusters were significantly different for CDH1_S/VIM expression (P = 1.49 ×10−5, Mann-Whitney U test), invasion (P = 0.002, Mann-Whitney U test) and migration (P = 0.009, Mann-Whitney U test). Clustering of the differentially expressed proteins separated the epithelial and mesenchymal cell lines into distinct groups that were significantly different for CDH1_S/VIM expression (P = 0.003, Mann-Whitney U test), invasion (P = 3.0 ×10−4, Mann-Whitney U test) and migration (P = 0.004, Mann-Whitney U test) and aggregation (P = 0.006, Mann-Whitney U test) (Figure 3E). The large number of upregulated proteins in mesenchymal cell lines includes many that have not been previously associated with EMT and thus would represent novel biomarkers. We selected several novel proteins (TCEAL4, FBXO2, FKBP65, and ISYNA1) and confirmed their increased expression in mesenchymal cell lines by Western blotting (Figure 3F).
Hybrid EMT states are distinguishable by molecular features linked to their phenotypes
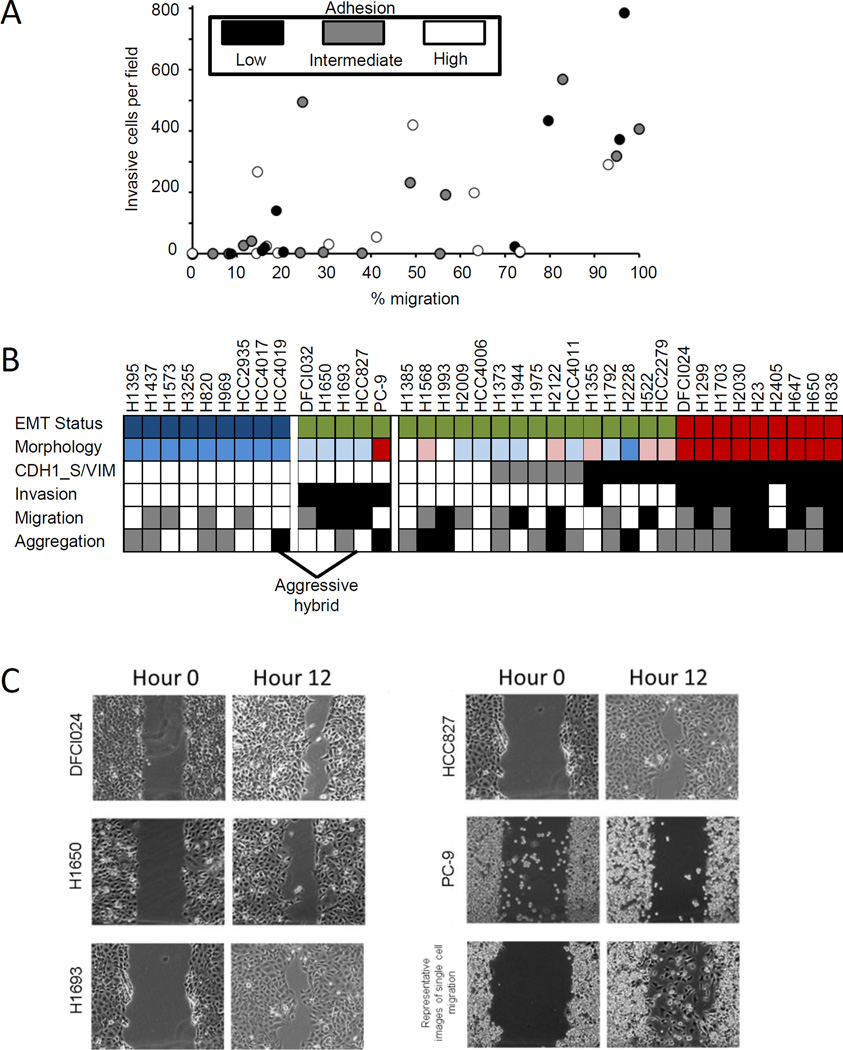
We observed phenotypic heterogeneity among the hybrid cell lines that was unrelated to their CDH1 and VIM expression. Cell lines in the hybrid group exhibited phenotypic traits of high invasion and migration that are a characteristic of a mesenchymal phenotype, together with high aggregation, a feature of epithelial cells (Figure 4A). A subset of hybrid cell lines (DFCI032, H1650, H1693, HCC827, and PC-9) exhibited high CDH1_S/VIM ratios but were also invasive and migratory (aggressive hybrid) (Figure 4B). Interestingly, while the mesenchymal cell lines migrated primarily as single cell, four of the five aggressive hybrid cell lines migrated by collective group migration (Figure 4C).
Figure 4. Identification of aggressive hybrid cell lines.
A. Plot of cell invasion against migration with cell-cell adhesion indicated by color. B. Cell lines are heterogeneous for phenotypic characteristics with hybrid epithelial-mesenchymal cells lines highlighted. CDH1_S/VIM protein ratios: □>1, and ■ < 1. Invasion: ■ >150 cells per field, □ <150 cells per field. Aggregation: ■ = diffuse,  = aggregates, □ = compact. Migration: □ ~ <33 % area covered,
= aggregates, □ = compact. Migration: □ ~ <33 % area covered,  >33% and <63% area covered, ■ >63% area covered. EMT Status: □ = epithelial,
>33% and <63% area covered, ■ >63% area covered. EMT Status: □ = epithelial,  = hybrid, ■ = mesenchymal. For cell morphology: red = mesenchymal, pink = mesenchymal-like, white = mixed mesenchymal/epithelial morphology, light blue = epithelial-like, blue = epithelial. For EMT status, mesenchymal cell lines are indicated by red, epithelial by blue, and hybrid cell lines by green. C. Migration of aggressive hybrid cell lines. Aggressive hybrid cell lines imaged at 0 and 12 hours during a scratch wound assay reveal collective cell migration in four of five cell lines.
= hybrid, ■ = mesenchymal. For cell morphology: red = mesenchymal, pink = mesenchymal-like, white = mixed mesenchymal/epithelial morphology, light blue = epithelial-like, blue = epithelial. For EMT status, mesenchymal cell lines are indicated by red, epithelial by blue, and hybrid cell lines by green. C. Migration of aggressive hybrid cell lines. Aggressive hybrid cell lines imaged at 0 and 12 hours during a scratch wound assay reveal collective cell migration in four of five cell lines.
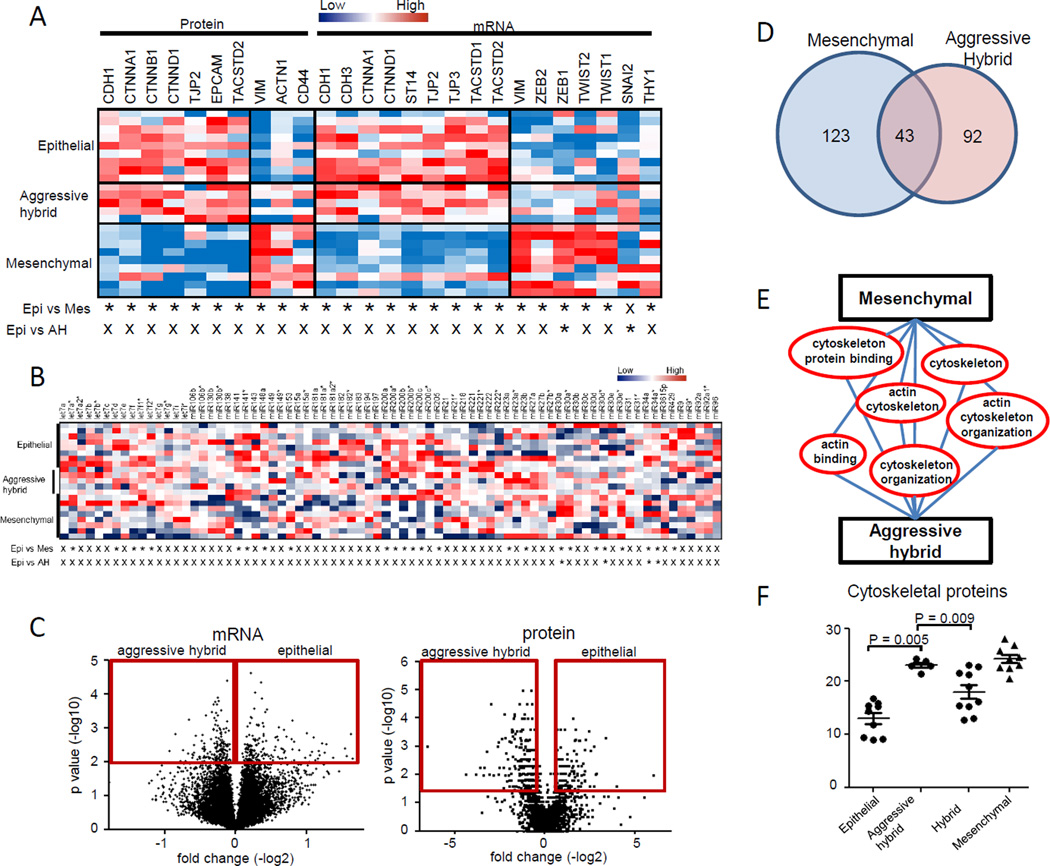
Recent work by Lu et al. proposed Zeb/Snail/mir-200/miR34 axis as regulators of hybrid phenotypes (25). We assessed transcript and protein expression of a large number of genes previously identified as regulators or markers of epithelial or mesenchymal cell types (Figure 5A for significantly expressed markers and Supplementary Table S3 for all markers). We identified up-regulation of ZEB1 (P = 0.042, Mann-Whitney U test) and SNAI2 (P = 0.041, Mann-Whitney U test) mRNA expression levels in the aggressive hybrid cell lines compared to epithelial cell lines. We observed increased expression of mir-34a in the aggressive hybrid cell lines compared to the epithelial cell lines, while expression of mir-200 family members was not significantly different (Figure 5B), concordant with the results that four of aggressive hybrid cell lines were clustered together with epithelial cell lines (Figure 2E). A comparative analysis between the epithelial and aggressive hybrid cell lines yielded 197 genes with up-regulated mRNA levels in the aggressive hybrid cell lines out of 20,598 total genes (Figure 5C). A set of 135 proteins were up-regulated in the aggressive hybrid cell lines compared with epithelial cell lines (Supplementary Table S4A). Interestingly, the 135 protein signature was enriched for GO terms associated with cytoskeleton, actin binding and organization, and significantly overlapped with the mesenchymal protein signature (P = 2.062×10−23 Fisher’s exact test) (Figures 5D, 5E and Supplementary Table S4B). Expression of cytoskeletal and actin-binding proteins in the aggressive hybrid cell lines was significantly higher than in the epithelial (P = 0.005, Mann-Whitney U test) or other hybrid cell lines (P = 0.009, Mann-Whitney U test) (Figure 5F) and were the primary discriminator of aggressive hybrid cell lines. Protein expression levels of TCEAL4 and ISYNA1 that are part of the mesenchymal protein signature were significantly elevated in the hybrid aggressive cell lines compared to epithelial type as determined by mass spectrometry, further supporting overlapping molecular characteristics of aggressive hybrid type and mesenchymal cell lines.
Figure 5. Molecular signatures for aggressive hybrid cell lines.
A. Expression of epithelial and mesenchymal markers at mRNA and protein levels in epithelial, aggressive hybrid and mesenchymal cell lines. A P value by t test under 0.05 is indicated with “*”, P value ≥ 0.05 with “X”. B. Expression of epithelial and mesenchymal related microRNAs in epithelial, aggressive hybrid and mesenchymal cell lines. A p value by t test under 0.05 is indicated with “*”, p value ≥ 0.05 with “X”. C. Volcano plot of the differential gene (left panel) and protein (right panel) in a comparison of epithelial and aggressive hybrid cell lines. Red box indicates significance cutoffs. D. Overlap of mesenchymal and aggressive hybrid protein signatures. E. Overlap of mesenchymal and aggressive hybrid protein significant Gene Ontology categories. F. Cytoskeletal proteins are upregulated in the mesenchymal and aggressive hybrid cell lines compared against epithelial cell lines.
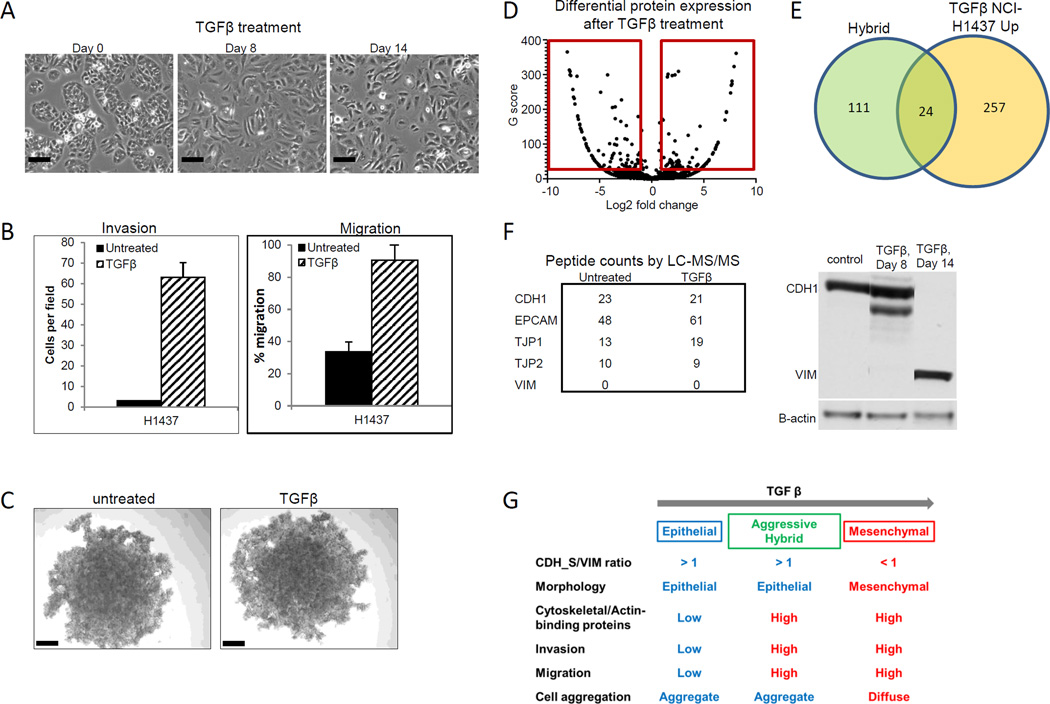
TGFβ induced up-regulation of cytoskeletal proteins precedes loss of E-cadherin
We next tested induction of EMT with TGFβ treatment to confirm that proteomic differences observed between the mesenchymal and epithelial cell lines were due to an EMT event. Following exposure to TGFβ, epithelial H1437 cells elongated and lost cell adhesion (Figure 6A). We further observed increased cell invasion and migration, but no changes in cell aggregation (Figures 6B and C). We next performed a proteomic analysis of H1437 cells after eight days of treatment with TGFβ in comparison with untreated cells, selecting differentially expressed proteins with > 1.5 fold change and G score greater than 3.85 (equivalent to a P value < 0.05) (Figure 6D and Supplementary Table S5A). Changes in protein expression after TGFβ treatment correlated significantly with the differentially expressed epithelial and mesenchymal proteins and the aggressive hybrid protein signature (P = 1.14 × 10−9 and P = 7.53 × 10−10 respectively, Spearman correlation) (Figure 6E). Gene Ontology analysis identified cytoskeletal and actin binding proteins, including AKAP12, as the most highly enriched group among the upregulated proteins (Supplementary Table S5B). Downregulated protein set with TGFβ treatment overlapped with the epithelial protein signature (P = 9.88 × 10−20 Fisher’s exact test). Down-regulated proteins were enriched for oxidation reduction and multiple categories related to metabolism and glycolysis. The morphological and cytoskeletal protein expression alterations preceded changes in EMT markers, as we did not observe changes in CDH1 and VIM proteins after 8 days of TGFβ treatment (Figure 6F) as confirmed by Western blotting. Loss of CDH1 expression and increase in VIM expression were observed at 14 days. The similarity between protein changes induced by TGFβ treatment of epithelial cells and the aggressive hybrid signature (invasive, migratory, high expression of cytoskeletal proteins) supports reorganization of cytoskeletal proteins preceding loss of CDH1 as an intermediate stage in EMT, with aggressive hybrid cell lines expressing epithelial markers (Figure 6G).
Figure 6. Proteomic analysis of TGFβ induced EMT in an epithelial cell line.
A. H1437 cells change from epithelial to mesenchymal morphology with TGFβ treatment. Scale = 40 µm. B. Invasion and migration of H1437 increase with TGFβ treatment. C. Aggregation of H1437 cells is unaffected by TGFβ treatment. Scale = 200 µm. Volcano plot of the differential protein expression analysis. Red box indicates significance cutoffs. E. Proteins upregulated in H1437_TGFβ cells overlap significantly with the aggressive hybrid protein signatures. F. Expression of epithelial or mesenchymal markers in TGFβ treated H1437 by LC-MS/MS or western blot. G. Alteration of molecular and functional characteristics during EMT via aggressive hybrid type in H1437 with TGFβ treatment.
Relevance of a mesenchymal signature enriched in genes encoding for cytoskeletal proteins to survival in early stage lung adenocarcinomas
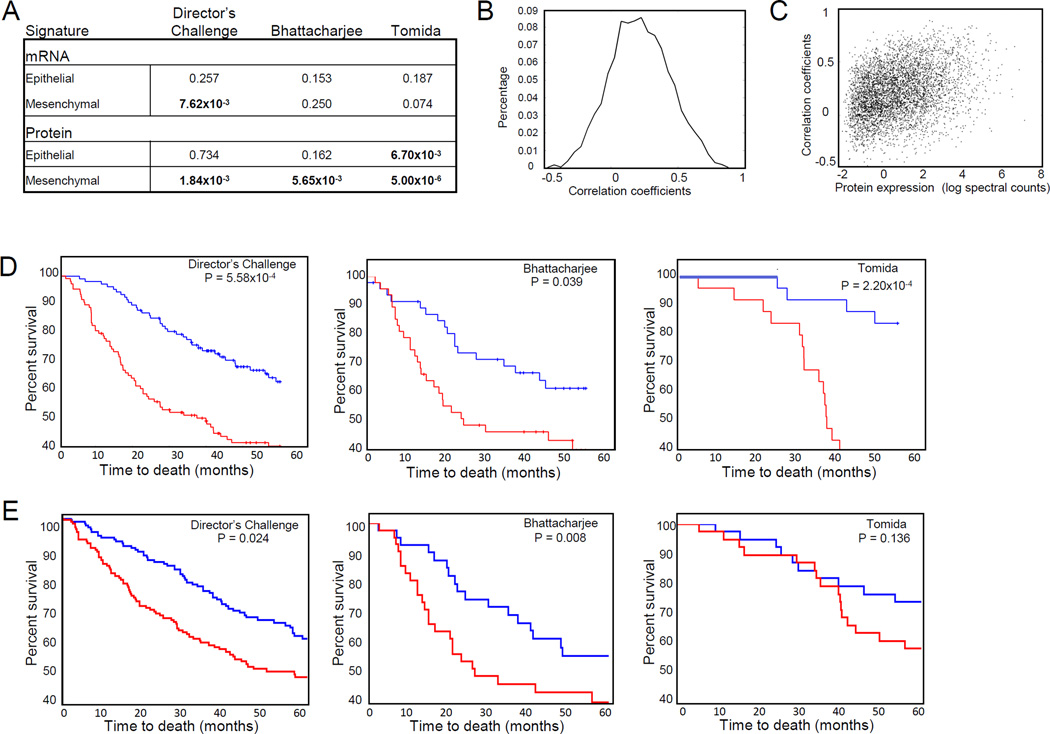
We next determined whether the mesenchymal gene signature we identified had predictive value in early stage lung adenocarcinoma by interrogating three independent gene expression datasets of lung adenocarcinoma annotated for outcome- Director’s Challenge, Bhattacharjee et al. and Tomida et al. data sets (32–34). Stage 1 and 2 tumors were ranked by their relative expression of genes in the signature and hazard ratios were calculated by Cox regression. The mesenchymal mRNA signature significantly predicted survival in the Director’s Challenge dataset (P = 7.62×10−3 Cox regression) (Figure 7A). Given the lack of databases for lung tumor protein expression annotated for survival, we next tested whether a set of mRNAs specifically encoding the mesenchymal proteomic signature had prognostic value. We first correlated protein abundance with mRNA expression by Spearman correlation and demonstrated significant correlation between transcript and protein expression with a mean of correlation coefficients of 0.216 (P = 1.0 ×10−16 based on permutation tests, Figure 7B). Transcript: protein expression increased with measures of protein abundance (Figure 7C). The set of mRNAs encoding the mesenchymal protein signature significantly predicted reduced survival in all three tumor datasets tested, whereas the epithelial signature was significantly associated with increased survival in one dataset (Figure 7A). The Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma tested 14 different methods for predicting survival in early stage non-small cell lung cancer with the best predictor found to be Model A, consisting of 13,830 genes produced from clustering (32). As a comparison, we tested Model A in the two other independent datasets and found that the mesenchymal protein signature had similar statistical significance as Model A (32). We next performed a Kaplan-Meier analysis and found that the mesenchymal signature significantly predicted survival in all three data sets tested (the Director’s Challenge, Bhattacharjee et al. and Tomida et al. data sets) (Figure 7D). Given that upregulation of cytoskeletal proteins was associated with both mesenchymal and aggressive hybrid phenotypes, we tested whether mRNAs representing the cytoskeletal proteins from the mesenchymal protein signature were predictive of survival. We found that this restricted signature also significantly predicted survival in the Director’s Challenge and Bhattacharjee et al. tumor sets (Figure 7E).
Figure 7. Mesenchymal and cytoskeletal protein signatures predict survival in NSCLC.
A. Cox regression of mRNA and protein signatures in three different gene expression datasets. B. Correlation of mRNA and protein expression in 38 cell lines. C. Correlation of mRNA and protein expression increases with protein abundance. D. Kaplan-Meier curves of mesenchymal protein signature in gene expression datasets. P values are derived from log-rank test. Red represents the top one-third of tumors ranked by expression of the mesenchymal signature; Blue represents the lower one-third of tumors by expression of the mesenchymal signature. E. Kaplan-Meier curves of cytoskeletal protein signature in gene expression datasets. P values are derived from log-rank test. Red represents the top one-third of tumors ranked by expression of the cytoskeletal signature; Blue represents the lower one-third of tumors by expression of the cytoskeletal signature.
DISCUSSION
We have undertaken proteomic, gene expression, microRNA, and DNA methylation analysis of lung adenocarcinoma cell lines representative of genomic heterogeneity in lung adenocarcinoma, together with their functional characterization. Subsets that emerged from the study encompassed epithelial, mesenchymal and an aggressive hybrid group with features of both epithelial and mesenchymal cell lines characterized by up-regulation of cytoskeleton and actin-binding proteins. Findings from TGFβ treatment of epithelial cells support the occurrence of an intermediate state during EMT with hybrid features. In consideration of the role that EMT plays in tumor progression, we sought to elucidate gene sets that may predictive outcome in early stage lung cancer based on biological functions and identified a signature enriched in cytoskeletal protein encoding genes predictive of survival.
Molecular profiling has revealed substantial tumor heterogeneity in many human cancers (35–38). The extensive molecular and phenotypic characterization of lung adenocarcinoma cell lines likewise has revealed substantial heterogeneity amongst the cell lines. Most cell lines could not be simply categorized as either mesenchymal or epithelial. We identified cell lines with features of both mesenchymal and epithelial cell types, substantiating the occurrence of a hybrid state in tumor cell populations (25). Co-stained E-cadherin and Vimentin in hybrid cell lines suggested the occurrence of hybrid transcriptional program. Partial EMT has also been described during development, wound healing in addition to tumorigenesis (39–42), while the existence of subpopulation of E-cadherin-positive and vimentin-positive cells has been recently indicated in head and neck cancer (43, 44), supporting the need to further characterize this group at the genomic and proteomic levels as we have undertaken in this study. In addition, our immunohistochemical studies of CDH1 and VIM protein were performed using non-serial section of lung adenocarcinoma tissues. On the basis of this potential limitation, further validation studies for the occurrence of hybrid type in tumors are also warranted. A subset of hybrid cell lines was highly invasive despite gene and protein expression of epithelial markers. Aggressive hybrid cell lines expressed a similar pattern of upregulated cytoskeletal and actin binding as mesenchymal cell lines. The occurrence of hybrid EMT states is supported by our analysis of TGFβ induction of EMT in the NCI-H1437 cell line in which we observed phenotypic alterations and up-regulation of cytoskeletal and actin binding proteins prior to changes in cadherin or vimentin expression. Lu et al. proposed a regulatory switch centered around miR-200/Zeb and Snail/miR34 that regulates hybrid states of EMT (25). Our findings support the importance of Zeb and miR-34a in the regulation of hybrid phenotypes. We observed limited concordance between mRNA and proteins for some genes, particularly for cells with a mesenchymal phenotype, suggesting an important role for post-transcriptional regulation affecting EMT.
Cytoskeletal rearrangements emerged as the dominant feature of mesenchymal and invasive cells based on both mRNA and protein analysis. This finding is further supported by up-regulation of cytoskeletal proteins following TGFβ induction of EMT in our and other studies (45–47) and provided a rationale to investigate the relationship between cytoskeletal gene rich signatures and survival. A set of 41 genes derived from the mesenchymal protein signature representing cytoskeletal and actin-binding proteins predicted survival in all three tumor datasets we tested. Moreover, while clearly a key feature of EMT, our study provides supporting evidence that cytoskeletal reorganization and invasiveness occur frequently in the absence of CDH1 loss.
We further assessed the relevance of the gene and protein signatures associated with EMT to survival in early stage lung adenocarcinoma in three independent sets. Reproducibility of statistical association of gene expression signatures with survival across independent datasets has been challenging (48). Association of the full set of mRNAs in the mesenchymal signature with survival was significant in the Director’s Challenge dataset. Remarkably, association of the more limited set of transcripts corresponding specifically to the mesenchymal proteomic signature was significant in all three independent datasets tested without initial training. Thus our findings emphasize the functional relevance of proteomics to integrated cancer molecular profiling, pointing to an association between cytoskeletal and actin binding proteins, a mesenchymal or aggressive hybrid EMT phenotype and invasive properties of lung adenocarcinomas. Byers et al. (49) identified an EMT gene signature consisting of 76 genes predictive of resistance to EGFR and PI3K/AKT inhibitors which partially overlapped with our epithelial and mesenchymal gene/protein signatures. We note that Byers et al. established their signature by selecting genes with significant correlation (both positive and negative) with gene expression of CDH1, VIM, CDH2, and FN1.In our study, we first defined epithelial and mesenchymal cell line properties based on cell morphology and expression at the protein level of CDH1 and VIM. Our results indicate aggressiveness among hybrid type of cell lines is not associated with expression levels of CDH1 and VIM.
In conclusion, an integrated systems approach that encompassed functional and molecular characterization, of lung adenocarcinoma cell lines, uncovered substantial heterogeneity with respect to epithelial and mesenchymal features among cell lines. Signatures were identified that distinguish epithelial and mesenchymal cells as well as signatures that were shared with cells with an intermediate/hybrid phenotype. Our findings point to an association between cytoskeletal and actin binding proteins, a mesenchymal or hybrid EMT phenotype and invasive properties of lung adenocarcinomas that impact survival.
Supplementary Material
Acknowledgments
We would like to thank members of the Hanash lab for their invaluable suggestions. Paul Schliekelman provided statistical support and advice on the manuscript.
Funding Support: This work was supported by the Department of Defense (DOD) Congressionally Mandated Lung Cancer Research Program, the National Cancer Institute Early Detection Program, the Canary Foundation and the Lungevity Foundation. M.T. was supported by the Canary Foundation. L.M. was supported by NIH grant R21/R33 CA-88245 and the Listwin Family Foundation.
Footnotes
Disclosure: The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Boyer B, Thiery JP. Epithelium-mesenchyme interconversion as example of epithelial plasticity. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 1993;101:257–268. doi: 10.1111/j.1699-0463.1993.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 2.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 3.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Developmental cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Kim SH, Kim JM, Shin MH, Kim CW, Huang SM, Kang DW, et al. Correlation of epithelial-mesenchymal transition markers with clinicopathologic parameters in adenocarcinomas and squamous cell carcinoma of the lung. Histology and histopathology. 2012;27:581–591. doi: 10.14670/HH-27.581. [DOI] [PubMed] [Google Scholar]
- 6.Prudkin L, Liu DD, Ozburn NC, Sun M, Behrens C, Tang X, et al. Epithelial-to-mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22:668–678. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi YS, Shim YM, Kim SH, Son DS, Lee HS, Kim GY, et al. Prognostic significance of E-cadherin and beta-catenin in resected stage I non-small cell lung cancer. European journal of cardio-thoracic surgery : official journal of the European Association for Cardio-thoracic Surgery. 2003;24:441–449. doi: 10.1016/s1010-7940(03)00308-7. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature reviews Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 9.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer research. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 10.Nawshad A, Lagamba D, Polad A, Hay ED. Transforming growth factor-beta signaling during epithelial-mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells, tissues, organs. 2005;179:11–23. doi: 10.1159/000084505. [DOI] [PubMed] [Google Scholar]
- 11.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 13.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. The Journal of biological chemistry. 2010;285:35293–35302. doi: 10.1074/jbc.M110.160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. The Journal of cell biology. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter K, Holcomb T, Januario T, Du P, Evangelista M, Kartha N, et al. DNA methylation profiling defines clinically relevant biological subsets of non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2360–2373. doi: 10.1158/1078-0432.CCR-11-2635-T. [DOI] [PubMed] [Google Scholar]
- 18.Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter HI, et al. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC research notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nature structural & molecular biology. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature reviews Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 22.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, et al. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. Journal of cellular physiology. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 23.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18144–18149. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 27.Taguchi A, Politi K, Pitteri SJ, Lockwood WW, Faca VM, Kelly-Spratt K, et al. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer cell. 2011;20:289–299. doi: 10.1016/j.ccr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gemmill RM, Roche J, Potiron VA, Nasarre P, Mitas M, Coldren CD, et al. ZEB1-responsive genes in non-small cell lung cancer. Cancer letters. 2011;300:66–78. doi: 10.1016/j.canlet.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng QF, Zhou CC, Su CX. Clinicopathological features and epidermal growth factor receptor mutations associated with epithelial-mesenchymal transition in non-small cell lung cancer. Respirology. 2009;14:371–376. doi: 10.1111/j.1440-1843.2009.01496.x. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akakura S, Gelman IH. Pivotal Role of AKAP12 in the Regulation of Cellular Adhesion Dynamics: Control of Cytoskeletal Architecture, Cell Migration, and Mitogenic Signaling. Journal of signal transduction. 2012;2012:529179. doi: 10.1155/2012/529179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Director's Challenge Consortium for the Molecular Classification of Lung A. Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nature medicine. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Budinska E, Popovici V, Tejpar S, D'Ario G, Lapique N, Sikora KO, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. The Journal of pathology. 2013;231:63–76. doi: 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fidler IJ. Biological heterogeneity of cancer: implication to therapy. Human vaccines & immunotherapeutics. 2012;8:1141–1142. doi: 10.4161/hv.19643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnen N, Francart ME, Thelen N, Cloes M, Thiry M. Evidence for a partial epithelial-mesenchymal transition in postnatal stages of rat auditory organ morphogenesis. Histochemistry and cell biology. 2012;138:477–488. doi: 10.1007/s00418-012-0969-5. [DOI] [PubMed] [Google Scholar]
- 40.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nature reviews Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 41.Nanba D, Nakanishi Y, Hieda Y. Changes in adhesive properties of epithelial cells during early morphogenesis of the mammary gland. Development, growth & differentiation. 2001;43:535–544. doi: 10.1046/j.1440-169x.2001.00596.x. [DOI] [PubMed] [Google Scholar]
- 42.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Current opinion in cell biology. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 43.La Fleur L, Johansson AC, Roberg K. A CD44high/EGFRlow subpopulation within head and neck cancer cell lines shows an epithelial-mesenchymal transition phenotype and resistance to treatment. PloS one. 2012;7:e44071. doi: 10.1371/journal.pone.0044071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasanen K, Sriswasdi S, Valiga A, Tang HY, Zhang G, Perego M, et al. Comparative secretome analysis of epithelial and mesenchymal subpopulations of head and neck squamous cell carcinoma identifies S100A4 as a potential therapeutic target. Molecular & cellular proteomics : MCP. 2013;12:3778–3792. doi: 10.1074/mcp.M113.029587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keshamouni VG, Schiemann WP. Epithelial-mesenchymal transition in tumor metastasis: a method to the madness. Future oncology. 2009;5:1109–1111. doi: 10.2217/fon.09.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer metastasis reviews. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 47.Levayer R, Lecuit T. Breaking down EMT. Nat Cell Biol. 2008;10:757–759. doi: 10.1038/ncb0708-757. [DOI] [PubMed] [Google Scholar]
- 48.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? Journal of the National Cancer Institute. 2010;102:464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.